Abstract
Smectite clays such as montmorillonite form complexes with a variety of biomolecules, including the nucleic acids DNA and RNA. Most previous studies of DNA adsorption onto clay have relied upon spectrophotometric analysis after separation of free nucleic acids from bound complexes by centrifugation. In the current work we demonstrate that such studies produce a consistent error due to (a) incomplete sedimentation of montmorillonite and (b) strong absorbance of the remaining clay at 260 nm. Clay sedimentation efficiency was strongly dependent upon cation concentration (Na+ or Mg2+) and on the level of dispersion of the original suspension. An improved clay:DNA adsorption assay was developed and utilized to assess the impact of metal counterions on binding of single-stranded DNA to montmorillonite. X-ray diffraction demonstrated, for the first time, formation of intercalated structures consistent with orientation of the DNA strands parallel to the clay surface. Observed gallery spacings were found to closely match values calculated utilizing atomistic modeling techniques.
Keywords: intercalate, montmorillonite, DNA, molecular modeling, smectite clay
Introduction
Clays are naturally occurring aluminosilicate materials composed primarily of alumina, silica and water with small amounts of metal cations such as Ca2+, Fe3+, K+, Mg2+ and Na+ also present.1,2 The most well-researched of these materials are the smectite clays (swelling clays), also called sheet or layered aluminosilicates. Many smectite clays, including the widely studied mineral montmorillonite, exist as sheet-like structures that are ∼ 1 nm thick and up to 1000 nm wide in the other dimensions, qualifying them as nanomaterials. Clays have interesting chemical and physical characteristics, e.g., montmorillonite has a high modulus, high cation exchange capacity, a large surface area to mass ratio, and the ability to form stable dispersions in aqueous solutions.2,3 The unique features of clays have led to their widespread use in materials developed for the automotive, medical, food and cosmetics industries.4,5
Montmorillonite sheets, or platelets, contain two outer layers of tetrahedral silica lattices surrounding a positively charged central octahedral layer composed of alumina (primarily Al3+ and oxygen, but with some substitution of cations such as Mg2+). The platelets normally stack on top of one another with water molecules adsorbed between them, forming tactoids.6,7 The stacks of sheets are turbostratic, having no crystallographic preference for how the plates orient relative to one another. This is evident in their X-ray diffraction pattern where no hkl reflections appear but only 001 and hk0 reflections are observed. The spacing between the plates is commonly referred to as the gallery, the size of which is dependent on a number of variables such as exchangeable cations, level of hydration, and intercalating species. The properties of clays vary depending on the exchangable cations that are present, such that, for example, homoionic Na- and Ca-montmorillonite have different characteristics.8 Many organic polymers are also able to enter the galleries, typically producing polymer-clay nanocomposites that have dramatically different properties from the original bulk polymers.1 Such organoclays may form intercalated, exfoliated or conventional composite structures.2
The interaction of clays with important biological polymers such as DNA and RNA has been extensively investigated. Determination of the mechanism by which nucleic acids interact with clays found naturally in soils is important for understanding how extracellular DNA might be preserved in the environment and also how it may be transferred between organisms over time.9 In addition, clays have been used successfully as vectors for delivery of DNA into cells in recent experiments.10-12 The binding of single-stranded or double-stranded DNA polymers to the large, negatively charged surfaces of montmorillonite particles is poorly understood, but is thought to be affected by pH and the concentration and nature of the metal cations that are present.9,13 Strands of DNA possess a strong negative charge due to the phosphate groups present on each nucleotide monomer. It has been proposed that exchangeable surface cations may form bridges between the phosphates on DNA and the negatively charged tetrahedral silica layer on the clay surface.13,14 Interactions of the phosphates with the narrow edges of platelets, which contain exposed Al3+ and Mg2+ ions at octahedral lattice breakpoints has also been suggested, though this mechanism is less likely because the edges are primarily terminated with hydroxyls rather than cations.1,2
Most quantitative studies of clay-DNA association have taken advantage of the fact that DNA has a strong absorbance peak at 260 nm to monitor its concentration in solution. In these experiments, DNA (nucleotides, single-stranded or double-stranded) and clay were mixed in aqueous solution and centrifuged at various speeds (typically between 15,000 and 40,000 × g) to pellet the clay plus any bound DNA molecules.9,11,13-24 Residual absorbance of the supernatant at 260 nm then permitted calculation of the concentration of unbound DNA. In the current study we demonstrate that the standard co-precipitation approach produces inaccurate results when applied to Ca or Na montmorillonite. The amount of free DNA in clay:DNA supernatants has been consistently overestimated because (a) montmorillonite absorbs ultraviolet light strongly between 220 and 270 nm, and (b) centrifugation, even at speeds as high as 40,000 × g, does not remove all clay particles from the solution. Clay precipitation efficiency was found to be poor in deoinized water, but could be enhanced by addition of cations such as sodium and magnesium. A modified version of the standard centrifugation assay was developed that takes factors into account that confounded previous studies. Formation of intercalated DNA structures was demonstrated experimentally, with the resulting structures exhibiting spacings consistent with insertion of molecules having the width of a single strand of DNA.
Materials and Methods
Clays and DNA
Homoionic sodium and calcium montmorillonite powders were obtained from Southern Clay Products, Inc. (Gonzales, TX). Stock solutions were prepared in double-deionized water at 2 or 5 mg/ml. Sonicated suspensions were prepared using a Vibracell VC501 sonicator from Sonics & Materials, Inc. (Newtown, CT). Samples were sonicated at amplitude 40 using a 3 mm probe tip for either 5 min (sodium clay) or 10 min (calcium clay). The oligonucleotide Pvu4a (sequence: AAATGAGTCACCCAGATCTAAATAA) was obtained from BioServe Biotechnologies (Laurel, MD).
Ultraviolet light spectroscopy
Spectroscopic measurements and scans were performed using a BioRad SmartSpec 3000 spectrophotometer. Scans were performed using a wavelength range of 200 to 450 nm. Na and Ca montmorillonite solutions were scanned utilizing 0.2 mg/ml suspensions in water and Pvu4a DNA was scanned using a 77 ug/ml solution. Aqueous FeCl3 solutions were scanned using a concentration of 50 ug/ml.
DNA-clay adsorption assays
Precipitation of clay and clay-DNA complexes was accomplished using either a Beckman-Coulter Microfuge 18 microcentrifuge spun at RT and 18,000 × g or a Beckman J2-21 centrifuge spun at 40,000 × g. Graphical analysis of centrifugation results utilized Prism GraphPad and Microsoft Excel software. For assessment of the absorbance of clay suspensions after centrifugation, 500 ul of each mixture was spun and 50 ul immediately removed from the top of the solution for analysis on the spectrophotometer.
For improved measurement of binding of single-stranded oligonucleotide DNA to montmorillonite, solutions containing sonicated clay, DNA and/or added salts were mixed in a total volume of 200 ul. Stock 2 mg/ml clay and 880 ug/ml Pvu4a DNA solutions were prepared in deionized water and aliquotted to achieve final concentrations of 0.5 mg/ml and 20 ug/ml, respectively. NaCl and MgCl2 stock solutions were diluted 10-fold to reach final concentrations of 0.1, 1.0, 10 or 100 mM. Samples were prepared, vortexed briefly and centrifuged at 18,000 × g for 15 min. The upper 50 ul of each sample was transferred to a microcuvette for determination of absorbance at 230 and 260 nm. Averages of three or four independent assays were calculated for each sample. The clay-to-DNA mass ratio employed for these experiments was 0.5 mg/ml vs. 20 ug/ml, or 25:1. The molar ratio of clay-to-DNA was estimated as 1.1 × 10-5 mM vs. 2.46 × 10-3 mM, or 1 clay platelet per 224 DNA molecules, assuming a platelet molecular weight of 4.44 × 107 as described in the Results.
X-ray diffractometry
For X-ray diffraction analysis, sonicated montmorillonite (2 mg) was added, with or without Pvu4a DNA (850 ug), to a solution containing 10 mM MgCl2 in a final volume of 4 ml. After brief vortexing, each mixture was centrifuged for 20 min at 18,000 × g at 20 °C. After removal of the supernatant, the wet pellets were analyzed by mounting the material onto glass slides and performing X-ray diffraction in a Bruker D-8 diffractometer. The diffractometer was equipped with a Cu KA X-ray tube and Solex detector. The scans were run from 1 to 20 degrees two theta with step size of 0.05 degrees and a 10 sec count time at each step.
Molecular modeling
Lowest energy structures of complexes containing single-stranded DNA bound to either sodium or calcium montmorillonite were calculated using the Cerius 2 commercial software package. For ease of modeling, a single strand of DNA contained in the software library having a sequence of CCTTCGCTGCTTTCCTCT was utilized in the calculations. The universal force field was utilized in all calculations. Charges on the DNA and clay were calculated in the OFFSETUP module utilizing the charge equilibration method of Rappe.25 The DNA strand was placed in the gallery between two clay plates. The atoms in the clay plates were constrained to their original positions to speed up the calculations. The gallery spacing was moved inwards slowly in small increments with several iterative cycles of minimization and molecular dynamics until a minimum energy was found. The minimization was conducted by starting with the steepest slope descent method followed by Adopted Basis Newton Raphson and Quasi-Newton methods and ending with the accurate truncated Newton method. The molecular dynamics were run under NVE conditions at 300 °K.
Results
Prior studies of DNA adsorption onto clay took advantage of the strong absorbance of DNA at 260 nm to quantitate its concentration in clay/DNA mixtures.9,11,13-24 In these experiments DNA was mixed with clay particles in aqueous solution and then centrifuged to pellet the clay plus any DNA molecules that had become adsorbed onto the platelets. Residual absorbance of the supernatant was then measured and presumed to represent the fraction of DNA that did not bind to the clay. As part of an investigation of the use of montmorillonite as a carrier molecule for gene delivery, we discovered a complication in this type of centrifugation experiment: iron-containing clays such as montmorillonite absorb light strongly at 260 nm and do not sediment completely after centrifugation at the speeds used in previous studies.
Montmorillonite suspensions absorb ultraviolet light at 260 nm
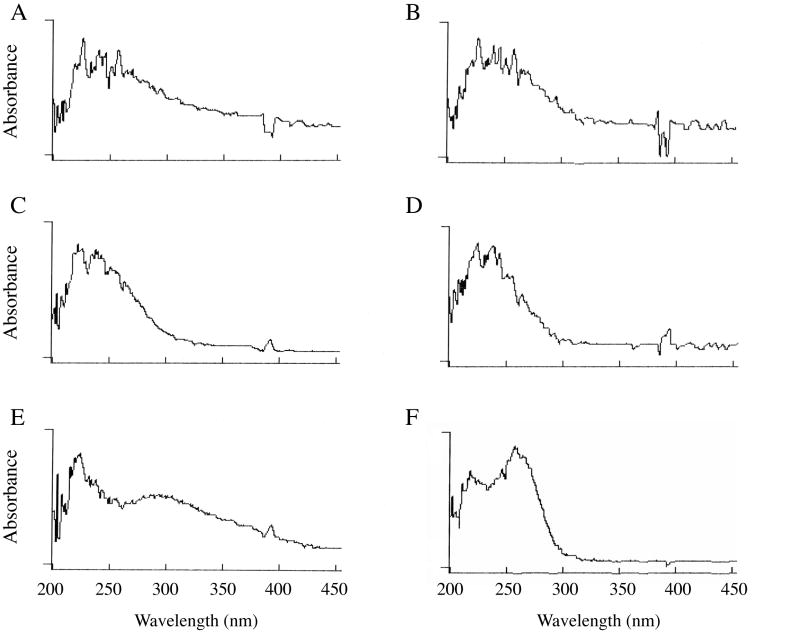
Absorbance scans of 0.2 mg/ml suspensions of homoionic Ca and Na montmorillonite in water are presented in Figure 1A and 1B, respectively. Each solution exhibits a broad absorbance peak from approximately 220 to 300 nm with a long right shoulder, especially apparent for the Ca clay. Absorbance of clays in the UV range has been reported previously6,26,27, and has been attributed to the presence of small amounts of iron (Fe III) in the octahedral alumina layer of each platelet.28,29 After Ca and Na clay solutions (2 mg/ml) were centrifuged for 15 min at 18,000 × g, similar to the forces used in most previous clay:DNA binding studies, the initially turbid solutions became transparent. Although absorbance of light by the solutions was quantitatively reduced (discussed below), the resulting supernatants retained the same broad absorbance band as before (Figures 1C and 1D). Interestingly, the right shoulders extending to near-UV and visible wavelengths were largely eliminated by the centrifugation. The simplest interpretation of this change is that much of the apparent absorbance of the original suspensions above 300 nm is caused by scattering of light by aggregates of clay platelets, or tactoids. Centrifugation caused the larger particles to sediment, leaving mostly single platelets in the supernatant. These smaller molecules still absorb ultraviolet light because of the presence of iron, but do not scatter light efficiently.
Figure 1.
Absorbance spectra for homoionic suspensions of Ca montmorillonite (A and B), Na montmorillonite (C and D), FeCl3 (E), and DNA (F). Spectra depicted in B and D were obtained after centrifugation of the solutions used for A and C, respectively, for 5 min at 18,000 × g.
Absorbance of a 50 ug/ml solution of FeCl3 is shown in Figure 1E for comparison. The metal also exhibits strong absorbance above 220 nm, though the pattern is not identical to that of the clays, a possible indication that the local environment within the clay influences absorbance at some wavelengths. Finally, an absorbance spectrum for a single-stranded DNA oligonucletide (25 nucleotides long) is presented in Figure 1F. Although the clay and DNA peak maxima are not identical, it is clear from the spectra that each of the clays has strong absorbance in the vicinity of the strong DNA absorbance peak at 260 nm.
Absorbances of Na and Ca montmorillonite at the peak wavelength are proportional to clay concentration and are differentially affected by sonication
The broad absorbance peaks observed for the two clays have an apparent maximum between 220 and 250 nm (Figure 1C and 1D). Using 230 nm as an approximation of peak absorbance, we observed that simple, unsonicated clay suspensions exhibited a linear relationship with respect to absorbance in the range of 0.05 – 1.0 mg/ml (Figure 2). The Beer's Law graph for the Na clay did not change substantially after the solution was sonicated for 5 min to break up tactoids (Figure 2A). In contrast, absorbance was increased after the Ca clay solution was sonicated, though the concentration:absorbance relationship remained linear (Figure 2B). This result suggests that the tactoids formed by Ca montmorillonite, which are larger than those of Na clay7,26,30, interfere with absorbance of UV light. Chen et al. noted that unsonicated Na montmorillonite absorbs UV light more strongly than unsonicated Ca clay (also visible in Figure 2 using a wavelength of 230 nm), a characteristic they attributed to destructive interference of light waves passing through the larger Ca clay tactoids though simple shielding may also be involved.29
Figure 2.
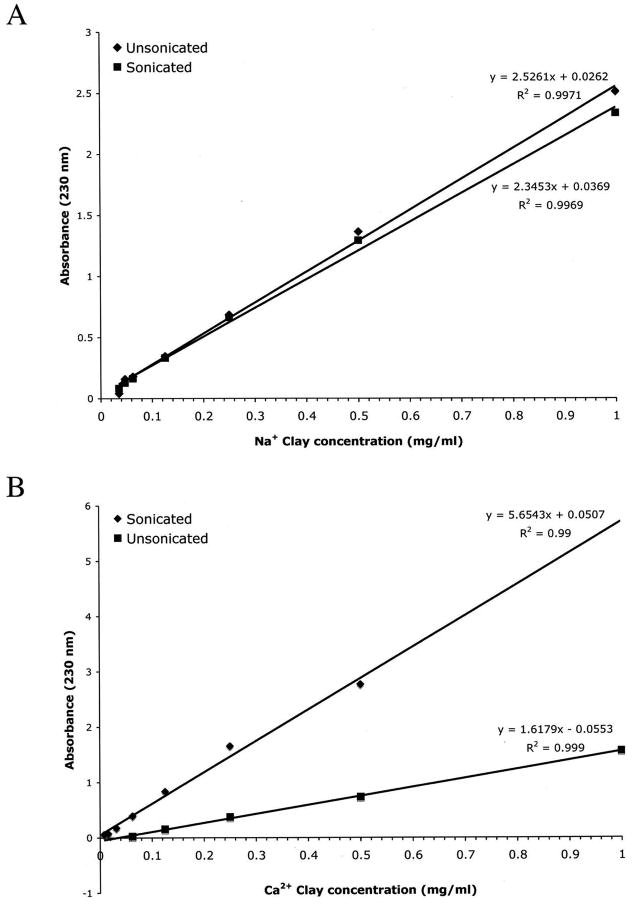
Absorbance of ultraviolet light (230 nm) by sonicated and unsonicated clay solutions follows Beer's Law.
Montmorillonite particles are sedimented inefficiently using conventional centrifugation methods
The Beer's Law equations derived for each clay were used to quantitate sedimentation efficiencies. Five mg/ml solutions of Na and Ca clay in water were centrifuged for 20 min at either 18,000 or 40,000 × g (the fastest speeds used in most previous clay:DNA binding studies) and the absorbances of the supernatants were monitored at 230 and 260 nm. The concentration of the Na clay was reduced from 5.0 to 0.85 mg/ml after centrifugation at 18,000 × g, indicating that almost 20% of the clay remained in suspension (Table 1). Ca clay was pelleted more efficiently, with the final concentration reduced to 0.11 mg/ml. Centrifugation at 40,000 × g improved sedimentation efficiencies, but the resulting supernatants still retained absorbances of 0.19 and 0.13 at 260 nm for the Na and Ca clay, respectively. The absorbance of DNA at 260 nm is linearly proportional to its concentration in a manner that is dependent on size and single- versus double-stranded structure. Using the widely employed relationship that an A260 of 1.0 corresponds to a concentration of 33 ug/ml for single-stranded DNA oligonucleotides31, DNA concentrations predicted for each supernatant are shown in the last column of Table 1. It is clear that the residual absorbance caused by unsedimented clay would result in a substantial overestimation of the free DNA if these solutions had been used for clay:DNA binding assays.
Table 1.
Fractions of Na and Ca montmorillonite clays that are resistant to precipitation by high speed centrifugation in unsonicated aqueous solutions.
| Supernatant |
||||||
|---|---|---|---|---|---|---|
| Homoionic clay | Initial [Clay] (mg/ml) | Centrifuge speed (× g) | A230 | Final [clay] a (mg/ml) | A260 | Predicted [dsDNA] b (μg/ml) |
| Na | 5 | 18,000 | 2.04 | 0.85 | 1.40 | 70.0 |
| Na | 5 | 40,000 | 0.34 | 0.13 | 0.19 | 9.5 |
| Ca | 5 | 18,000 | 0.65 | 0.11 | 0.29 | 14.5 |
| Ca | 5 | 40,000 | 0.24 | 0.03 | 0.13 | 6.4 |
Residual clay concentrations were calculated based on A230 values according to the Beer's law relationships determined in Figure 2. Samples were centrifuged for 40 min at 18,000 or 40,000 × g.
Predicted concentration of free double-stranded DNA (not bound by clay particles) if employed for clay:DNA binding study. Theoretical DNA concentrations were calculated based on the standard relationship: A260 of 1.00 = 50 μg/ml DNA.31
Sonication is often employed to break up clay tactoids into individual platelets prior to mixing with polymers for preparation of nanocomposites. While this is advantageous because it creates more surface area for potential interactions, it exacerbates the problem of unsedimented clay molecules that has plagued past clay:DNA adsorption studies. As shown in Table 2, residual Na clay in the supernatant was 2.5 fold higher when a 2 mg/ml suspension was sonicated before centrifuging at 18,000 × g. Even more striking, sonicated Ca clay supernatants retained 13 times more unsedimented clay than unsonicated solutions (0.27 mg/ml vs. 0.02 mg/ml final concentrations). The supernatants of sonicated solutions also exhibited stronger absorbances at 260 nm, 2-fold (Na) and 8-fold (Ca) higher than unsonicated suspensions (Table 2).
Table 2.
Impact of sonication on sedimentation of Na and Ca montmorillonite clays
| Homoionic clay | Initial [clay] | Supernatant |
||||
|---|---|---|---|---|---|---|
| A230 | [clay] a | A260 | ||||
| Na (unsonicated) | 2 mg/ml | 0.61 | 0.24 mg/ml | 0.52 | ||
| Na (sonicated) | 2 mg/ml | 1.46 | 0.61 mg/ml | 1.22 | ||
| Ca (unsonicated) | 2 mg/ml | 0.17 | 0.02 mg/ml | 0.16 | ||
| Ca (sonicated) | 2 mg/ml | 1.60 | 0.27 mg/ml | 1.36 | ||
Residual clay concentrations were calculated using the equations derived in Figure 2. Sonicated or unsonicated clay suspensions were centrifuged for 15 min at 18,000 × g.
The effects of altering concentrations of salts such as NaCl or MgCl2 on the affinity of DNA for clay surfaces has been investigated using centrifugation methods17,21,22, but their effect on precipitation of the clay particles is unknown. The possibility that salt concentration might differentially affect clay sedimentation was investigated in Figure 3. Absorbances of sonicated clay suspensions (0.5 mg/ml) at 230 nm were determined before and after centrifugation for 15 min at 18,000 × g in the presence of various concentrations of NaCl or MgCl2. The presence of the salts did not affect absorbance of uncentrifuged clay suspensions, but did exert a concentration-dependent effect on pelleting efficiency during centrifugation. With both Na- and Ca-montmorillonite, addition of NaCl had only slight effects at concentrations from 0.1 to 10 mM, but demonstrated a strong reduction of residual A230 above this threshold at 100 mM NaCl. In contrast, addition of MgCl2 caused precipitation at much lower concentrations (1 mM and above) for each type of montmorillonite (Figure 3A and 3B).
Figure 3.
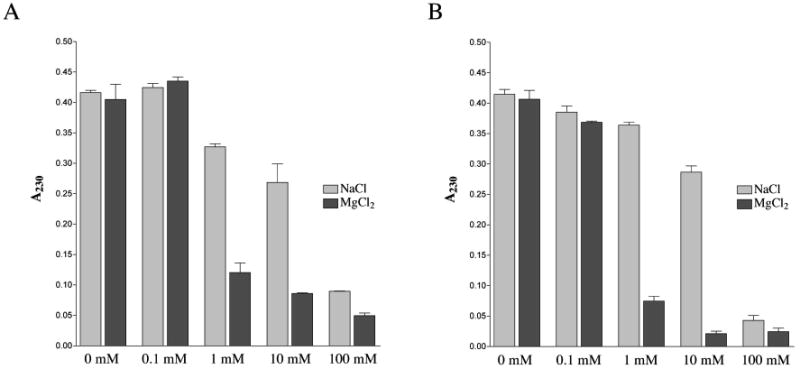
Effect of salt on the sedimentation efficiencies of Na and Ca montmorillonite. Absorbance at 230 nm was monitored after centrifugation of clay suspensions for 15 min at 18,000 × g. Error bars indicate standard deviations.
Individual montmorillonite platelets vary in size, with most sheets ∼ 1 nm thick and over 100 nm in each of the other two dimensions.1,2 Based on a typical sheet size of 1 × 200 × 200 nm, the molecular weight of a clay platelet is ∼ 4.44 × 107 (calculated based on the regular 2:1 layered structure of the clay, excluding the small contribution from Na or Ca ions [2-4% of total mass] and the water content of the dry powder). In the experiment cited above the 0.5 mg/ml clay solutions (1.1 × 10-5 mM) exhibited a threshold effect whereby transition from 10 mM to 100 mM NaCl strongly increased precipitation efficiency. This indicates that increasing the sodium-to-clay molar ratio from approximately 900,000 to 9,000,000 ions per platelet achieved a critical concentration of positive charges that could facilitate aggregation and precipitation of the particles. In the case of MgCl2 salt, increasing the magnesium-to-clay molar ratio from only 9,000 to 90,000 ions per sheet (0.1 mM to 1 mM in Figure 3) produced the same strong increase in aggregation and precipitation. This increased sensitivity of clay to Mg2+ is most likely due to the higher charge density of the ion and its ability to promote formation of tactoids.
A new DNA adsorption assay reduces problems due to overestimation of unbound DNA
Although montmorillonite absorbs light at 260 nm and is incompletely sedimented, even at high g-forces, it is possible to design an improved version of the standard clay:DNA binding assay as long as these factors are taken into consideration. The binding of Na montmorillonite to a single-stranded DNA oligonucleotide that is 25 nucleotides long (Pvu4a) was analyzed by a new version of the centrifugation method (Table 3). For these experiments, solutions containing sonicated clay only (0.5 mg/ml) or clay plus 20 ug/ml DNA were mixed and immediately centrifuged at 18,000 × g for 15 min. The A260 of each supernatant was determined and the values for the clay-only samples subtracted from those of the clay+DNA suspensions, allowing calculation of the free DNA concentration. All assays were performed with and without addition of mono- or divalent cations (NaCl or MgCl2). Three or four samples were tested at each salt concentration and results were averaged.
Table 3.
Impact of sodium and magnesium counterions on binding of single-stranded oligonucleotide DNA to sonicated homoionic Na montmorillonite
| Assay Components | Supernatant A260 | (Clay+DNA - Clay) Δ A260 | Free [DNA] (ug/ml) | Bound [DNA] | % DNAi bound |
|---|---|---|---|---|---|
| NaCl: | |||||
| 0 mM + DNA only | .685 | N/A | 22.6 | N/A | N/A |
| 0 mM + clay | .354 | ||||
| 0 mM + clay+DNA | .842 | .488 | 16.1 | 6.5 | 29% |
| 0.1 mM + clay | .315 | ||||
| 0.1 mM + clay+DNA | .853 | .538 | 17.8 | 4.8 | 22% |
| 1.0 mM + clay | .330 | ||||
| 1.0 mM + clay+DNA | .837 | .507 | 16.7 | 5.9 | 26% |
| 10 mM + clay | .339 | ||||
| 10 mM + clay+DNA | .818 | .479 | 15.8 | 6.8 | 30% |
| 100 mM + clay | .263 | ||||
| 100 mM + clay+DNA | .739 | .476 | 15.7 | 6.9 | 30% |
| MgCl2: | |||||
| 0 mM + DNA only | .685 | N/A | 22.6 | N/A | N/A |
| 0 mM + clay | .359 | ||||
| 0 mM + clay+DNA | .878 | .519 | 17.0 | 5.0 | 22% |
| 0.1 mM + clay | .344 | ||||
| 0.1 mM + clay+DNA | .847 | .503 | 16.6 | 6.0 | 26% |
| 1.0 mM + clay | .302 | ||||
| 1.0 mM + clay+DNA | .409 | .107 | 3.5 | 19.1 | 85% |
| 10 mM + clay | .283 | ||||
| 10 mM + clay+DNA | .338 | .055 | 1.8 | 20.8 | 92% |
| 100 mM + clay | .241 | ||||
| 100 mM + clay+DNA | .300 | .059 | 1.9 | 20.6 | 91% |
Clay suspensions with or without added NaCl or MgCl2 were centrifuged for 15 min at 18,000 × g. Bound DNA = initial DNA concentration ([DNA]i) minus free (supernatant) DNA concentration expressed as ug/ml. N/A, not applicable.
Addition of NaCl to final concentrations of 0.1, 1.0, 10 or 100 mM had no effect on association of oligonucleotide DNA with the clay particles, with bound DNA ranging from 22 – 30% under all conditions (Table 3). This result indicates that the interaction between the DNA and the exchangeable sodium cation on the surface of the clay is only moderately strong and, since there are only a limited number of exchange sites, that the changes in sodium ion concentration in the surrounding solution would not be expected to change the sorption of the DNA. Results with MgCl2 were similar at 0.1 mM (26.5% bound), but the fraction of DNA bound to the clay jumped to 85, 92 and 91% at 1.0, 10 and 100 mM MgCl2, respectively. It is important to note that the the single-stranded 25-mer DNA does not precipitate under the conditions used here. The A260 of solutions containing DNA only was not reduced by centrifugation in water or in the presence of the highest concentrations of NaCl or MgCl2 employed in these studies (100 mM).
The cation-dependence of binding of single-stranded DNA to Ca-montmorillonite was assessed in Table 4. DNA binding was only moderately promoted at high concentrations of sodium, peaking at 58% in the presence of 100 mM NaCl. As observed for the Na clay, association with DNA was increased in the presence of MgCl2, reaching 93-95% bound when Mg2+ concentrations of 1 mM and above were used. This is reasonable since the higher charge density of the magnesium will allow it to readily bridge the clay to the DNA.
Table 4.
Impact of sodium and magnesium counterions on binding of single-stranded oligonucleotide DNA to sonicated homoionic Ca montmorillonite
| Salt added | Supernatant A260 | (Clay+DNA - Clay) Δ A260 | Free [DNA] (ug/ml) | Bound [DNA] | % DNAi bound |
|---|---|---|---|---|---|
| NaCl | |||||
| 0 mM + DNA only | .813 | N/A | 26.8 | N/A | N/A |
| 0 mM + clay | .536 | ||||
| 0 mM + clay+DNA | 1.088 | .552 | 18.2 | 8.6 | 32% |
| 0.1 mM + clay | .590 | ||||
| 0.1 mM + clay+DNA | 1.098 | .508 | 16.7 | 10.1 | 38% |
| 1.0 mM + clay | .843 | ||||
| 1.0 mM + clay+DNA | 1.247 | .404 | 13.3 | 13.5 | 50% |
| 10 mM + clay | .762 | ||||
| 10 mM + clay+DNA | 1.201 | .440 | 14.5 | 12.3 | 46% |
| 100 mM + clay | .659 | ||||
| 100 mM + clay+DNA | .996 | .337 | 11.1 | 15.7 | 58% |
| MgCl2 | |||||
| 0 mM + DNA only | .791 | N/A | 26.1 | N/A | N/A |
| 0 mM + clay | .494 | ||||
| 0 mM + clay+DNA | .931 | .437 | 14.4 | 11.7 | 45% |
| 0.1 mM + clay | .563 | ||||
| 0.1 mM + clay+DNA | .778 | .215 | 7.1 | 19.0 | 73% |
| 1.0 mM + clay | .447 | ||||
| 1.0 mM + clay+DNA | .499 | .052 | 1.7 | 24.4 | 93% |
| 10 mM + clay | .441 | ||||
| 10 mM + clay+DNA | .482 | .041 | 1.4 | 24.7 | 95% |
| 100 mM + clay | .395 | ||||
| 100 mM + clay+DNA | .440 | .046 | 1.5 | 24.6 | 94% |
Ca-clay suspensions with or without added sodium or magnesium chloride were centrifuged for 15 min at 18,000 × g. Bound DNA = initial DNA concentration ([DNA]i) minus free [DNA] expressed as ug/ml. N/A, not applicable.
To directly compare results of the new approach with previous methods, the DNA adsorption assay was repeated exactly as described for Tables 3 and 4 with MgCl2 except that unsonicated clay was used and the absorbance of the unsedimented clay was not subtracted from the samples containing clay plus DNA. The fractions of DNA bound to Na clay and to Ca clay in the presence of increasing concentrations of MgCl2 are shown in Figure 4A and 4B, respectively. For both Na and Ca montmorillonite, the deduced fraction bound was smaller with the unsonicated clays due to overestimation of free DNA in the supernatants after centrifugation.
Figure 4.
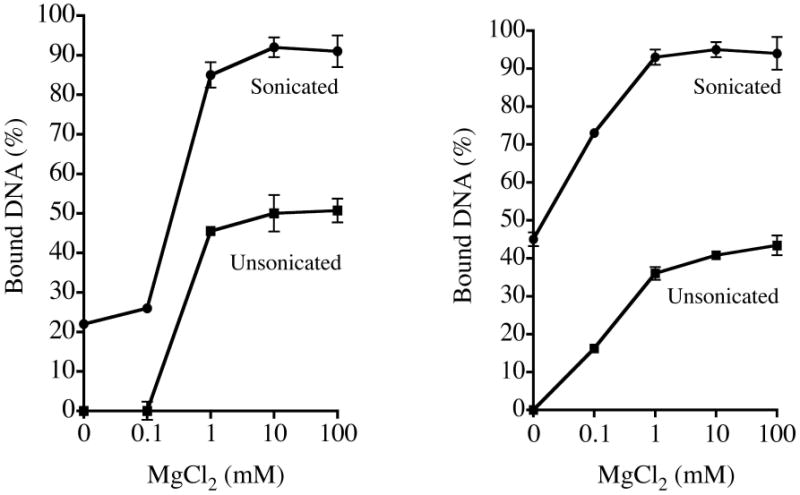
Comparison of DNA adsorbed onto (A) Na montmorillonite and (B) Ca montmorillonite with or without MgCl2. Experiments utilized previously described methods with unsonicated clay (squares) or a new protocol employing sonicated clay and subtraction of A260 contributions due to unsedimented clay (circles).
Adsorption of single-stranded oligonucleotide DNA onto sonicated montmorillonite produces intercalated structures
As given in the experimental section, the Cerius 2 software was utilized to calculate the expected d spacing between clay galleries if the DNA were to intercalate. This program has been previously used to analyze gallery spacing within polymer:clay nanocomposites, demonstrating strong agreement with experimentally determined distances.32,33 The program was used to analyze simple platelet:DNA model structures formed by each of the two clays with oligonucleotide DNA. After multiple rounds of energy minimization a lowest energy structure was predicted. For the structure depicted in Figure 5A, the clay plates have been portrayed as polyhedrons to increase contrast between the plates and the DNA. The mode of bonding in the gallery that was predicted involves cations on the surface of the clay (shown in purple) forming bridges between the phosphate groups of the DNA and the clay plates. This bonding is illustrated in detail in Figure 5B, where a sodium cation that bridges between the silica surface and a phosphate can be seen. The sodium ion in this case is 0.252 and 0.239 nm away from the two free oxygen atoms of the phosphate group, with the bonds illustrated in the figure by thin, dark gray lines connecting the sodium to the two oxygens. In some locations a sodium only associates with one oxygen on the phosphate and in these interactions the bonding distance is reduced to 0.218 nm. Predicted gallery d-spacings are shown in Figure 5C. The nanocomposite structures were predicted to exhibit spacings of 2.03 and 1.84 nm for the Na and Ca clay:DNA complexes, respectively.
Figure 5.
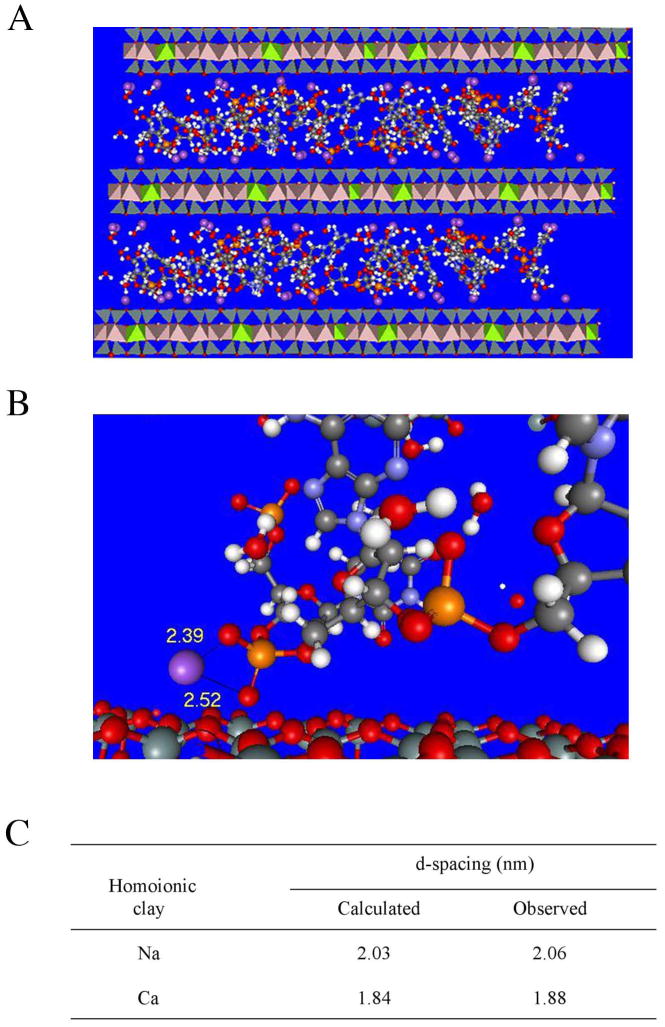
(A) Atomic structure of Na montmorillonite platelets associated with intercalated single-stranded DNA predicted by Cerius 2 molecular modeling program after energy minimization iterations. Purple spheres, sodium ions; red, oxygen; white, hydrogen; grey, carbon; orange, phosphorus; silver, silicon. (B) Detailed view of cation bridging interactions with bond distances between a representative sodium atom and two oxygen atoms shown (given by the program in Angstroms). (C) Gallery spacings within montmorillonite:single-stranded DNA nanocomposites measured using X-ray diffraction analysis are compared with spacings predicted by modeling.
Structural analysis of the molecules formed by binding of oligonucleotide DNA to each of the sonicated clays in the presence of 10 mM MgCl2 was performed using X-ray diffractometry. In these experiments the clay-to-DNA mass ratio was set at 2:1 in order to enhance the likelihood that any intercalate formed would be readily detected. Both clays formed intercalated structures, with spacing of 2.06 and 1.88 nm for Na and Ca montmorillonite, respectively, corresponding to differences of only 1.5% and 2.1% from the predicted values (Figure 5C). These spacings correspond to increases of 1.06 and 0.88 nm over the inter-platelet distances observed in each clay alone when it is dehydrated. The expansion of the galleries caused by the presence of the single-stranded DNA is consistent with the diameter of an individual strand of DNA, which is approximately 1 nm.
Discussion
In the current study we have analyzed the spectrophotometric and sedimentation properties of aqueous suspensions of sodium and calcium montmorillonite. Results of these experiments were used to develop improved assays for quantitating the binding of single-stranded DNA molecules to clay surfaces. The approach described here should be generally applicable to studies of the interaction of clays with many other biopolymers and organic molecules.
Spectrophotometric scanning of Na and Ca clays revealed strong absorbance peaks for each suspension near 230 nm. Chen et al. and Karickhoff and Bailey demonstrated that the broad absorbance of montmorillonite under 300 nm is due to the presence of Fe(III) within the clay.28,29 Loss of transmittance at higher wavelengths (above 300 nm) observed in the current study could be attributed primarily to light scattering caused by tactoid complexes in the solution.
The absorbances of sonicated and unsonicated Ca clays monitored in the Beer's Law experiments were strikingly different. Ca montmorillonite forms tactoids in aqueous suspensions whose average sizes are larger than those of homoionic Na or Li clays.7,26,29,30 The greater absorbance of the Ca clay solution after sonication (reducing most particles to platelet monomers) is consistent with prior studies demonstrating increased absorbance in solutions with smaller tactoids, e.g., in unsonicated Li or Na versus Ca clay suspensions. Karickhoff and Bailey suggested that shielding in the larger tactoids reduced their ability to absorb photons, though Chen et al. attributed the reduction in absorbance to destructive interferences between light waves passing through the inter- and intraparticle spaces.28,29 Na montmorillonite particles form relatively small tactoid sizes in water.7,26,30 We observed little difference in the Beer's Law absorbances of the sonicated Na clay versus the unsonicated Na clay, presumably because of the similarity in size distributions.
Many past studies of the association of nucleic acids with clay surfaces have used centrifugation to separate clay particles and clay:nucleic acid complexes from unbound nucleic acids.9,11,13-24 However, we observed that sedimentation of montmorillonite was inefficient, even at forces up to 40,000 × g. Sedimentation was particularly poor with sonicated clay solutions, which are preferred as starting material because they are composed primarily of individual platelet molecules, and was also strongly dependent on salt concentration. MgCl2 supplementation stimulated clay precipitation at lower concentrations than NaCl. This is likely a reflection of its greater ability to form stable cation bridges between the silica surfaces, leading to aggregation of the platelets into higher molecular weight species.
The data presented here suggest that the combination of two factors, strong absorbance of montmorillonite at 260 nm and inefficient precipitation of the clay by centrifugation, produced a consistent overestimation of free DNA concentration in past studies. The problem was exacerbated in some experiments because of the high clay concentrations used, e.g., four studies employed clay solutions between 8 mg/ml and 50 mg/ml.9,13,16,20 A possible exception to this trend was the report by Poly et al., which detected unsedimented DNA using a diphenylamine-based calorimetric assay.20 However, the assay used 8 mg/ml montmorillonite solutions and required measurement of absorbance at 590 nm, where clay scatters light, and it is unclear if unprecipitated clay particles may have influenced the outcome. To improve the assay, we developed a new approach that allows subtraction of absorbance due to unsedimented clay from the overall absorbance at 260 nm. The new methodology was employed to assess binding of Na and Ca montmorillonite to single-stranded oligonucleotide DNA molecules that are 25 nucleotides long. Addition of NaCl to clay+DNA solutions did not promote strong adsorption, even at concentrations as high as 100 mM. In contrast, addition of MgCl2 at concentrations of only 1.0 mM (for Na montmorillonite) or 0.1 mM (for Ca montmorillonite) stimulated strong adsorption of the DNA strands to the clay. These observations are qualitatively consistent with previous studies demonstrating that divalent metal cations promote DNA:clay association.17,21,22
X-ray diffraction demonstrated that the nanocomposites formed by mixing Na or Ca clays with single-stranded DNA in the presence of 10 mM MgCl2 formed intercalated structures. The resulting d-spacings were consistent with an expansion of the galleries due to the presence of inserted DNA strands. The observed spacings were also in accord with distances predicted using the Cerius 2 atomistic modeling program, with the program predicting essentially a 1 nm spacing in the galleries. This value is close to what was determined experimentally and is also in agreement with theoretical predictions since it is half of the 2 nm diameter of conventional B-form helical double-stranded DNA.
The structure of clay:DNA complexes has been investigated previously using methods such as X-ray diffraction, transmission and scanning electron microscopy, and FT-IR.9,14,20 These experiments suggested that all nucleic acids tested, including double-stranded DNA (linear and circular) as well as single-stranded molecules, adsorb primarily to the surfaces and external edges of the clay without intercalation. Thus, intercalated structures were readily detected in the current work, but not in the earlier studies. The most likely differences would appear to be that (a) clay platelets used in the current study were strongly dispersed by sonication prior to mixing with the DNA and (b) the mass ratio of clay to DNA employed in the earlier experiments was much higher than the ratio of 2:1 used in this study, and detection of an intercalated system would have been unlikely or impossible under such conditions. Montmorillonite and halloysite have shown promise recently as vectors for gene delivery into cells, with potential applications in cancer and gene therapy.10,11,12 The determination here, for the first time, that clays are capable of forming nanocomposite materials containing intercalated DNA is important as the use of clays in biochemistry and medicine continues to expand.
Acknowledgments
KL was supported in part by National Institutes of Health grant 1R15AG028520-01A1 and a departmental research grant from the Welch Foundation.
References and Notes
- 1.Pinnavaia TJ, Beall GW, editors. Polymer-clay nanocomposites. Wiley Press; Chichester, UK: 2000. [Google Scholar]
- 2.Chen B. Brit Ceramic Trans. 2004;103:241–249. [Google Scholar]
- 3.Carrado KA. Appl Clay Sci. 2000;17:1–23. [Google Scholar]
- 4.Carretero MI. Appl Clay Sci. 2002;21:155–163. [Google Scholar]
- 5.Choy JH, Choia SJ, Oha JM, Park T. Appl Clay Sci. 2007;36:122–132. [Google Scholar]
- 6.Schramm LL, Kwak JCT. Clays Clay Min. 1982;30:40–48. [Google Scholar]
- 7.Whalley WR, Mullins CE. Clay Minerals. 1991;26:11–17. [Google Scholar]
- 8.Vaia RA, Jandt KD, Kramer EJ, Giannelis EP. Chem Materials. 1996;8:2628–2635. [Google Scholar]
- 9.Franchi M, Bramanti E, Bonzi LM, Gallori E. Orig Life Evol Biosph. 1999;29:297–315. doi: 10.1023/a:1006557832574. [DOI] [PubMed] [Google Scholar]
- 10.Kawase M, Hayashi Y, Kinoshita F, Yamato E, Miyazaki J, Yamakawa J, Ishida T, Tamura M, Yagi K. Biol Pharm Bull. 2004;27:2049–2051. doi: 10.1248/bpb.27.2049. [DOI] [PubMed] [Google Scholar]
- 11.Lin FH, Chen CH, Cheng WT, Kuo TF. Biomaterials. 2006;27:3333–3338. doi: 10.1016/j.biomaterials.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 12.Shamsi MH, Geckeler KE. Nanotechnology. 2008;19:1–5. doi: 10.1088/0957-4484/19/7/075604. [DOI] [PubMed] [Google Scholar]
- 13.Pietramellara G, Franchi M, Gallori E, Nannipieri P. Biol Fertil Soils. 2001;33:402–409. [Google Scholar]
- 14.Khanna M, Yoder M, Calamai L, Stotzky G. Sci Soils. 1998;3:1–10. [Google Scholar]
- 15.Banin A, Lawless JG, Mazzurco J, Church FM, Margulies L, Orenberg JB. Orig Life Evol Biosph. 1985;15:89–101. doi: 10.1007/BF01809491. [DOI] [PubMed] [Google Scholar]
- 16.Ferris JP, Ertem G, Agarwal VK. Orig Life Evol Biosph. 1989;19:153–164. doi: 10.1007/BF01808149. [DOI] [PubMed] [Google Scholar]
- 17.Paget E, Monrozier LJ, Simonet P. Microbiology Letters. 1992;97:31–40. [Google Scholar]
- 18.Khanna M, Stotzky G. Appl Environ Microbiol. 1992;58:1930–1939. doi: 10.1128/aem.58.6.1930-1939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallori E, Bazzicalupo M, Dal Canto L, Fani R, Nannipieri P, Vettori C, Stotzky G. FEMS Microbiology Ecology. 1994;15:119–126. [Google Scholar]
- 20.Poly F, Chenu C, Simonet P, Rouiller J, Monrozier LJ. Langmuir. 2000;16:1233–1238. [Google Scholar]
- 21.Franchi M, Ferris JP, Gallori E. Orig Life Evol Biosph. 2003;33:1–16. doi: 10.1023/a:1023982008714. [DOI] [PubMed] [Google Scholar]
- 22.Cai P, Huang Q, Zhang X. Appl Clay Sci. 2006;32:147–152. [Google Scholar]
- 23.Cai P, Huang Q, Chen W, Zhang D, Wang K, Jiang D, Liang W. Soil Biol Biochem. 2007;39:1007–1013. [Google Scholar]
- 24.Cai P, Huang Q, Zhu J, Jiang D, Zhou X, Ron X, Liang W. Colloids Surfaces B: Biointerfaces. 2007;54:53–59. doi: 10.1016/j.colsurfb.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Rappe AK, Goddard WA., III J Phys Chem. 1991;95:3358. [Google Scholar]
- 26.Banin A, Lahav N. Nature. 1968;217:1146–1147. [Google Scholar]
- 27.Bishop JL, Louris SK, Rogoff DA, Rothschild LJ. Intl J Astrobiol. 2006;5:1–12. [Google Scholar]
- 28.Karickhoff SW, Bailey GW. Clays Clay Min. 1973;21:59–70. [Google Scholar]
- 29.Chen Y, Shaked D, Banin A. Clay Min. 1979;14:93–102. [Google Scholar]
- 30.Yariv S, Lapides I. Clays Clay Min. 2003;51:23–32. [Google Scholar]
- 31.Sambrook J, Russell SW. Molecular cloning: a laboratory manual. 3rd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 32.Bartels J, Beall GW, Grah M, Jin K, Speer D, Yarbrough J. J Applied Polymer Sci. 2008;3:1908–1916. [Google Scholar]
- 33.Beall GW, Goss M. Applied Clay Science. 2004;27:3–4. [Google Scholar]