Abstract
Distinct from other spirochetes, cells of Leptospira interrogans contain orthologues of all the Escherichia coli lpx genes required for lipid A biosynthesis, but they synthesize a modified form of lipopolysaccharide that supposedly activates toll-like receptor 2 (TLR2) instead of TLR4. The recent determination of the L. interrogans lipid A structure revealed an unprecedented O-methylation of its 1-phosphate group. The enzymatic activity responsible for selective 1-phosphate methylation has not been previously explored. A membrane enzyme that catalyzes the transfer of a methyl group from S-adenosylmethionine (SAM) to the 1-phosphate moiety of E. coli Kdo2-[4′-32P]lipid A has now been discovered. The gene encoding this enzyme was identified based on the hypothesis that methylation of a phosphate group is chemically analogous to methylation of a carboxylate moiety at a membrane-water interface. Database searching revealed a candidate gene (renamed lmtA) in L. interrogans showing distant homology to the yeast isoprenylcysteine carboxyl methyltransferase, encoded by sterile-14, which methylates the a-type mating factor. Orthologues of lmtA were not present in E. coli, the lipid A of which normally lacks the 1-phosphomethyl group, or in other spirochetes, which do not synthesize lipid A. Expression of the lmtA gene behind the lac promoter on a low copy plasmid resulted in the appearance of SAM-dependent methyltransferase activity in E. coli inner membranes and methylation of about 30% of the endogenous E. coli lipid A. Inactivation of the ABC transporter MsbA did not inhibit methylation of newly synthesized lipid A. Methylated E. coli lipid A was analyzed by mass spectrometry and NMR spectroscopy to confirm the location of the phosphomethyl group at the 1-position. In human cells, engineered to express the individual TLR subtypes, 1-phosphomethyl-lipid A purified from lmtA-expressing E. coli potently activated TLR4 but not TLR2.
The outer membrane of Gram-negative bacteria is an asymmetric lipid bilayer. The inner monolayer consists of glycerophospholipids, whereas the outer monolayer consists of lipopolysaccharide (LPS).1 LPS is composed of a saccharolipid anchor (1) termed lipid A, a nonrepeating oligosaccharide core, and a distal polysaccharide (or O-antigen). The minimal LPS required for growth in Escherichia coli consists of lipid A and two 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) residues, designated Kdo2-lipid A. Lipid A is a potent immunostimulant in animals that activates the toll-like receptor 4 (TLR4) (2-4), and it is implicated in Gram-negative septic shock (5). The Lpx enzymes responsible for the assembly of Kdo2-lipid A have been fully characterized in E. coli (5). Several of these enzymes are attractive targets for the design of new antibiotics (6, 7). The lpx genes are well conserved among Gram-negative bacteria, despite some variations in lipid A structure. Additional modifying enzymes, not present in E. coli, are responsible for generating most of the structural diversity, and they generally function late in the biosynthetic pathway (8-13).
One example of a bacterium with an unusual, modified lipid A is Leptospira interrogans, a spirochete responsible for causing leptospirosis in humans. This disease is a problem in highly populated, tropical urban centers, and its clinical presentations range from flu-like symptoms to fatal kidney, liver, or pulmonary damage (14, 15). Typical O-antigen gene clusters and orthologues of nearly all of the E. coli lpx genes are present in the genomes of various Leptospira (16, 17). The presence of LPS is a major distinguishing feature that sets L. interrogans apart from the other spirochetes (such as Treponema pallidum, Treponema denticola, and Borrelia burgdorferi), perhaps explaining why L. interrogans is easily cultivated outside of its host.
The structure of L. interrogans lipid A has been elucidated recently using mass spectrometry, NMR spectroscopy, and biochemical analysis (18). Fig. 1 illustrates the key structural differences between E. coli and L. interrogans lipid A. E. coli lipid A is a β,1′-6-linked disaccharide of glucosamine that is phosphorylated at the 1- and 4′-positions and is acylated with (R)-3-hydroxymyristate at the 2-, 3-, 2′-, and 3′-positions (Fig. 1A) (5). The 2′- and 3′-linked fatty acyl chains are further esterified with secondary laurate and myristate chains, respectively. The structure of L. interrogans lipid A is a β,1′-6-linked disaccharide, consisting of the glucosamine analogue 2,3-diamino-2,3-dideoxy-D-glucopyranose (Fig. 1B) (18). L. interrogans lipid A is acylated with R-3-hydroxylaurate at the 3- and 3′-positions and with R-3-hydroxypalmitate at the 2- and 2′-positions. The secondary acyl chains most likely are 12 or 14 carbons in length, and each contains one double bond. As in many strains of Rhizobium and Francisella, the 4′-phosphate group is missing in L. interrogans lipid A. However, the most unusual property of L. interrogans lipid A is the presence of a methylated 1-phosphate moiety.
Fig. 1. Comparison of E. coli lipid A, L. interrogans lipid A, and 1-phosphomethyl-lipid A from E. coli expressing LmtA.
Four key differences in the structures of E. coli (A) and L. interrogans (B) lipid A are the presence in the latter of four N-linked acyl chains instead of two, two unsaturated secondary acyl chains, a methylated 1-phosphate group, and the absence of the 4′-phosphate moiety. The structure of 1-phosphomethyl-lipid A from E. coli expressing LmtA is shown in C.
The proposed biosynthetic pathway for the assembly of L. interrogans Kdo2-lipid A is diagrammed in Fig. 2. For the most part, the pathway is catalyzed by orthologues of the E. coli lpx gene products. However, there are at least four additional genes that are required in the L. interrogans system. The first two, gnnA and gnnB, were originally discovered in Acidithiobacillus ferrooxidans because of their location between lpxA and lpxB (12). Together, these gene products function to synthesize the sugar nucleotide UDP-2-acetamido-3-amino-2,3-dideoxy-α-D-glucose (UDP-GlcNAc3N). GnnA catalyzes the oxidation of the glucosamine 3-OH of UDP-GlcNAc, and GnnB catalyzes the subsequent transamination to form UDP-GlcNAc3N. LpxA from L. interrogans is absolutely specific for UDP-GlcNAc3N versus UDP-GlcNAc (19). E. coli LpxA uses both UDP-GlcNAc and UDP-GlcNAc3N in vitro, but it cannot synthesize the latter, because it lacks the gnnA and gnnB genes. Consequently, L. interrogans lipid A contains four N-linked acyl chains, whereas E. coli has only two.
Fig. 2. Proposed biosynthetic pathway for L. interrogans Kdo2-lipid A.
Most of these reactions are catalyzed by orthologues of the E. coli Lpx enzymes. However, GnnA and GnnB act at the beginning of the pathway to make the unique sugar nucleotide, UDP-GlcNAc3N, whereas the methyltransferase and the 4′-phosphatase function in the later stages. GlcN3N, 2,3-diamino-2,3-dideoxy-D-glucopyranose.
The chemical structure of L. interrogans lipid A (Fig. 1B) implies that two additional lipid A-processing enzymes must be present in this organism. A 1-methyltransferase and a 4′-phosphatase are proposed to methylate the 1-phosphate group and dephosphorylate the 4′-position, respectively (Fig. 2). Methylated phosphate residues are relatively uncommon in biology (20, 21). There is only one well characterized example of a methylated phospholipid, a methylated phosphatidylglycerophosphate analogue found in the halophile Halobacterium salinarium (22). In the field of lipid A biochemistry, a methylated phosphate moiety is without precedent (5).
The significance of the distinct lipid A structure seen in L. interrogans is unknown. A recent study suggested that leptospiral LPS might activate an alternative TLR as compared with the LPS from E. coli and most other Gram-negative bacteria (23). In the case of E. coli (2, 3), LPS first interacts with the LPS-binding protein, which delivers the LPS to the GPI-linked peripheral membrane protein, CD14. LPS is then brought into contact with the integral membrane protein, TLR4, and the accessory protein, MD-2. Upon activation, MyD88 is recruited to the cytoplasmic tail of TLR4, which in turn triggers a series of events that culminates in the translocation of NF-κB to the nucleus and the transcriptional activation of numerous cytokine genes. However, Werts et al. (23) reported that L. interrogans LPS instead activates TLR2.
It is tempting to speculate that the apparent differences in TLR activation between L. interrogans and E. coli are due to the structural characteristics of their respective lipid A molecules. Identification of the L. interrogans genes and enzymes responsible for some of these structural variations should provide helpful tools for investigating this hypothesis. Here, it is reported that a novel L. interrogans membrane enzyme, designated LmtA, catalyzes the selective transfer of a methyl group in vitro from SAM to the 1-phosphate residue of Kdo2-lipid A. When LmtA is expressed in E. coli, a modified lipid A species is synthesized in vivo, which is shown to be the 1-phosphomethyl derivative of E. coli lipid A. Lipid A methylation probably occurs on the cytoplasmic face of the inner membrane, since it is independent of MsbA function. The addition of the methyl group to E. coli lipid A does not alter its potent, TLR4-specific bioactivity.
EXPERIMENTAL PROCEDURES
Materials
γ-32P]ATP, 32Pi, [glycerol-U-14C]phosphatidic acid, and [glycerol-U-14C]glycerol-3-phosphate were purchased from PerkinElmer Life Sciences, and Silica Gel 60 (0.25-mm) TLC plates were obtained from Merck. Tryptone and yeast extract were from Difco, whereas chloroform, ammonium acetate, and sodium acetate were from EM Science. Triton X-100 and the BCA protein determination kit were purchased from Pierce. LPS derived from E. coli strain O111:B4 was purchased from Sigma and reextracted by phenol chloroform as described (24). Pam2CysK4 was from EMC Microcollection GmbH (Tubingen, Germany), and human IL-1β was purchased from Peprotech (Rocky Hill, NJ). All other reagent grade chemicals were obtained from Sigma or Mallinckrodt.
Bacterial Strains and Growth Conditions
All bacterial strains used in this study are described in Table I. L. interrogans serovar icterohemeorrhagiae (strain Verdun) cell pellets were kindly provided by Catherine Werts (Institut Pasteur) (18). E. coli strains were grown at 37 °C in LB medium containing 10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter (25). Overnight cultures were grown from a single colony and used to inoculate LB cultures of varying volume (initial A600 = 0.01) containing 1 mM isopropyl-1-thio-β-D-galactopyranoside. Cultures were grown to A600 = 1 before harvesting. Ampicillin was added at 100 μg/ml when required for plasmid selection.
Table I.
Relevant bacterial strains and plasmids
| Strain/Plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| L. interrogans | ||
| Strain Verdun | Avirulent variant of serovar icterohaemorrhagiae | Ref. 23 |
| E. coli | ||
| XL1 Blue-MR | mcrABC recA1 endA1 gyrA96 relA1 supE44 thi-1 lac | Stratagene |
| W3110 | Wild-type, F-, λ- | E. coli Genetic Stock Center (Yale) |
| W3110A | Wild-type, F-, λ-, aroA::Tn10 | Ref. 37 |
| WD2 | W3110, aroA::Tn10 msbA (A270T) | Ref. 36 |
| Plasmids | ||
| pET23a | Expression vector, T7lac promoter, ampr | Novagen |
| pMBH8 | pET23a expressing lmtA | This work |
| pWSK29 | Low copy expression vector, lac promoter, ampr | Ref. 27 |
| pLmtA | pWSK29 expressing lmtA | This work |
Molecular Biology Protocols
Plasmids were isolated using the Qiagen Spin Prep Kit, and DNA fragments were recovered from agarose gels using the QIAquick Gel Extraction Kit. Pfu DNA polymerase (Stratagene), T4 DNA ligase (Invitrogen), restriction endonucleases (New England Biolabs), and shrimp alkaline phosphatase (U. S. Biochemical Corp.) were used according to the manufacturers’ instructions. The Duke University DNA Analysis Facility sequenced double-stranded DNA with an ABI Prism 377 instrument. All primers were obtained from MWG-Biotech. Chemically competent cells for transformations were prepared by the method of Inoue et al. (26).
Cloning of lmtA from L. interrogans Genomic DNA
The L. interrogans lmtA gene was cloned into pET23a (Novagen) behind the T7lac promoter to generate pMBH8. First, the gene was amplified by PCR from L. interrogans serovar icterohemeorrhagiae (strain Verdun) genomic DNA, kindly provided by Catherine Werts (Institut Pasteur). The primers were designed based on the DNA sequence of L. interrogans serovar lai (strain 56601), which is 99.7% identical to serovar icterohemeorrhagiae (strain Verdun) at the DNA level and 100% at the protein level for LmtA. The forward primer consisted of a clamp region and an NdeI site (underlined) that overlaps with the first 28 base pairs of the lmtA gene (start codon in boldface type). The reverse primer contained a clamp region, a BamHI site (underlined), and the last 24 base pairs of lmtA (stop codon in boldface type). Sequences of the forward and reverse primers were 5′-GGCCATATGGCTTTGATCGAAGAATTTGAATCTC-3′ and 5′-GGCGGATCCTTAACGACCATCTACATGTAAAAG-3′, respectively. The PCR consisted of 100 ng of genomic DNA template, 250 ng of each primer, 200 μM each of dNTPs, 1× Pfu buffer (20 mM Tris-HCl, pH 8.8, 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1 mg/ml bovine serum albumin (BSA), 0.1% Triton X-100), and 5 units of Pfu DNA polymerase in a total reaction volume of 100 μl. The reaction conditions were as follows: 94 °C denaturation for 1 min followed by 25 cycles of 94 °C (denaturation) for 1 min, 50 °C (annealing) for 1 min, and 72 °C (extension) for 1 min. This was followed by a 10-min run-off at 72 °C. The gel-purified PCR product was digested with NdeI and BamHI and ligated into a NdeI/BamHI-digested and shrimp alkaline phosphatase-treated pET23a vector. The resulting pMBH8 was transformed into XL-1 Blue cells (Stratagene). The accession number for the lmtA DNA coding sequence is DQ097086.
The lmtA gene was also cloned into the lac-inducible, low copy expression vector, pWSK29 (27). Using XbaI and BamHI, the fragment containing lmtA and the upstream ribosome-binding site was excised. This fragment was ligated into XbaI/BamHI-digested and shrimp alkaline phosphatase-treated pWSK29. This plasmid, pLmtA, was then transformed into E. coli W3110.
Preparation of Cell-free Extracts and Washed Membranes
L. interrogans was provided as a frozen cell pellet. E. coli W3110 cells, harboring either pLmtA or pWSK29, were grown in 100-ml cultures that were harvested by centrifugation (3500 × g, 20 min, 4 °C). Cells were resuspended in 4 ml of ice-cold 50 mM HEPES, pH 7.5, and lysed by two passages through a French pressure cell at 10,000 p.s.i. The lysate was cleared by centrifugation at 10,000 × g for 20 min at 4 °C. A small portion of the resulting supernatant (cell-free extract) was saved at -80 °C. Membranes were prepared from the remaining supernatant by ultracentrifugation at 100,000 × g for 60 min at 4 °C, and the resulting high speed supernatant (cytosol) was saved at -80 °C. The membranes were washed in 8 ml of 50 mM HEPES, pH 7.5, and subjected to an additional ultracentrifugation step. The final pellet was resuspended in 750 μl of 50 mM HEPES, pH 7.5, and stored at -80 °C. The BCA assay was used to determine protein concentration.
Preparation of Lipid Substrates
The radiolabeled substrates, [4′-32P]Kdo2-lipid A, [4′-32P]lipid IVA, and [4′-32P]-Kdo2-lipid IVA, were prepared in vitro following a published procedure (28, 29). Phosphatidyl-[U-14C]glycerophosphate was prepared by enzymatic synthesis using CDP-diacylglycerol and [glycerol-U-14C]-glycerol-3-phosphate (30). Unlabeled Kdo2-lipid A was isolated from the heptose-deficient E. coli strain WBB06, as described (31). Unlabeled lipid IVA and Kdo2-lipid IVA were obtained following a published procedure (32).
Methyltransferase Assay
The activity of LmtA in either cell-free extracts, cytosol, or membranes was assayed under optimized conditions in a 10–35-μl reaction volume with the substrates Kdo2-lipid A and SAM. Assays included 50 mM HEPES, pH 7.5, 0.1% Triton X-100, 5mM EDTA, 1 mg/ml BSA, 10 mM SAM, 500 cpm/μl [4′-32P]Kdo2-lipid A, and 1 μM unlabeled Kdo2-lipid A. Assays conducted with E. coli membranes did not include 5 mM EDTA in order to minimize the formation of the PagP product, palmitoyl-Kdo2-lipid A (33). Reactions were incubated at 30 °C for varying times and terminated by spotting 4-μl portions onto Silica Gel 60 TLC plates. Substrates and products were separated in chloroform/methanol/water/acetic acid (25:15:4:4, v/v/v/v) and detected using a Amersham Biosciences PhosphorImager system (STORM 840) equipped with ImageQuant software.
Mild Acid Hydrolysis of Methyltransferase Products
The assay was performed as described above for varying times. However, to terminate the reaction, 10-μl aliquots were added to 170 μl of 12.5 mM sodium acetate, pH 4.5, containing 1% SDS. The mixture was then boiled at 100 °C for 30 min to hydrolyze the Kdo residues. The resulting solution was converted into a two-phase Bligh/Dyer system by adding 200 μl each of chloroform and methanol, and it was centrifuged for 5 min. The lower phase (containing the free lipid A species) was removed, dried under vacuum, and resuspended in 10 μl of chloroform/methanol (2:1, v/v). The entire sample was spotted onto a TLC plate, and the lipid A molecules were resolved in chloroform, pyridine, 88% formic acid, water (50:50:16:5, v/v/v/v) and analyzed as described above.
Preparation of Protein Samples for SDS-PAGE Analysis
Cell-free extracts or membrane samples (20 μg of protein) were incubated at 40 °C for 30 min in 50 mM Tris-HCl, pH 6.8, 12.5% glycerol, 3% SDS, 50 mM dithiothreitol, and 0.02% bromphenol blue. The samples were loaded onto a 12% SDS-polyacrylamide gel and subjected to electrophoresis at 150 V for 60 min.
Inner and Outer Membrane Separation
Membranes derived from either W3110/pLmtA or W3110/pWSK29 were separated by isopycnic sucrose gradient centrifugation (34). A 2-ml membrane suspension, containing 5 mg of protein, was layered on top of a seven-step gradient (30–60% sucrose, w/w) (35) and ultracentrifuged for 18 h at 35,000 rpm in a Beckman SW40.1 rotor at 4 °C. A set of 22 0.5-ml fractions were collected and analyzed for protein concentration, NADH oxidase activity (inner membrane marker), and phospholipase A activity (outer membrane marker) (36-38). Each fraction was also assayed for LmtA activity.
In Vivo Labeling of E. coli Cells Expressing LmtA
Lipid A species were radiolabeled in 20-ml cultures of W3110/pLmtA or W3110/pWSK29 containing 5 μCi/ml 32Pi. Cells were collected by centrifugation and washed once with 5 ml of phosphate-buffered saline, pH 7.4. Next, the cells were resuspended in 3 ml of a single-phase Bligh/Dyer solution (chloroform/methanol/water, 1:2:0.8, v/v/v) and incubated at room temperature for 60 min in order to extract the phospholipids. The sample was centrifuged, and the pellet (containing the LPS, proteins, and DNA) was washed two times with 3 ml of a single-phase Bligh/Dyer mixture. Then the pellet was boiled at 100 °C in 3 ml of 12.5 mM sodium acetate, pH 4.5, containing 1% SDS, for 30 min to release the lipid A from the LPS core residues. The lipid A molecules were extracted by conversion into a two-phase Bligh/Dyer system (chloroform/methanol/water, 2:2:1.8, v/v/v). The lower phase, containing the lipid A, was removed after centrifugation, and the remaining upper phase was washed once with preequilibrated lower phase. Lower phases were pooled and dried under a stream of N2. The dried lipid was redissolved in 200 μl of chloroform/methanol (2:1, v/v), and 5000 counts were spotted onto a TLC plate. The lipid A species were separated in chloroform, pyridine, 88% formic acid, water (50:50:16:5, v/v/v/v) and detected using a PhosphorImager system as described above.
The labeling experiment performed with the temperature-sensitive mutant WD2 (36) was modified slightly from the above. In this case, overnight cultures grown at 30 °C were used to inoculate 5-ml LB cultures (initial A600 = 0.01) containing 1 mM isopropyl-1-thio-β-D-galactopyranoside. Once these cultures reached an A600 of 1.0, they were diluted into 15 ml of LB containing 1 mM isopropyl-1-thio-β-D-galactopyranoside. After 30 min of growth at the appropriate temperature, the cells were labeled with 5 μCi/ml 32Pi for 15 min. Cells were harvested, and lipid A was extracted as described for the W3110 strains above.
Purification of 1-Phosphomethyl-lipid A from E. coli Cells Expressing LmtA
To obtain sufficient quantities of modified lipid A from W3110/pLmtA for mass spectrometry, NMR spectroscopy, and cellular activation studies, a large scale preparation and purification was performed. Unmodified lipid A from W3110/pWSK29 was also purified for activation studies. Cells from a 1-liter culture were harvested by centrifugation (5000 rpm, 20 min, 4 °C). The lipid A extraction protocol follows that described above, only on a larger scale.
Anion exchange chromatography was used to purify the putative methylated lipid A from the nonmethylated lipid A species. The dried lipid was dissolved in 10 ml of chloroform/methanol/water (2:3:1, v/v/v) and loaded onto a 2-ml DEAE-cellulose column (Whatman DE-52), equilibrated as the acetate form in the 2:3:1 solvent mixture. The column was then washed with five column volumes of the 2:3:1 solvent mixture. The elution profile consisted of five column volumes each of chloroform/methanol/ammonium acetate (2:3:1, v/v/v) in which the concentration of ammonium acetate was increased stepwise (60, 120, 240, 360, and 480 mM). Then 20 μl of each 2-ml fraction was spotted on a TLC plate, which was developed in chloroform, pyridine, 88% formic acid/water (50:50:16:5, v/v/v/v). After drying, the plates were sprayed with 10% sulfuric acid in ethanol and charred to detect the lipids. Fractions containing the desired lipid A species were pooled, and the lipid was extracted by conversion into a two-phase Bligh/Dyer system. The lower phase was dried under a stream of N2.
An additional chromatography step was performed on the lipid samples subjected to TLR activation studies. The dried lipids obtained from the DEAE-cellulose column were redissolved in 5 ml of 50% acetonitrile in water (Solution A) and 5 ml of 85% isopropyl alcohol in water (Solution B), containing 1 mM tetrabutylammonium phosphate. The sample was loaded onto a 1 ml octadecyl-bonded silica gel column equilibrated in solution A/solution B (1:1, v/v) with 1 mM tetrabutylammonium phosphate. The lipid was eluted in five steps: 6 ml of A/B (1:1), 12 ml of A/B (1:2), 6 ml of A/B (1:3), 6 ml of A/B (1:4), and 6 ml of A/B (1:6). Each solution contained 1 mM tetrabutylammonium phosphate. Lipids were detected by spotting 20 μl of each 1-ml fraction on a TLC plate, which was developed in chloroform, pyridine, 88% formic acid, water (50:50:16:5, v/v/v/v) and charred. To remove the tetrabutylammonium phosphate, 2 ml of chloroform/methanol/water (2:3:1, v/v/v) was added to each 1-ml fraction that contained the desired lipid. These fractions were pooled and loaded onto a 1-ml DEAE-cellulose column equilibrated as described above. The column was washed with 5 ml of chloroform/methanol/water (2:3:1, v/v/v) before eluting with 8 ml of chloroform, methanol, 480 mM ammonium acetate (2:3:1, v/v/v). Fractions were collected in 1-ml volumes, and TLC analysis determined which fractions contained the desired lipid. These fractions were pooled, and the lipid was extracted as described above. The total amount of lipid A isolated was determined by phosphate analysis (39).
MALDI-TOF Mass Spectrometry Analysis
Lipids were analyzed using an AXIMA-CFR (Kratos Analytical, Manchester, UK) matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometer utilizing 20-kV extraction voltage and time-delayed extraction. Dried lipid A samples were dissolved in chloroform/methanol (4:1, v/v) and prepared for MALDI-TOF analysis by depositing 0.3 μl of the sample solution on the sample plate. An equal volume of matrix solution was deposited immediately on top of the sample spot, and the two solutions were allowed to dry together at room temperature. The matrix utilized for all analyses was a saturated solution of 6-aza-2-thiothymine in 50% acetonitrile and 10% tribasic ammonium citrate (9:1, v/v). Spectra for both positive and negative ions were acquired in linear mode. Each spectrum represents the average of 100 laser shots.
NMR Spectroscopy Analysis
A 0.3-mg sample of 1-phosphomethyl-lipid A was dissolved in 0.35 ml of CDCl3/CD3OD/D2O (2:3:1, v/v/v) in a 3-mm NMR tube. Proton chemical shifts are reported relative to tetramethylsilane at 0.00 ppm. The 2H signal of CD3OD was used as a field frequency lock. NMR spectra were obtained on Varian Inova 800 or 500-MHz NMR spectrometers, each equipped with a Sun Ultra 10 computer and 5-mm Varian probe. 1H NMR spectra at 800 MHz were obtained with a 7.2-kHz spectral window, a 67° pulse field angle (4.5 μs), a 4.5-s acquisition time, and a 1-s relaxation delay. The spectra were digitized using 64,000 points to obtain a digital resolution of 0.225 Hz/pt. Directly detected 1H-decoupled 31P NMR spectra were recorded at 202.37 MHz with a spectral window of 12,143.3 Hz digitized into 25,280 data points (digital resolution of 1 Hz/point or ∼0.005 ppm/point), a 60° pulse flip angle (8 μs), and a 1.6-s repeat time. 31P chemical shifts were referenced to 85% H3PO4 at 0.000 ppm. Inverse decoupled difference spectra were recorded as 1H-detected 31P-decoupled heteronuclear NMR experiments as previously described (18, 40-42).
TLR Activation Studies
HEK293 cells stably expressing fluorescently tagged TLR2 or TLR4/MD2 were described previously (43). Cellular activation of HEK293 cells that express TLR2YFP, TLR4YFP/MD2, or empty vector was assessed by an NF-κB-luciferase reporter assay (43).
RESULTS
Methyltransferase Activity Associated with L. interrogans Membranes
Both cell-free extracts and membranes isolated from L. interrogans appear to modify 1 μM Kdo2-lipid A in vitro in the presence of the methyl donor SAM (10 mM). When the reaction is monitored by TLC, a new SAM-dependent product band appears, which migrates faster than Kdo2-[4′-32P]lipid A (Fig. 3A). This substance is not formed when high speed supernatant (cytosol) is used as the enzyme source, suggesting that the putative methyltransferase is membrane-bound. The more rapid migration of the SAM-dependent product on the TLC plate suggests the addition of a hydrophobic moiety, such as the methyl group found in leptospiral lipid A (Fig. 1B) (18). Product formation by L. interrogans membranes was linear for 30 min at 0.25 mg/ml protein (specific activity of 0.064 nmol/min/mg).
Fig. 3. SAM-dependent methyltransferase activity localized in L. interrogans membranes.

A, cell-free extracts, membranes, and cytosol (0.25 mg/ml) derived from L. interrogans cells were assayed for methyltransferase activity. Assays were conducted at 30 °C for 30 min and included 50 mM HEPES, pH 7.5, 0.1% Triton X-100, 5 mM EDTA, 1 mg/ml BSA, 10 mM SAM, 500 cpm/μl [4′-32P]Kdo2-lipid A, and 1 μM unlabeled Kdo2-lipid A. Products were separated in chloroform/methanol/water/acetic acid (25:15:4:4, v/v/v/v) and detected using a PhosphorImager system. Formation of the methylated product by 0.25 mg/ml membranes is linear with time under the assay conditions described in A. B, the methyl group is transferred to the lipid A portion of the Kdo2-lipid substrate. Membranes (0.25 mg/ml) were assayed as in A. At the indicated time points, the Kdo residues were hydrolyzed in 12.5 mM sodium acetate, pH 4.5, containing 1% SDS, for 30 min at 100 °C. The resulting lipid A molecules were extracted, separated by TLC in chloroform/pyridine/88% formic acid/water (50:50:16:5, v/v/v/v), and detected using a PhosphorImager.
The Addition of the Presumed Methyl Group to the Lipid A Portion of the Molecule
To rule out the possibility that the methyl group is added to the Kdo residues of Kdo2-lipid A, further analysis was required. L. interrogans membranes were assayed for various time intervals, and then the Kdo residues were removed by hydrolysis at 100 °C in 12.5 mM sodium acetate, pH 4.5, containing 1% SDS. The released lipid A molecules were extracted and analyzed by TLC. As illustrated in Fig. 3B, the more rapidly migrating component is present in the lipid A portion of the modified Kdo2-[4′-32P]lipid A, verifying that the putative methylation is occurring on the lipid A moiety. No lipid A modification occurs in the absence of SAM.
Identification and Cloning of the Candidate Methyltransferase Gene, lmtA
A possible gene encoding the lipid A methyltransferase was identified using bioinformatics. There are no available sequences for enzymes that methylate phosphate residues with which to search the L. interrogans genome. However, it was reasoned that the chemical environment required for the methylation of a phosphate group at a membrane surface might share some common features with the methylation of a carboxylate moiety. The latter reaction is catalyzed by the Saccharomyces cerevisiae isoprenylcysteine carboxyl methyltransferase, Ste14p (44, 45), a member of the same enzyme family that methylates Ras. A BLAST search revealed a distant orthologue of Ste14p in L. interrogans serovar lai (E value of 10-8), which had been annotated as a “putative protein-S-isoprenylcysteine methyltransferase” (17). Several properties of this protein suggested that it might actually be the leptospiral lipid A methyltransferase, now designated LmtA. First, orthologues were not found in E. coli, which lacks the 1-phosphomethyl group. Likewise, no orthologues were detected in the other spirochetes, which do not synthesize lipid A. Like Ste14p, leptospiral LmtA was predicted to have six transmembrane segments.
L. interrogans lmtA was expressed behind the lac promoter on the plasmid pWSK29 in E. coli W3110. The SDS-PAGE gel in Fig. 4A clearly demonstrates successful heterologous expression. An additional protein with the expected molecular mass (∼27 kDa) is seen in both cell-free extracts and membranes of E. coli W3110 harboring pLmtA, but not in the vector controls (W3110/pWSK29). Membranes of W3110/pLmtA catalyzed time- and SAM-dependent synthesis of the presumed methyltransferase product with a specific activity of 2.95 nmol/min/mg when Kdo2-lipid A was used as the substrate (Fig. 4B). Methyltransferase activity was linear with respect to protein (data not shown). The identical migration during TLC of the product band generated by W3110/pLmtA membranes compared with leptospiral membranes strongly suggests that LmtA is indeed the relevant lipid A methyltransferase. W3110/pLmtA membranes were also assayed under identical conditions using alternative lipid substrates (lipid IVA, Kdo2-lipid IVA, phosphatidylglycerophosphate, and phosphatidic acid). Specific activities were less than 1% of that reported for Kdo2-lipid A (data not shown).
Fig. 4. Expression and assay of LmtA in E. coli W3110 membranes.
A, SDS-PAGE analysis of cell-free extracts and membranes prepared from isopropyl-1-thio-β-D-galactopyranoside-induced E. coli W3110 cells harboring either pLmtA or the vector control (pWSK29). In each lane, 20 μg of protein was loaded. Lane 1, W3110/pLmtA cell-free extracts; lane 2, W3110/pWSK29 cell-free extracts; lane 3, W3110/pLmtA membranes; lane 4, W3110/pWSK29 membranes. B, membranes (0.01 mg/ml) from cells harboring pLmtA or pWSK29 were assayed at 30 °C in the presence of 50 mM HEPES, pH 7.5, 0.1% Triton X-100, 1 mg/ml BSA, 10 mM SAM, 500 cpm/μl [4′-32P]Kdo2-lipid A, and 1 μM unlabeled Kdo2-lipid A. L. interrogans membranes (0.25 mg/ml) served as the positive control. Products were separated in chloroform/methanol/water/acetic acid (25:15:4:4, v/v/v/v) and detected using a PhosphorImager system.
LmtA Expressed in E. coli Is Located in the Inner Membrane
The outer and inner membranes of E. coli W3110 expressing LmtA were separated by isopycnic sucrose gradient centrifugation. Protein concentration, outer membrane phospholipase A, and inner membrane NADH oxidase assays revealed that the membranes were well resolved (Fig. 5). The profile of LmtA activity mirrors that of the NADH oxidase, revealing that LmtA is an inner membrane protein when expressed in E. coli. Given its amino acid sequence, it also is likely to be an inner membrane enzyme in L. interrogans. A similar membrane separation performed with the vector control W3110/pWSK29 did not yield fractions with measurable methyltransferase activity (data not shown).
Fig. 5. LmtA is an inner membrane enzyme.

Membranes were separated into inner and outer fractions using isopycnic sucrose gradient centrifugation. Outer membrane phospholipase A (triangles), inner membrane NADH oxidase (squares), and total protein concentration (diamonds) were measured and expressed as the percentage of the total. Each fraction was also assayed for LmtA activity (circles). Assays were conducted at 30 °C for 30 min and included 50 mM HEPES, pH 7.5, 0.1% Triton X-100, 1 mg/ml BSA, 10 mM SAM, 500 cpm/μl [4′-32P]Kdo2-lipid A, and 1 μM unlabeled Kdo2-lipid A.
Modification of Lipid A in E. coli Cells Expressing LmtA
A 32P labeling study was conducted to determine whether expression of LmtA modifies E. coli lipid A in living cells. Both W3110/pLmtA and W3110/pWSK29 were grown in the presence of 32Pi, and crude LPS was recovered in the residue of a single-phase Bligh/Dyer extraction of the cell pellet. Lipid A was released from the Kdo residues by hydrolysis at pH 4.5 in the presence of SDS, extracted, and analyzed by TLC. As shown in Fig. 6A, an additional modified lipid A species is observed in E. coli cells harboring pLmtA, but not in the empty vector control. The higher migration of the extra band is consistent with a methylated lipid A species, which accounts for ∼30% of the total. Unmodified hexa-acylated lipid A and its 1-diphosphate variant are also present.
Fig. 6. Expression of LmtA in E. coli results in the formation of a modified lipid A species.
A, cells harboring either pLmtA or the vector control (pWSK29) were grown in the presence of 32Pi. Lipid A species were extracted, separated in chloroform/pyridine/88% formic acid/water (50:50:16:5, v/v/v/v), and detected using a PhosphorImager. B, the active site of LmtA faces the cytoplasm. Either pLmtA or the empty vector control (pWSK29) was expressed in the E. coli temperature-dependent msbA mutant WD2 or in E. coli W3110A. Cells were grown at 30 °C for 30 min before labeling with 32Pi for 15 min at either 30 °C or 44 °C, as described under “Experimental Procedures.” Lipid A species were extracted, separated, and detected as described in A.
LmtA Activity Is MsbA-independent
MsbA is an essential ABC transporter that acts as the LPS flippase within the inner membrane (36, 37). The temperature-sensitive strain WD2, harboring a point mutation in MsbA, accumulates lipid A on the inner face of its inner membrane at the nonpermissive temperature (37). An in vivo labeling study, similar to that described in Fig. 6A, was used to analyze the dependence of LmtA activity on MsbA. In doing so, it is possible to determine whether the active site of LmtA faces the cytoplasmic or periplasmic surface of the inner membrane. LmtA was expressed in WD2, and its lipid A species were labeled at both the permissive (30 °C) and nonpermissive (44 °C) temperatures. Control labeling studies were performed at 44 °C with W3110A/pLmtA and W3110A/pWSK29, which possess wild-type MsbA. The vector control, WD2/pWSK29, was also labeled in parallel. Fig. 6B shows the TLC analysis of the lipid A species extracted from each culture. Comparable amounts of the putative methylated lipid A species are present in WD2/pLmtA at both temperatures. Therefore, even when MsbA is not functioning to flip newly synthesized lipid A to the periplasmic face of the inner membrane at 44 °C, LmtA is still active. Consequently, it is very likely that LmtA methylates lipid A on the cytoplasmic face of the inner membrane. The W3110A/pLmtA control likewise synthesizes modified lipid A at the elevated temperature, whereas neither strain harboring the vector control produces modified lipid A.
Demonstration of a Methyl Group at the 1-Position of Lipid A by Mass Spectrometry
Based on the published structure of L. interrogans lipid A (18), it is presumed that LmtA is catalyzing the addition of a methyl group solely to the 1-position of Kdo2-lipid A. However, detailed structural analysis was needed to verify this hypothesis. The modified lipid A from W3110/pLmtA was isolated from 1 liter of cells grown to late log phase. A DEAE-cellulose column was used to separate the putative methylated lipid A from the unmodified lipid A species. Negative ion MALDI-TOF mass spectroscopy revealed a prominent [M – H]- ion at m/z 1812.3 (Fig. 7A), consistent with the presence of one extra methyl group in the modified lipid A (Fig. 1C). Additional structural information is revealed in the positive ion mode spectrum (Fig. 7B). The B1+ oxonium ion, derived from the distal sugar unit, is formed by cleavage of the glycosidic linkage, and the B2+ ion is formed by the loss of the substituent attached to the 1-position in the proximal sugar (Fig. 1C). The B1+ ion at m/z 1086.6 and the B2+ ion at m/z 1701.0 do not differ from those observed with unmodified wild-type E. coli lipid A (data not shown). Therefore, the methylphosphate moiety must be located at the 1-position. These results confirm the tentative identification of LmtA as the lipid A methyltransferase.
Fig. 7. Mass spectrometry reveals that LmtA-expressing E. coli produce 1-phosphomethyl-lipid A.

A, MALDI-TOF spectrum acquired in the negative mode. The molecular weight of E. coli lipid A with a methyl group attached at the 1-position is 1812.4. B, spectrum acquired in the positive mode, demonstrating that the methyl group is attached to the lipid A 1-phosphate moiety.
NMR Spectroscopy of the Methylated Lipid A Species
The 800-MHz 1H NMR spectrum of the putative 1-phosphomethyllipid A from W3110/pLmtA, dissolved in CDCl3/CD3OD/D2O (2:3:1, v/v/v), reveals sharp and well resolved resonances in the sugar (3.5–5.5 ppm) region (Fig. 8A), similar to previous 500-MHz NMR spectra of unmodified E. coli lipid A in the same solvent system (40, 41). However, the spectrum of the modified lipid A shows an unusual doublet at 3.57 ppm, which integrates to three protons and shows a splitting of 11.0 Hz. A similar doublet signal, which arises from the 1-phosphomethyl group, is observed for lipid A of L. interrogans (18). The 3.57-ppm doublet signal in the 1H NMR spectrum of the modified E. coli lipid A, like that of L. interrogans lipid A, did not manifest any cross-peaks from homonuclear coupling in COSY, ZQCOSY, or TOCSY spectra (data not shown). 31P NMR spectroscopy at 202 MHz of the modified E. coli lipid A revealed two 31P resonances, one at 0.149 and the other at 1.325 ppm (data not shown). To investigate the possibility of heteronuclear coupling of the doublet arising from the presumed methyl group to one of the phosphorus atoms, selective inverse decoupling difference spectroscopy was implemented (40, 41). In this analysis, the difference spectra generated by subtracting on- and off-resonance 31P-decoupled 1H NMR lipid A spectra reveal the protons that are spin-coupled to the irradiated phosphate group. As noted above, the expanded sugar region of the modified lipid A in the absence of 31P decoupling (control) clearly reveals a doublet suggestive of a methyl group at 3.57 ppm (Fig. 8A). The difference spectrum of two selective, 31P-decoupled, 1H spectra (on- and off-resonance) for the 1.325-ppm 31P signal shows the simplification of the H-4′ signal (4.14 ppm) into a triplet (Fig. 8B). Therefore, the 1H observed, 31P-decoupled experiments give direct evidence that the 1.325-ppm 31P signal arises from the 4′-phosphate group in the modified lipid A. On the other hand, taking the difference spectrum of two selective, 31P-decoupled, 1H spectra (on- and off-resonance) for the 0.149-ppm 31P signal shows the collapse of the putative methyl resonance from a doublet to a singlet (Fig. 8C). In addition, this difference spectrum shows the simplification of the H-1 signal (5.42 ppm) into a doublet. These experiments give direct unequivocal evidence that the 0.149-ppm 31P signal arises from the 1-phosphate group in the modified lipid A. This phosphate group must therefore be a bridging monophosphodiester moiety between the C-1 of the proximal glucosamine and the capping methyl group. An analysis of the key chemical shifts and coupling constants of the 1-phosphomethyl-lipid A isolated from W3110/pLmtA is shown in supplemental Table I. The corresponding numbering scheme is shown in supplementary Fig. 1.
Fig. 8. NMR spectroscopy confirms that LmtA-expressing E. coli produce 1-phosphomethyl-lipid A.
A, partial 800-MHz 1H NMR spectrum showing the sugar proton region. A distinct 3-proton methyl doublet is observed at ∼3.57 ppm. The other resonances integrate to 14 sugar and four β-hydroxymethines, consistent with the presence of two hexose sugars and four β-hydroxyacyl chains. The intensity of the H-1′ doublet is reduced due to a presaturation pulse used to eliminate the water solvent signal. B, difference 500-MHz 1H NMR spectrum obtained from subtraction of 1H spectra with selective on- and off-resonance decoupling of the 31P NMR signal at 1.325 ppm. The difference spectrum reveals the collapse of the 4.14-ppm multiplet to a triplet (H-4′). This pattern confirms that this phosphorus group is a monophosphomonoester at C-4′ of the distal sugar. C, difference 500-MHz 1H NMR spectrum obtained from subtraction of 1H spectra with selective on- and off-resonance decoupling of the 31P NMR signal at 0.149 ppm. The difference spectrum reveals the simultaneous collapse of the 3.57-ppm methyl doublet to a singlet and the appearance of the doublet at 5.42 ppm (H-1). This pattern confirms that the phosphorus group is a bridging monophosphodiester between C-1 of the proximal lipid A sugar and the methyl group.
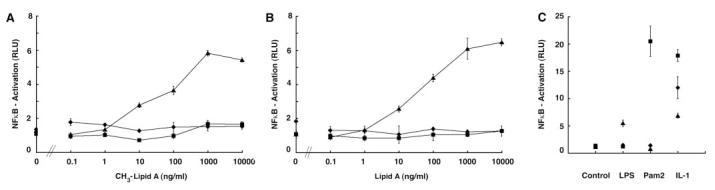
1-Phosphomethyl-lipid A Activates TLR4 but Not TLR2
HEK293 cells stably transfected with TLR2YFP or TLR4YFP/MD2 were used in a luciferase reporter assay, as reported previously (43). Both the 1-phosphomethyl-lipid A and the unmodified E. coli lipid A showed dose-dependent activation of TLR4YFP/MD2-expressing cells (Fig. 9, A and B, respectively). Neither lipid A preparation activated TLR2YFP-expressing cells or cells harboring the empty vector. Furthermore, the cell lines responded to their respective positive control ligands as expected (Fig. 9C).
Fig. 9. Purified 1-phosphomethyl-lipid A does not activate TLR2-expressing HEK cells.
HEK293 cells expressing TLR2YFP (squares), TLR4YFP/MD2 (triangles), or empty vector (diamonds) were stimulated with increasing concentrations of 1-phosphomethyl-lipid A (A) or unmodified lipid A (B). LPS (10 ng/ml), Pam2CysK4 (2.5 nM), and recombinant human IL-1β (10 ng/ml) were used as positive controls (C). Cellular activation was measured by luciferase activity.
DISCUSSION
Four significant differences are apparent when the structure of L. interrogans lipid A (18) is compared with that of E. coli lipid A (Fig. 1). Due to the presence of GnnA, GnnB, and a highly selective LpxA acyltransferase, leptospiral lipid A contains four N-linked hydroxyacyl chains (Fig. 2), whereas that of E. coli has only two. Furthermore, L. interrogans lipid A has two unsaturated secondary acyl chains and lacks the 4′-phosphate moiety. Last, the lipid A of L. interrogans contains a methylphosphate group at the 1-position, leading to the proposal that L. interrogans lipid A biosynthesis must include a novel methylation reaction (Fig. 2). This structural variation has not been observed in the lipid A of any other Gram-negative bacteria (5). In fact, there are few well characterized examples of methylated phosphate groups in all of biology. The N-linked oligosaccharides on the lysosomal enzymes of Dictyostelium discoideum contain mannose 6-phosphomethyl residues (20), and a γ-monomethyl-phosphate cap at the 5′ end of 7SK, B2, and U6 small RNAs in eukaryotes has also been reported (21). The only well documented example in lipid biochemistry is the methylated analogue of phosphatidylglycerophosphate seen in H. salinarium (22). The relevant methyltransferases and their structural genes have not been identified in any of these systems. The L. interrogans enzyme transfers the methyl group of SAM to the 1-phosphate group of E. coli Kdo2-lipid A in vitro. Presumably, the Kdo2-lipid A analogue with the structure shown in Fig. 2, synthesized by the leptospiral orthologues of the E. coli lpx gene products (17), functions as the natural substrate for LmtA in L. interrogans.
Based on the assumption that phosphate group methylation is chemically analogous to carboxylate methylation, bioinformatics was used to identify the L. interrogans lipid A methyltransferase gene. Database searching revealed a distant orthologue of S. cerevisiae Ste14p in L. interrogans. This yeast membrane metalloenzyme (45) is a member of the isoprenylcysteine carboxyl methyltransferase (ICMT) family and performs the last step of CAAX box protein processing, which involves C-terminal carboxylmethylation of farnesylated or geranylgeranylated substrates (44, 45). A detailed topological and genetic analysis predicted that Ste14p is composed of six transmembrane helices, with both the N and C termini facing the cytosol (44). The fifth and sixth transmembrane segments are proposed to form a helix-turn-helix (helical hairpin) within the membrane. Orthologues of Ste14p are found in the genomes of Schizosaccharomyces pombe, Xenopus laevis, Caenorhabditis elegans, mice, rats, and humans. Sequence alignments revealed a consensus sequence at the C terminus (44). Interestingly, this consensus motif also appears in yeast phosphatidylethanolamine N-methyltransferases, certain ergosterol biosynthetic enzymes (presumed to be methyltransferases), and open reading frames of unknown function from various bacteria. The sequence of LmtA had not yet been deposited in the data base when this consensus motif was first reported (44).
Fig. 10 shows the primary sequence alignment of LmtA with several members of the ICMT family obtained using ClustalW. Numerous regions of similarity are observed and are spread throughout the sequence, including in the C-terminal consensus sequence (dashed arrow in Fig. 10). Like Ste14p, LmtA is predicted to have six transmembrane segments, which approximately coincide in the sequence alignment (Fig. 10, black lines). The two C-terminal transmembrane segments of LmtA are predicted to be more distinct from each other than those of Ste14p, suggesting that the helical hairpin motif has a larger connecting loop in LmtA. Although the mechanistic significance of the C-terminal region in the ICMT family members has not been established, it might include one or more substrate binding sites. Members of this family lack the usual tripartite consensus sequences believed to comprise the SAM binding sites found in the large majority of SAM-dependent enzymes (46). An alternative SAM binding site might exist in the cytoplasmic regions of the ICMT consensus sequence (residues 136–175 and 207–239 of Ste14p in Fig. 10). Mutagenesis studies of both the ICMTs and LmtA are needed to determine the significance of the conserved C-terminal region. Within the ICMT family, however, there is significantly more similarity within the consensus region than when compared with LmtA. This finding is understandable, considering the structural differences between the substrates.
Fig. 10. Sequence alignment of LmtA with members of the isoprenylcysteine carboxyl methyltransferase family.
Shown are alignments of S. cerevisiae Ste14p, human pcCMTp, X. laevis Xmam4p, C. elegans open reading frame (accession number U80450), S. pombe mam4p, and L. interrogans LmtA, obtained using ClustalW software (available on the World Wide Web at www.ebi. ac.uk/clustalw/). Red boxes indicate amino acid identity, blue boxes indicate conserved amino acid substitutions, and green boxes indicate semiconserved amino acid substitutions. The bars represent the predicted transmembrane domains of Ste14p (44). The dashed arrow indicates the start of the C-terminal consensus sequence.
Direct evidence that the methyl group added to Kdo2-lipid A by LmtA resides on the 1-phosphate moiety could not be achieved until after the identification of the leptospiral lmtA gene. Expression of LmtA in wild-type E. coli led to the production of a putative methylated lipid A species in vivo (Fig. 6A), which could be purified in 1-mg quantities for use in structural studies. Positive mode MALDI-TOF mass spectrometry (Fig. 7) and selective inverse decoupling difference NMR spectroscopy (Fig. 8) confirmed unequivocally the 1-position of the phosphomethyl group. Subcellular fractionation (Fig. 5) and MsbA inactivation (Fig. 6B) experiments revealed that LmtA is an inner membrane enzyme with its active site facing toward the cytoplasm. These results were expected due to the likely cytoplasmic location of the SAM substrate, and they are consistent with the topology of the ICMTs. The effect of MsbA inactivation has previously been used to determine the sidedness of other lipid A-modifying enzymes. For instance, the Salmonella typhimurium LpxO active site faces the cytoplasm (37), whereas that of F. novicida LpxE faces the periplasmic space (13).
Radioactive labeling studies showed that LmtA methylates only about 30% of the total lipid A when expressed in E. coli (Fig. 6A). It is likely that the optimal substrate of LmtA is not present in these experiments due to the other structural differences between L. interrogans and E. coli lipid A (Figs. 1 and 2). One or more of these structural modifications could play an important role in the ability of LmtA to bind its substrate. Alternatively, the relatively low amount of methylated lipid A in E. coli expressing LmtA might be due to rapid lipid A export by the ABC transporter, MsbA, which is responsible for flipping lipid A from the inner face to the outer face of the inner membrane (36, 37). If E. coli MsbA preferentially flips lipid A with an unmodified 1-phosphate group, it might compete with LmtA for substrate utilization. The effects of the 1-phosphate group on MsbA function are unknown. Interestingly, most known modifications of the 1-phosphate group occur on the periplasmic surface of the inner membrane (13, 47, 48).
A BLAST search with the protein sequence of LmtA against all organisms revealed one significant orthologue in Mesorhizobium sp. BNC1 (E value of 10-40). Like LmtA, this protein is currently annotated as a “putative protein-S-isoprenylcysteine methyltransferase.” However, given the current results, it is more likely that it functions as a lipid A phosphomethyltransferase. Nothing is known about the lipid A structure of Mesorhizobium sp. BNC1. The next best E value in the data base is ∼10-10, belonging to a Ralstonia metallidurans protein that is annotated as a “cobalt-zinc-cadmium resistance protein.”
The function of the 1-phosphomethyl group on lipid A is not understood. It has been reported that leptospiral LPS activates TLR2 (23), suggesting that perhaps structural differences in lipid A can alter receptor specificity during the innate immune response. The 1-phosphomethyl-lipid A, purified from E. coli cells expressing LmtA, was used to investigate this issue in mammalian cells. This compound is E. coli lipid A with an extra methyl group on the 1-phosphate (Fig. 1C). It activated only TLR4, not TLR2 (Fig. 9). One or more of the other structural variations (such as the lack of the 4′-phosphate group, the presence of four N-linked fatty acyl chains, or the presence of unsaturated secondary acyl chains) may be responsible singly (or in combination with 1-methylation) for the TLR2 specificity of leptospiral LPS. The only demonstration thus far of selective TLR2 activation was achieved with complete leptospiral LPS (43). Therefore, the core sugar residues of L. interrogans LPS may also play an important role. Alternatively, the apparent TLR2 activation observed with whole L. interrogans LPS might yet be explained by the presence of a potent lipopeptide impurity. In any event, the availability of a recombinant lipid A 1-phosphomethyltransferase will facilitate the preparation of novel lipid A analogues with possible utility as TLR4 activators or antagonists.
Supplementary Material
Acknowledgments
We thank Dr. Catherine Werts for providing the L. interrogans cells and DNA. We also thank Drs. Xiaoyuan Wang, David Six, and Mark Karbarz for providing the lipid IVA, Kdo2-lipid IVA, and phosphatidylglycerophosphate substrates, respectively.
Footnotes
This research was funded by National Institutes of Health (NIH) Grants 1-F32-GM073251-01 (to M. B. H.), GM-51310 and GM-51796 (to C. R. H. R.), GM-54882 (to R. J. C.), and GM-54060 and AI057784 (to D. T. G.). The Duke NMR Center is partially supported by NCI, NIH, Grant P30-CA-14236. Duke NMR Center instrumentation was funded by the National Science Foundation, the NIH, the North Carolina Biotechnology Center, and Duke University. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank ™/EBI Data Bank with accession number(s)DQ097086.
S The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table I.
- LPS
- lipopolysaccharide
- BSA
- bovine serum albumin
- ICMT
- isoprenylcysteine carboxyl methyltransferase
- Kdo
- 3-deoxy-D-manno-oct-2-ulosonic acid
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- SAM
- S-adenosylmethionine
- TLR
- toll-like receptor
- UDP-GlcNAc3N
- UDP-2-acetamido-3-amino-2,3-dideoxy-α-D-glucose
REFERENCES
- 1.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr., Murphy RC, Raetz CRH, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, Vannieuwenhze MS, White SH, Witztum JL, Dennis EA. J. Lipid Res. 2005;46:839–862. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B, Hoebe K, Du X, Ulevitch RJ. J. Leukocyte Biol. 2003;74:479–485. doi: 10.1189/jlb.0203082. [DOI] [PubMed] [Google Scholar]
- 3.Gioannini TL, Teghanemt A, Zhang D, Coussens NP, Dockstader W, Ramaswamy S, Weiss JP. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller SI, Ernst RK, Bader MW. Nat. Rev. Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 5.Raetz CRH, Whitfield C. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onishi HR, Pelak BA, Gerckens LS, Silver LL, Kahan FM, Chen M-H, Patchett AA, Galloway SM, Hyland SA, Anderson MS, Raetz CRH. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 7.Coggins BE, Li X, McClerren AL, Hindsgaul O, Raetz CRH, Zhou P. Nat. Struct. Biol. 2003;10:645–651. doi: 10.1038/nsb948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brozek KA, Kadrmas JL, Raetz CRH. J. Biol. Chem. 1996;271:32112–32118. [PubMed] [Google Scholar]
- 9.Basu SS, Karbarz MJ, Raetz CRH. J. Biol. Chem. 2002;277:28959–28971. doi: 10.1074/jbc.M204525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Que-Gewirth NLS, Karbarz MJ, Kalb SR, Cotter RJ, Raetz CRH. J. Biol. Chem. 2003;278:12120–12129. doi: 10.1074/jbc.M300379200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karbarz MJ, Kalb SR, Cotter RJ, Raetz CRH. J. Biol. Chem. 2003;278:39269–39279. doi: 10.1074/jbc.M305830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweet CR, Ribeiro AA, Raetz CRH. J. Biol. Chem. 2004;279:25400–25410. doi: 10.1074/jbc.M400596200. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CRH. J. Biol. Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira MM, Matsuo MGS, Bauab AR, Vasconcelos SA, Moraes ZM, Baranton G, Girons I. Saint. J. Clin. Microbiol. 2000;38:450–452. doi: 10.1128/jcm.38.1.450-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levett PN. Clin. Microbiol. Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulach DM, Kalambaheti T, de la Pena-Moctezuma A, Adler B. J. Mol. Microbiol. Biotechnol. 2000;2:375–380. [PubMed] [Google Scholar]
- 17.Ren S-X, Fu G, Jiang X-G, Zeng R, Miao Y-G, Xu H, Zhang Y-X, Xiong H, Lu G, Lu L-F, Jiang H-Q, Jia J, Tu Y-F, Jiang J-X, Gu W-Y, Zhang Y-Q, Cai Z, Sheng H-H, Yin H-F, Zhang Y, Zhu G-F, Wan M, Huang H-L, Qian Z, Wang S-Y, Ma W, Yao Z-J, Shen Y, Qiang B-Q, Xia Q-C, Guo X-K, Danchin A, Girons I. Saint, Somerville RL, Wen Y-M, Shi M-H, Chen Z, Xu J-G, Zhao G-P. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 18.Que-Gewirth NLS, Ribeiro AA, Kalb SR, Cotter RJ, Bulach DM, Adler B, Girons I. Saint, Werts C, Raetz CRH. J. Biol. Chem. 2004;279:25420–25429. doi: 10.1074/jbc.M400598200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweet CR, Williams AH, Karbarz MJ, Werts C, Kalb SR, Cotter RJ, Raetz CRH. J. Biol. Chem. 2004;279:25411–25419. doi: 10.1074/jbc.M400597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeze HH, Hindsgaul O, Ichikawa M. J. Biol. Chem. 1992;267:4431–4439. [PubMed] [Google Scholar]
- 21.Shimba S, Reddy R. J. Biol. Chem. 1994;269:12419–12423. [PubMed] [Google Scholar]
- 22.Kates M, Moldoveanu N, Stewart LC. Biochim. Biophys. Acta. 1993;1169:46–53. doi: 10.1016/0005-2760(93)90080-s. [DOI] [PubMed] [Google Scholar]
- 23.Werts C, Tapping RI, Mathison JC, Chuang T-H, Kravchenko V, Girons I. Saint, Haake DA, Godowski PJ, Hayashi F, Ozinsky A, Underhill DM, Kirschning CJ, Wagner H, Aderem A, Tobias PS, Ulevitch RJ. Nat. Immunol. 2001;2:346–352. doi: 10.1038/86354. [DOI] [PubMed] [Google Scholar]
- 24.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. J. Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 25.Miller JR. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 26.Inoue H, Nojima H, Okayama H. Gene (Amst.) 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 27.Wang RF, Kushner SR. Gene (Amst.) 1991;100:195–199. [PubMed] [Google Scholar]
- 28.Reynolds CM, Kalb SR, Cotter RJ, Raetz CRH. J. Biol. Chem. 2005;280:21202–21211. doi: 10.1074/jbc.M500964200. [DOI] [PubMed] [Google Scholar]
- 29.Basu SS, York JD, Raetz CRH. J. Biol. Chem. 1999;274:11139–11149. doi: 10.1074/jbc.274.16.11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karbarz MJ. Biochemistry of Endotoxin in Rhizobium leguminosarum: Characterization of a Family of Lipid Phosphatases Specific for the 1-Position of Lipid A. Ph.D. thesis. Duke University Medical Center; Durham, NC: 2004. [Google Scholar]
- 31.Doerrler WT, Raetz CRH. J. Biol. Chem. 2002;277:36697–36705. doi: 10.1074/jbc.M205857200. [DOI] [PubMed] [Google Scholar]
- 32.Brozek KA, Hosaka K, Robertson AD, Raetz CRH. J. Biol. Chem. 1989;264:6956–6966. [PubMed] [Google Scholar]
- 33.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CRH. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osborn MJ, Munson R. Methods Enzymol. 1974;31:642–653. doi: 10.1016/0076-6879(74)31070-1. [DOI] [PubMed] [Google Scholar]
- 35.Guy-Caffey JK, Rapoza MP, Jolley KA, Webster RE. J. Bacteriol. 1992;174:2460–2465. doi: 10.1128/jb.174.8.2460-2465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doerrler WT, Reedy MC, Raetz CRH. J. Biol. Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- 37.Doerrler WT, Gibbons HS, Raetz CRH. J. Biol. Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CRH. J. Biol. Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez-Romo P, Sanchez-Nieto S, Gavilanes-Ruiz M. Anal. Biochem. 1992;200:235–238. doi: 10.1016/0003-2697(92)90458-j. [DOI] [PubMed] [Google Scholar]
- 40.Ribeiro AA, Zhou Z, Raetz CRH. Magn. Reson. Chem. 1999;37:620–630. [Google Scholar]
- 41.Zhou Z, Ribeiro AA, Raetz CRH. J. Biol. Chem. 2000;275:13542–13551. doi: 10.1074/jbc.275.18.13542. [DOI] [PubMed] [Google Scholar]
- 42.Que NLS, Ribeiro AA, Raetz CRH. J. Biol. Chem. 2000;275:28017–28027. doi: 10.1074/jbc.M004009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA, Golenbock DT, Espevik T. J. Biol. Chem. 2002;277:47834–47843. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- 44.Romano JD, Michaelis S. Mol. Biol. Cell. 2001;12:1957–1971. doi: 10.1091/mbc.12.7.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson JL, Frase H, Michaelis S, Hrycyna CA. J. Biol. Chem. 2005;280:7336–7345. doi: 10.1074/jbc.M410292200. [DOI] [PubMed] [Google Scholar]
- 46.Kagan RM, Clarke S. Arch. Biochem. Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- 47.Tran AX, Karbarz MJ, Wang X, Raetz CRH, McGrath SC, Cotter RJ, Trent MS. J. Biol. Chem. 2004;279:55780–55791. doi: 10.1074/jbc.M406480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H, Hsu FF, Turk J, Groisman EA. J. Bacteriol. 2004;186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.