Abstract
Amelogenin is a unique protein that self-assembles into spherical aggregates called “nanospheres” and is believed to be involved in controlling the formation of the highly anisotropic and ordered hydroxyapatite crystallites that form enamel. The adsorption behavior of amelogenin onto substrates is of great interest because protein-surface interactions are critical to its function. We report studies of the adsorption of amelogenin onto self-assembled monolayers containing COOH end group functionality as well as single crystal fluoroapatite, a biologically relevant surface. We found that although our solutions contained only nanospheres of narrow size distribution, smaller structures such as dimers or trimers were observed on the hydrophilic surfaces. This suggests that amelogenin can adsorb onto surfaces as small structures that “shed” or disassemble from the nanospheres that are present in solution.
Keywords: amelogenin, nanospheres, quaternary structure
Introduction
The amelogenin protein is involved in the formation of the highly controlled hydroxyapatite crystals found in tooth enamel.1 These crystals have unusually high aspect ratios, much higher than those found for bone or man-made apatite, and are assembled into ordered bundles called “prisms” resulting in the formation of a biomineral with exceptional mineral content and mechanical hardness. Although the function of amelogenin is not completely understood, roles in nucleation, growth, regulation of crystal size and shape, and control of crystal–crystal aggregation have been proposed.1–3 Amelogenin is a unique biomineralization protein because it self-assembles to form supramolecular quaternary structures called “nanospheres,” spherical aggregates of amelogenin monomers typically 20–60 nm in diameter.4 Nanospheres have been detected in solution by dynamic light scattering (DLS)5 and nanospheres have been observed in vivo, within growing enamel,6 suggesting their importance in the proper development of enamel. Although the nanosphere quaternary structure has been observed in solution, the quaternary structure of amelogenin adsorbed onto surfaces is of special interest because the function of amelogenin is thought to depend on its interactions with surfaces. Previous studies have shown that amelogenin can adsorb onto enamel,7 fluoroapatite (FAP),8 and HAP9 substrates, consistent with the premise that amelogenin's role involves interactions with surfaces.
We report studies of the adsorption of amelogenin onto self-assembled monolayers (SAMs) containing COOH end group functionality as well as single crystal FAP. SAMs have highly controlled chemistry and structures, making them ideal model systems for the study of protein interactions,10 and FAP surfaces have important biological relevance. Both systems can be made molecularly smooth, greatly aiding the experimental determination of quaternary structure. The quaternary structures of the protein in solution as determined by DLS were compared with the quaternary structures of the protein physisorbed onto surfaces as studied by atomic force microscopy (AFM).
Results
Solution Studies
Studies were done using the histidine-tagged amelogenin, rp(H)M180, and native amelogenin, rM179, with no significant difference in surface structure. The protein was prepared in solutions adjusted to pH 8 at 158 μg/ml (see experimental details in the Supp. Info.). Figure 1 showed size distributions of the protein solutions determined by DLS from the autocorrelation function (Figure S1a). The particle size distribution was bimodal with one peak at 42 ± 7 nm and one peak at 204 ± 32 nm diameter. The 42 nm peak was consistent with a previously determined DLS size for rp(H)M180 that was attributed to the diameter of individual nanospheres.5 Protein solutions were mixed with a paraformaldehyde/glutaraldehyde fixative and dropped onto freshly cleaved mica. This technique was thought to preserve the integrity of the nanospheres in solution.11 If we exposed mica to the protein solutions without mixing with fixative we observed small subnanosphere-sized structures similar to those shown in Figure 2. AFM images of the fixed nanospheres (Figure 1b) showed that the ∼200 nm structures observed by DLS were clusters of several nanospheres. Nanosphere diameters determined by analysis of the AFM images were 24 ± 5 nm. Discrepancies between the sizes of the smallest structures determined by DLS and other methods such as transmission electron microscopy (TEM), AFM, and small angle X-ray scattering (SAXS) have been seen previously5,11–13 and have been attributed to extension of C-terminal domains to form an outer shell.13 We believe the larger DLS sizes may be due to a small degree of clustering of nanospheres in the small size fraction (Figure S1b). The DLS data showed that the nanospheres had a narrow size distribution with standard error of ∼17% and that there was no evidence for the presence of smaller structures such as monomers and dimers.
FIGURE 1.
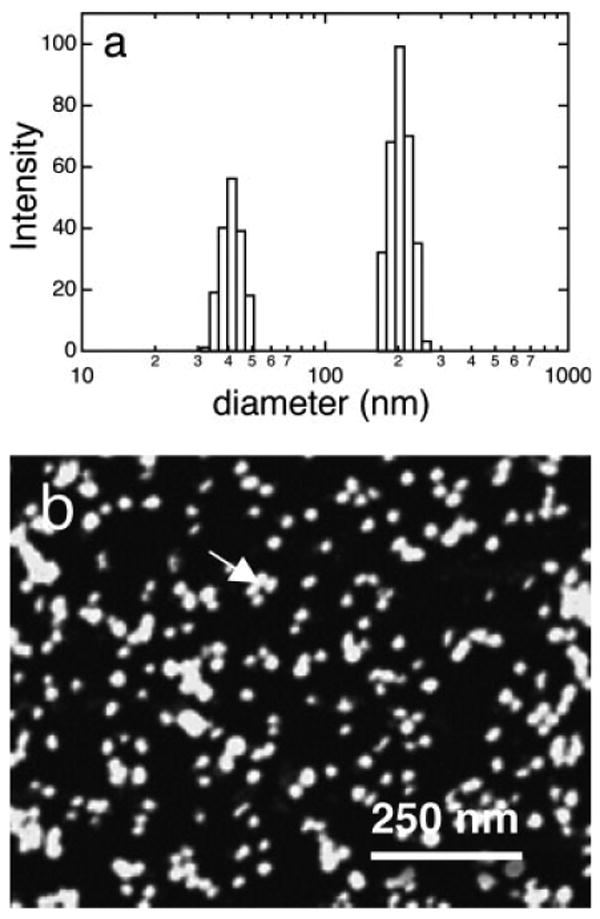
(a) Size distributions of amelogenin in pH 8 solutions at 158 μg/ml by DLS showing peaks at 42 and 204 nm (attributed to clusters of nanospheres) and (b) AFM image of protein solutions mixed with a fixative and dropped onto mica showing individual nanospheres and clusters of nanospheres (see arrow). The fixative method preserves the structures of the nanospheres in solution.
FIGURE 2.
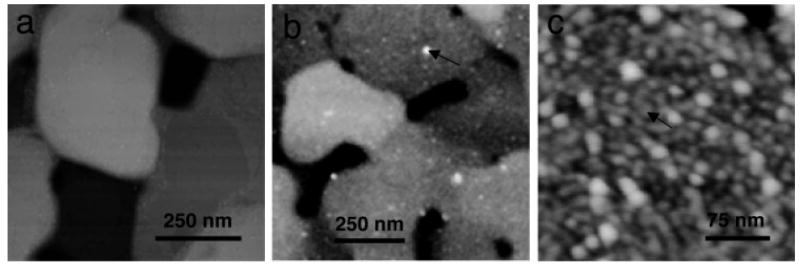
Tapping mode AFM images of (a) COOH SAMs on atomically smooth gold terraces on mica, (b) amelogenin adsorbates onto COOH SAMs showing a high coverage of oligomers and just a few remnant nanospheres (arrow), (c) higher resolution 300 nm scan of adsorbates on COOH3 SAMs (arrow points to amelogenin dimer/trimer). The adsorption of oligomers is suggested by the ellipsometric thicknesses of 7.5–8.0 nm. Although the AFM diameters of the oligomers were large because of the tip broadening effect (16 nm), these structures had much smaller AFM heights (1.2 nm) and were clearly much smaller than the sparse remnant nanospheres (∼21 nm × 7 nm).
Surface Studies
SAMs with COOH terminal groups and diamond polished (100) and (110) faces of FAP were exposed to the nanosphere containing solutions, removed, rinsed with water, and dried. Figure 2a showed a 1 μm scan of a COOH SAM on gold on mica revealing large 500–800 nm atomically smooth gold terraces separated by 1–5 nm step edges. The images of protein adsorbates on COOH SAMs (Figures 2b and 2c) showed several large nanosphere-like structures overlying smaller adsorbates at much higher coverage. The smaller adsorbates were difficult to resolve in the 1 μm scan but can be seen more clearly in the 300 nm scan of Figure 2c. Ellipsometric measurements of the protein layers resulted in thicknesses of 7.5–8.0 nm. Because the amelogenin monomers is estimated to be 4.4–4.6 nm in diameter and amelogenin oligomers such as dimers, trimers, and hexamers are calculated to be 7.0–9.6 nm in diameter,12 we believe the small adsorbates were small amelogenin oligomers.
Unfortunately, AFM did not accurately measure the size of the small adsorbate structures because AFM overestimated the diameter of structures that were smaller than the radius of curvature of the tip (∼10–20 nm) by several times due to the well-known tip broadening effect.14 The large tip exaggerates the lateral dimensions as it traces over a structure smaller than the radius of the tip. We believe that the AFM diameters of the much larger solution nanospheres in Figure 2b were fairly accurate because the size is larger then the radius of curvature of most tips and was similar to sizes obtained by TEM.12 Also, AFM greatly underestimated the height of soft protein structures by as much as six times because of the nonlinear dynamic response of the oscillating cantilever.15 Although AFM height and diameter measurements of the small adsorbates and height measurements of the large adsorbates were inaccurate, we found they were useful for comparison purposes and could be calibrated with the ellipsometry and X-ray photoelectron spectroscopy (XPS) results. The small adsorbates averaged 16.4 nm × 1.2 nm. The diameter overestimated the oligomer size by a factor of two and the height underestimated the size by a factor of six. Adsorption of the protein onto the COOH SAMs was also evidenced by the presence of infrared vibrational modes for the peptide bond at 1677 cm−1 (amide I) and 1544 cm−1 (amide II) as determined by external reflectance infrared spectroscopy and by changes in the advancing contact angle of water from 20° to 54°.
The polished FAP surfaces (Figure 3a) had root mean square (rms) roughnesses of 0.2–0.3 nm, indicating they were molecularly smooth. AFM images of protein adsorbates on FAP (Figures 3b and 3c) showed large nanospheres overlying small adsorbates at high coverage. Protein adsorption onto the FAP surfaces was also evidenced by the presence of C1s and N1s peaks as determined by XPS (Table S1) and changes in advancing contact angle of water from 15° to 50°. The adsorbate thicknesses determined by XPS (by the change in intensity of the substrate peaks due to the overlayer)16 were ∼8.0 nm suggesting that the small structures were dimer/trimer in thickness, similar to the COOH surfaces. The average AFM diameter was 15.5 nm, also consistent with the COOH surfaces. Like the COOH surfaces, the AFM diameter overestimated the size of the structures by a factor of two. The large nanosphere-like structures averaged 19.0 nm × 7.0 nm in size (diameter × height), smaller than the AFM determined size of the original nanospheres (24.3 nm × 12.5 nm). The large structures were partial nanospheres, therefore, remnants of the original nanospheres, and appeared to adsorb as multilayers over the underlying oligomer layers.
FIGURE 3.
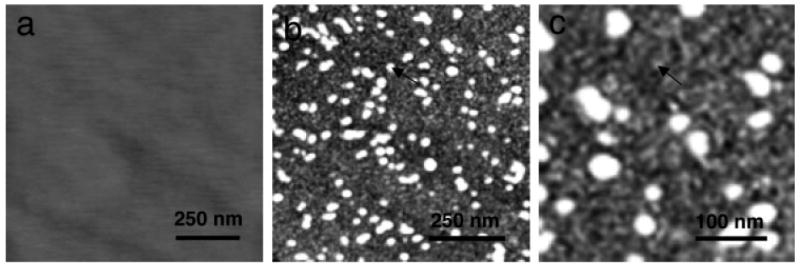
Tapping mode AFM images of (a) bare molecularly smooth single crystal FAP substrate, (b) 1 μm scan of amelogenin adsorbates onto FAP showing a low coverage of remnant nanospheres (arrow) overlying a high coverage of primarily dimer/trimer thickness adsorbates, and (c) a higher magnification view of the image in b showing the small oligomeric adsorbates (arrow). Oligomers are suggested by the 8 nm thicknesses determined by XPS.
Discussion
The AFM images and ellipsometry measurements indicate that the amelogenin structures binding to the surfaces are not intact nanospheres but much smaller structures that were dimers to hexamers in thickness. It is possible that these structures were present in the solutions and not detected by DLS, however, numerous other studies have also only found nanospheres in amelogenin solutions at pH 8 using DLS as well as SANS and SAXS.5,11–13 Monomers and dimers have only been detected in acidic solutions less than pH 413 and in nonpolar solvents such as 60% acetonitrile in water.12 Given the fact only nanospheres were observed in our solutions, we propose that the subnanosphere-sized amelogenin structures “disassemble” or “shed” from the nanospheres onto surfaces. The removal of amelogenin from the nanospheres was also evidenced by the partial nanospheres adsorbed onto FAP surfaces as multilayers. This “shedding” phenomenon is very similar to the “fusion” of spherical phospholipid vesicles onto surfaces to form planar lipid monolayers.17 In that work, the interactions between the phospholipid and the surface are stronger than the interactions holding the vesicles together, “breaking up” the vesicles.
The amelogenin protein rpH(M180) is a ∼21 kD protein with a large, highly hydrophobic central region and charged regions in the C-terminal domain and N-terminus. It has been suggested that amelogenin monomers assemble into dimers, trimers, and hexamers which in turn assemble into the larger nanosphere.12 In our study, small protein structures were disassembled from the nanospheres in solution onto surfaces, reversing the process of nanosphere assembly. Since nanosphere formation and disassembly is a reversible process in going from one solvent condition to another, we believe it is reasonable to expect that surfaces could also promote disassembly. Amelogenin disassembled onto hydrophilic surfaces similar to the way that vesicles can disassemble onto hydrophilic surfaces promoted by electrostatic interactions between polar head groups of lipids and hydrophilic surfaces such as glass and mica.18
In our work, it is expected that the charged C-terminal domain containing both positively and negatively charged residues (DKTKREEVD-COOH) would promote adsorption onto the charged COOH and FAP surfaces (see Supp. Info.). TEM studies have shown that the nanospheres consist of substructures ∼4–8 nm in diameter.12 The 8 nm substructures have been suggested to be amelogenin dimers or trimers with the C-terminus located on the surface of the oligomer and the hydrophobic domains oriented toward the center of the oligomer. The charged C-terminal domains of the oligomers may interact with the hydrophilic COOH and FAP surfaces resulting in the breaking away of the entire 8 nm oligomer, overcoming the interactions holding the oligomers together within the nanosphere. We believe this is a reasonable mechanism to propose but ongoing research on the internal structure of the nanospheres and the structure of the adsorbed oligomers will be necessary to improve our understanding of the disassembly mechanism.
Previous studies have used AFM to report the adsorption of nanosphere-like structures onto enamel crystals and FAP.7,8 Our work would suggest that the nanosphere-like structures are remnant nanospheres and may overlie smaller disassembled adsorbates. We found that it was difficult to detect the small adsorbates on rough surfaces such as SAMs on polycrystalline gold so that these structures may have gone undetected on the rougher enamel crystals.7 The underestimation of protein heights by AFM contributed to the difficulty in detecting these structures (1–2 nm in height relief) without the use of molecularly smooth substrates. Also, the nanospheres in previous studies were present at a high coverage and may have masked the underlying disassembled adsorbates. Small structures were present in AFM images of amelogenin adsorbed onto FAP surfaces by Habelitz et al. (Figure 1f)9 and may have been disassembled structures.
This work reveals that amelogenin can exist at interfaces as relatively small units such as dimers, trimers, and hexamers. The nanosphere quaternary structure has been observed in vivo and may have important biological function. Our work suggests that amelogenin may also exist as small oligomers in vivo. To our knowledge, there has been very little work to look for these types of structures within developing enamel, understandably a very challenging experimental task. It would be very interesting if the various quaternary structures allow amelogenin to have several biological functions. For example, since nanospheres have been seen in rows between enamel crystals,11 the protein in the nanosphere quaternary structure may aid in promoting the self-assembly of mineral crystals, acting as “cushions” between the mineral crystals, guiding their spacing and organization. Our adsorption studies onto FAP suggest that amelogenin in the small oligomer quaternary structure may function by adsorbing onto specific faces of apatite, controlling crystal growth. The ability of amelogenin to adopt several quaternary structures by assembling into nanospheres and disassembling onto surfaces makes it a very interesting biomineralization protein, one that offers a unique opportunity to explore relationships between protein structure and function.
Acknowledgments
Contract grant sponsor: NIH-NIDCR, Contract grant number: DE-015347
Footnotes
Additional Supporting Information may be found in the online version of this article.
This research was performed at Pacific Northwest National Laboratory, operated by Battelle for the US-DOE. A portion of the research was performed in the EMSL, a national scientific user facility sponsored by the DOE-OBER at PNNL.
Reviewing Editor: David Case
References
- 1.Fincham AG, Moradian-Oldak J, Simmer JP. J Struct Biol. 1999;136:270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 2.Moradian-Oldak J. Matrix Biol. 2001;20:293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- 3.Moradian-Oldak J, Tan J, Fincham AG. Biopolymers. 1998;46:225–238. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 4.Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. Biopolymers. 1994;34:1339–1347. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- 5.Moradian-Oldak J, Paine ML, Lei YP, Fincham AG, Snead ML. J Struct Biol. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 6.Fincham AG, Moradian-Oldak J, Diekwisch TGH, Lyaruu DM, Wright JT, Bringas P, Jr, Slavkin HC. J Struct Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 7.Wallwork ML, Kirkham J, Zhang J, Smith DA, Brookes SJ, Shore RC, Wood SR, Ryu O, Robinson C. Langmuir. 2001;17:2508–2513. [Google Scholar]
- 8.Habelitz S, Kullar A, Marshall SJ, DenBesten PK, Balooch M, Marshall GW, Li W. J Dent Res. 2004;83:698–702. doi: 10.1177/154405910408300908. [DOI] [PubMed] [Google Scholar]
- 9.Bouropoulos N, Moradian-Oldak J. Calcif Tissue Int. 2003;72:599–603. doi: 10.1007/s00223-002-1099-1. [DOI] [PubMed] [Google Scholar]
- 10.Prime KL, Whitesides GM. Science. 1991;252:1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 11.Wen HB, Fincham AG, Moradian-Oldak J. Matrix Biol. 2001;20:387–395. doi: 10.1016/s0945-053x(01)00144-5. [DOI] [PubMed] [Google Scholar]
- 12.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 13.Aichmayer B, Margolis HC, Sigel R, Yamakoshi Y, Simmer JP, Fratzl P. J Struct Biol. 2005;151:239–249. doi: 10.1016/j.jsb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Garcia R, Perez R. Surf Sci Rep. 2002;47:197–301. [Google Scholar]
- 15.Round AN, Miles MJ. Nanotechnology. 2004;15:S176–S183. [Google Scholar]
- 16.Cumpson PJ, Seah MP. Surf Interface Anal. 1997;25:430–446. [Google Scholar]
- 17.Meuse CW, Niaura G, Lewis ML, Plant A. Langmuir. 1998;14:1604–1611. [Google Scholar]
- 18.Richter R, Mukhopadhyay A, Brisson A. Biophys J. 2003;85:3035–3047. doi: 10.1016/S0006-3495(03)74722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


