Abstract
Accurate inheritance of genomic content during cell division is dependent on synchronized changes in cellular organization and chromosome dynamics. Elucidating how these events are coordinated is necessary for a complete understanding of cell proliferation. Previous in vitro studies have suggested that the nuclear pore protein Nup153 is a good candidate for participating in mitotic coordination. To decipher whether this is the case in mammalian somatic cells, we reduced the levels of Nup153 in HeLa cells and monitored consequences on cell growth. Reduction of Nup153 resulted in a delay during the late stages of mitosis accompanied by an increase in unresolved midbodies. Depletion of Nup153 to an even lower threshold led to a pronounced defect early in mitosis and an accumulation of cells with multilobed nuclei. Although global nucleocytoplasmic transport was not significantly altered under these depletion conditions, the FG-rich region of Nup153 was required to rescue defects in late mitosis. Thus, this motif may play a specialized role as cells exit mitosis. Rescue of the multilobed nuclei phenotype, in contrast, was independent of the FG-domain, revealing two separable roles for Nup153 in the execution of mitosis.
INTRODUCTION
The nuclear pore complex (NPC) bridges the inner and outer nuclear membranes to form a conduit for both active transport of large molecules and diffusion of smaller molecules between the nucleus and cytoplasm (Terry et al., 2007). Approximately 30 proteins (termed nucleoporins), each present in multiple copies, comprise this macromolecular structure (Hetzer et al., 2005), providing a vital link between intracellular environments. In higher eukaryotes, once chromosomes undergo condensation, nuclear membranes and nuclear pore complexes are disassembled. A series of hallmark mitotic events ensue, including segregation of chromosomal content, rapid rebuilding of nuclei and, finally, abscission near the midbody, a structure that connects the daughter cells until cytokinesis is complete. Nucleoporins were originally thought to be stored inertly once released from the NPC at the entry into mitosis until once again incorporated into pores during late mitosis; however, recent evidence suggests that certain nucleoporins perform important mitotic functions independently of their roles during interphase. For example, the Nup107-160 complex as well as Nup358 (RanBP2) localize in part to kinetochores after their release from the NPC, and they function in chromosome congression (Zuccolo et al., 2007), maintenance of tension between sister kinetochores (Zuccolo et al., 2007), and sister chromatid segregation (Salina et al., 2003; Joseph et al., 2004; Dawlaty et al., 2008). Other mitotic functions of nucleoporins include roles in proper spindle assembly (Nup107-160 complex; Orjalo et al., 2006) and anaphase onset via regulation of APC/C (Nup98; Jeganathan et al., 2005).
Disassembly of the NPC occurs in coordination with nuclear membrane dispersal (Burke and Ellenberg, 2002; Hetzer et al., 2005; Prunuske and Ullman, 2006; Antonin et al., 2008); thus, the availability of nucleoporins for mitotic roles coincides with nuclear remodeling events that are important prerequisites for chromosome dynamics in prometaphase and metaphase. Before full dispersal, the NPC itself is remodeled (Lenart et al., 2003), suggesting that nucleoporins could also take on new roles during this brief window of transition. Indeed, distinct cell cycle-driven functions for nucleoporins can couple morphological remodeling of the nucleus and NPC with regulation of chromosomal inheritance and other mitotic processes. The precedents to date indicate that understanding the role of individual nucleoporins throughout the cell cycle is important to deciphering the network of interactions that orchestrate accurate cell division in mammalian cells.
Studies performed in Xenopus egg extracts, a system that recapitulates embryonic cell division, have implicated Nup153 in the process of nuclear envelope breakdown (Liu et al., 2003; Prunuske et al., 2006), indicating that this nucleoporin plays a specialized role at mitosis. Although mammalian Nup153 has been suggested to be essential for cell survival (Harborth et al., 2001), previous studies in which it was targeted for small interfering RNA (siRNA)-mediated depletion in cultured cells (Harborth et al., 2001; Hase and Cordes, 2003; Sabri et al., 2007) did not address a role for Nup153 in proper mitotic execution. More recently, Nup153 was identified in an unbiased genome-wide screen for proteins with roles in early mitosis (Rines et al., 2008). Defects in the absence of Nup153 were not characterized in detail, but the discovery of Nup153 in this context suggests a link to mitosis in mammalian cells. To gain insight into the mitotic role of Nup153, we have used graded levels of depletion to reveal functions of this nucleoporin that become impaired at different thresholds of expression. Additionally, using a rescue strategy, we have found that Nup153 plays distinct roles that impact both early and late stages of mitosis.
MATERIALS AND METHODS
Plasmid Constructs and siRNA Oligonucleotides (Oligos)
Full-length Nup153, cloned into the pEGFP-N2 vector, was modified by polymerase chain reaction (PCR) to introduce seven silent point mutations in the region targeted by the 153-1 siRNA oligo. Other Nup153 constructs were generated from either the parental Nup153-EGFP-N2 clone or the siRNA-resistant clone as follows: N+Z (amino acids 1-879), N+C (1-657 + 875-1475), N1-339+C (1-339 + 875-1475), N (1-657), Z (658-879), and C (880-1475). The histone H2B-cyan fluorescent protein (CFP) plasmid was a gift from Mary Dasso (National Institutes of Health, Bethesda, MD). All siRNA oligos were obtained from Ambion (Austin, TX): 153-1 (Harborth et al., 2001), 153-2 (GGACUUGUUAGAUCUAGUU), and a scrambled version of 153-1, Scr-1 (GCAAAUCUCCGAUCGUAGA), which was used as a control treatment.
Cell Culture, RNA Interference (RNAi) Depletion of Nup153, and Stable Cell Line Generation
HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS). Transfection with siRNA was achieved using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, except that oligo 153-1 was used at either 0.17 nM (153-1lo) or 10 nM, whereas 153-2 and Scr-1 oligos were used at 10 nM. Cells were harvested 48 h after transfection for further analysis. Stable cell lines were generated by transfecting the Nup153 constructs into HeLa cells by using Lipofectamine LTX (Invitrogen). Twenty-four hours after transfection, cells were split 1:30 and incubated with 1 mg/ml Geneticin (Invitrogen) for 10 d, after which single colonies were purified and expanded. The histone H2B-CFP cell line was generated in a similar manner, except that individual clones were not isolated.
Immunoblotting and Immunofluorescence
Whole cell extracts were prepared by cell lysis in radioimmunoprecipitation assay buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) and clarified by centrifugation. Samples were resolved by SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, probed with the appropriate antibodies, and developed using the Western Lightning Chemiluminescence Reagent (PerkinElmer Life and Analytical Sciences, Boston, MA). For immunofluorescence analysis, cells were fixed either in methanol for 5 min at −20°C or in 4% paraformaldehyde for 10 min on ice. In the latter case, cells were then permeabilized by 5-min incubation in 0.5% Triton X-100 in phosphate-buffered saline (PBS) on ice. Samples were blocked by incubation in 3% bovine serum albumin, 0.02% Triton X-100 (in PBS), and then incubated with the appropriate antibodies. DNA was detected by incubation with 0.2 μg/ml Hoechst 33258 (Calbiochem, San Diego, CA) during the final wash. Coverslips were mounted using ProLong Gold anti-fade (Invitrogen).
Antibodies used in this study include anti-nucleoporins (mAb414; Covance Research Products, Princeton, NJ), anti-α-tubulin (YL1/2; Accurate Chemical & Scientific, Westbury, NY), anti-green fluorescent protein (GFP) (JL-8; Clontech, Mountain View, CA; and ab290, Abcam, Cambridge, MA), anti-Nup153-Z (Liu et al., 2003), anti-Nup153 (SA-1; a gift from Brian Burke, University of Florida, Gainesville, FL), anti-lamin B2 (LN43; Abcam), anti-CENP-E (1H12; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Nup133 (gift from D. Forbes, University of California, San Diego, La Jolla, CA), and anti-Nup62 (BD Biosciences, San Jose, CA). All secondary antibodies were obtained from Invitrogen.
Nuclear Transport and mRNA Export Assays
Twenty-four hours after siRNA treatment, HeLa cells grown in DMEM + 10% charcoal-stripped FBS were transfected (Lipofectamine LTX; Invitrogen) with a plasmid encoding a chimeric Rev-GFP-Glucocorticoid Receptor protein (RGG; Love et al., 1998; provided by M. Dasso), which can be induced to import into the nucleus upon addition of dexamethasone and to export to the cytoplasm upon removal of dexamethasone. An additional 24 h later, cells were treated with 250 nM dexamethasone and fixed at the indicated times. For protein export assays, cells treated for 120 min with 250 nM dexamethasone were washed and incubated in fresh culture medium for the indicated times. Export of poly(A)+ RNA was assessed by fluorescence in situ hybridization by using a biotinylated oligo(dT) 50mer probe as described previously (Bastos et al., 1996).
Live Cell Time-Lapse Imaging and Analysis
Forty-eight hours after siRNA transfection, cells stably expressing histone H2B-CFP were imaged in 24-well plastic dishes in a stage-top incubator (OKO Lab, Ottaviano, Italy), which maintained appropriate temperature, humidity, and CO2 levels. Images (CFP and phase contrast) were acquired every 6–10 min from five to 15 different stage positions for 15–45 h by using a 40× PLANAPO numerical aperture 0.9 objective (with correction collar) on an Olympus IX81 microscope equipped with a motorized XY stage and a Hamamatsu Orca ER camera, all controlled using the MetaMorph software (Molecular Devices, Sunnyvale, CA). Time-lapse image stacks were converted to QuickTime movies by using MatLab-based software developed at the University of Utah Fluorescence Microscopy Core Facility (Salt Lake City, UT). Statistical analysis was performed using the GraphPad Prism software (GraphPad Software, San Diego, CA).
RESULTS
Nup153 Depletion Leads to Two Mitotic Phenotypes
To probe for mitotic function of Nup153 in mammalian somatic cells, we reduced levels of this protein in HeLa cells by using two independent siRNA oligonucleotides. Similar reduction was achieved after transfecting either 0.17 nM oligo 153-1 (referred to as 153-1lo) or 10 nM oligo 153-2, compared with control treatment (Scr-1; Figure 1, A and B). Using detection of tubulin to monitor mitotic alterations, we found that Nup153 reduction at these levels resulted in an approximately threefold increase in the number of unresolved cytokinetic midbodies (Figure 1, C and F). Interestingly, we found that increased depletion of Nup153 resulted in a strikingly different phenotype. Using 10 nM of the more effective oligo (153-1; Figure 1A) resulted in many cells (∼30%) with severely multilobed nuclei (Figure 1G), whereas the number of midbodies was unaffected (Figure 1F). The distinctive appearance of the nuclei was readily observed by tracking either DNA (Figure 1D) or the nuclear lamina (Figure 1E). Although this nuclear morphology is somewhat reminiscent of apoptosis, Nup153-depleted cells showed no increase in activated caspase-3 (data not shown) and displayed viability over time in live imaging (Supplemental Video 4). Thus, these alterations in nuclear architecture likely reflect a role for Nup153 directly in forming nuclei or in influencing chromosomal dynamics during mitosis; deregulation of the latter process has previously been found to result in a very similar nuclear appearance (Draviam et al., 2007; Stegmeier et al., 2007; Ohsugi et al., 2008).
Figure 1.
Nup153 depletion by RNAi leads to two mitotic phenotypes. HeLa cells were transfected with 0.17 nM oligo 153-1 (153-1lo), 10 nM oligo 153-2, 10 nM oligo 153-1, or 10 nM of Scr-1, a scrambled version of 153-1, and analyzed after 48 h. (A) Western blot of extracts from siRNA-treated cells probed with mAb414 to detect Nup153 and Nup62 or anti-α-tubulin. (B–E) siRNA-treated cells stained with anti-Nup153-Z (B), anti-α-tubulin (C and D), or anti-lamin B2 (E). Examples of unresolved midbodies are indicated by yellow arrowheads (C). The regions indicated in white boxes are shown at higher magnification in D illustrating the multilobed nuclear morphology. (F) Quantification of the number of midbodies after Nup153 siRNA treatment relative to control (Scr-1) treatment. (G) Percentage of cells displaying multilobed nuclei after the indicated siRNA treatments. Error bars indicate the SD of four independent experiments in which >900 cells were counted for each treatment. The significance of differences between Scr-1 and Nup153 siRNA treatments was assessed by calculating p values and those <0.005 are indicated: *p < 0.0001, **p < 0.0005. Bars, 15 μm (B, D, and E) and 50 μm (C).
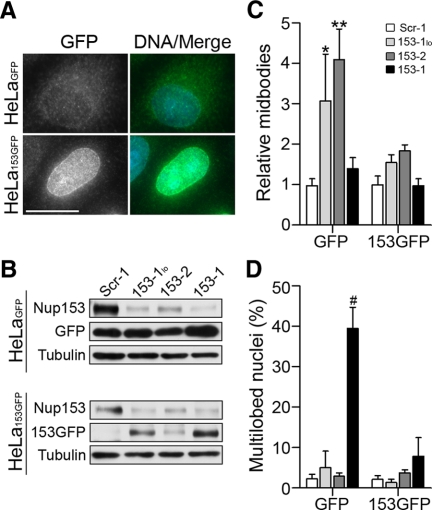
To verify that the observed phenotypes are attributable specifically to disruption of Nup153 protein levels and not to off-target effects, we developed stable cell lines expressing either GFP or siRNA-resistant Nup153-GFP. Nup153-GFP was targeted to the nuclear rim and the nucleoplasm, consistent with the endogenous localization of Nup153 and with previous reports (compare Figure 2A with Figure 1B; Bastos et al., 1996). The results of Nup153 depletion in HeLaGFP cells mirrored those obtained with the parental HeLa cells: a three- to fourfold increase in the number of unresolved midbodies upon reduction of Nup153 and a dramatic increase in the number of multilobed nuclei upon further depletion (Figure 2, C and D). In contrast, when HeLa153GFP cells were transfected with siRNA oligonucleotides directed against endogenous Nup153, very little increase in either midbodies or multilobed nuclei was observed (Figure 2, C and D), indicating that these phenotypes result specifically from alterations in the level of Nup153.
Figure 2.
Expression of exogenous Nup153 rescues accumulation of midbodies and multilobed nuclei. (A) Indirect immunofluorescence with an anti-GFP antibody was used to detect expression of GFP and Nup153GFP in HeLa cells stably transfected with these respective expression cassettes. (B) HeLaGFP and HeLa153GFP were transfected with siRNA as described in Figure 1. Western blot of extracts from siRNA-treated cells were probed with mAb414 to detect Nup153 and Nup153GFP, anti-GFP, to detect GFP, and anti-α-tubulin. (C) Quantification of the number of midbodies relative to control (Scr-1) treatment in each cell line. (D) Percentage of cells displaying multilobed nuclei following the indicated siRNA treatments in each cell line. Error bars indicate the SD of four independent experiments in which >900 cells were counted for each treatment. p values were calculated and reported as in Figure 1: *p < 0.003, **p < 0.0001, and #p < 0.0003. Bar, 15 μm.
Conditions of Nup153 Depletion Here Do Not Impair Global Transport Function of the NPC
Given the central role of the NPC in nucleocytoplasmic transport and the role that Nup153 has been characterized to play in this process (Shah and Forbes, 1998; Walther et al., 2001; Sabri et al., 2007), we next assessed nuclear import and export in Nup153-depleted cells. siRNA-treated cells were transfected with a plasmid encoding a nucleocytoplasmic transport reporter (RGG) (Love et al., 1998). Treatment with dexamethasone induces RGG to translocate to the nucleus and concentrate in nucleoli, and only a subtle delay in protein import was seen between control and Nup153-depleted cells (Figure 3A). After removal of dexamethasone, efficient protein export was observed in both treatments (Figure 3B).
Figure 3.
Nup153 depletion has little effect on nucleocytoplasmic transport of protein or mRNA. (A) Nuclear import was tracked using the glucocorticoid responsive RGG construct (see Materials and Methods for more detailed information). Shown are representative images after treatment with dexamethsone at the indicated time points. (B) Export of the RGG substrate was induced by removal of dexamethasone and monitored at the indicated time points. (C) Distribution of poly(A)+ RNA was detected by fluorescence in situ hybridization. Samples were costained with the pan-Nup antibody mAb414, which reacts with Nup358, Nup214, Nup153, and Nup62. Note the decreased intensity of nuclear rim staining as well as some cytoplasmic mislocalization of these nuclear pore proteins. (D) Nup133 and Nup62 were tracked by indirect immunofluorescence with specific antibodies. Expression and localization for both proteins at the nuclear rim are not significantly affected by Nup153 depletion. Similar to the pattern with mAb414, some Nup62 was detected in perinuclear cytoplasmic foci in the Nup153 depletion conditions. Bars, 20 μm.
To assess mRNA export, we analyzed distribution of poly(A)+ RNA by fluorescence in situ hybridization. mAb414-reactive nucleoporins (Nups 358, 214, 153, and 62) were detected by indirect immunofluorescence to delineate the nucleocytoplasmic boundary. Little difference in poly(A)+ RNA distribution was observed between cells treated with control oligo (Scr-1) and cells in which Nup153 was reduced (153-1lo, Figure 3C; 153-2; data not shown). A slight decrease in cytoplasmic poly(A)+ RNA was seen when Nup153 levels were further depleted (153-1, Figure 3C), but this observation is difficult to fully interpret due to concomitant alterations in cell and nuclear morphology. The intensity of mAb414 reactivity at the nuclear rim was reduced under both knockdown conditions (Figure 3C, left), which may reflect the lower Nup153 levels. Staining with a Nup62-specific antibody indeed showed little reduction of Nup62 at the nuclear rim (Figure 3D, middle). Nup133 was also present at the nuclear rim (Figure 3D, left), indicating that core NPC structure remains intact under these depletion conditions. Together, these results suggest that the phenotypes observed in this study are unlikely to be downstream consequences of a global alteration in nucleocytoplasmic trafficking (discussed further below).
Live Imaging Reveals Distinct Effects on Cell Cycle Timing That Correspond to the Level of Nup153 Depletion
To gain additional insight into the mitotic roles of Nup153, we performed time-lapse imaging of cells expressing histone H2B-CFP. Lowering the levels of Nup153 prolonged the total duration of mitosis, from an average time of 86 min (Scr-1) to 106 or 112 min (153-2 and 153-1lo, respectively; Figure 4A, Supplemental Table 1, and Supplemental Videos 1–3). Not only was the time in mitosis extended upon Nup153 reduction, but there was notably higher variation in mitotic timing, consistent with miscoordination of mitotic progression (Meraldi et al., 2004). Live imaging also revealed an increase in cortical blebbing during interphase. However, this was much more pronounced with 153-1lo treatment compared with 153-2 (Supplemental Videos 2 and 3), indicating that blebbing activity at the cell periphery does not correlate with changes in mitotic timing. We further analyzed hallmark events during mitosis to determine whether the mitotic delay was due to an overall lengthening of mitosis or whether it was specific to certain mitotic stages. Very little difference was found between control and Nup153-depleted cells during prophase/prometaphase (Figure 4B) or metaphase (Figure 4C). Progression from anaphase through cytokinesis, however, was markedly prolonged in Nup153-depleted cells (153-1lo and 153-2; Figure 4D; Supplemental Table 1).
Figure 4.
Moderate depletion of Nup153 leads to a mitotic delay during the later stages of mitosis. (A–D) Time-lapse imaging of live cells was performed 48 h after siRNA transfection. Images were acquired every 10 min for 18–45 h in three independent experiments. For this analysis, prophase/prometaphase was measured as the time between the first frame in which DNA condensation and rounded cells were apparent until DNA alignment along an equatorial metaphase plate; metaphase was measured by tracking the time between metaphase plate alignment and chromosome segregation; and anaphase/telophase/cytokinesis was measured as the time from the first indication of chromosome segregation until both daughter cells were flattened. Examples of time-lapse imaging can be found in Supplemental Videos 1–3. Frequency distribution plots illustrating a skew-normal distribution of the total length of time in mitosis (A; Scr-1, n = 417 cells; 153-1lo, n = 395 cells; 153-2, n = 390 cells), time in prophase/prometaphase (B; Scr-1, n = 242 cells; 153-1lo, n = 301 cells; 153-2, n = 228 cells), time in metaphase (C), and time in anaphase/telophase/cytokinesis (D). Cells analyzed in C and D are the same as in B. The connecting lines were smoothed using a spline function in GraphPad Prism 5.
Live imaging of cells treated with the higher concentration of the 153-1 oligo revealed a much more dramatic and pleiotropic mitotic phenotype (Supplemental Video 4). Whereas all of the control cells that initiated mitosis resulted in successful divisions (Figure 5, A and E), only 26% of the Nup153-depleted cells that initiated mitosis were successful in dividing (n = 116 cells; Figure 5, B and E), and merely 7% progressed within the range of mitotic kinetics seen in 90% of control cells. Additionally, 24% of cells that attempted to divide displayed oscillations of the DNA between the presumptive daughter cells during cytokinesis and/or failed to complete cytokinesis (Figure 5C). Strikingly, 50% of the Nup153-depleted cells that attempted to undergo cell division remained in a rounded state that persisted for several hours and was accompanied by waves of nonoriented cortical blebbing (Figure 5D). Of the Nup153-depleted cells that entered mitosis, a majority displayed delayed progression from the onset of mitosis to the end of metaphase (Figure 5F), demonstrating that depletion of Nup153 to a low threshold leads to a significant delay during early mitosis. Early aberrancies in mitosis may contribute to abnormal or failed cytokinesis. These latter defects could also reflect further impairment of the late mitotic role detected in the lower level knockdown.
Figure 5.
More robust depletion of Nup153 leads to profound defects in early mitosis. Fluorescence and phase contrast images of cells stably expressing histone H2B-CFP were acquired every 6 min for 15 or 45 h, in two independent experiments, overlaid, and assembled into montages. (A) All (100%) of Scr-1 treated cells that initiated mitosis underwent normal cell division, with an average time of 81 min (n = 118 cells). (B) Twenty-six percent of mitotic 153-1–treated cells (n = 116 cells) exhibited successful division, although with rather delayed kinetics (average time, 151 min; n = 30). (C) Twenty-four percent of cells that initiated mitosis displayed either failed or abnormal cytokinesis. (D) Half (50%) of mitotic 153-1–treated cells underwent prolonged periods of persistent rounding (up to 45 h in some cases) with extensive cortical blebbing. (E) Summary of mitotic phenotypes. (F) Of the Nup153-depleted cells that completed mitosis, including those that underwent abnormal cytokinesis (n = 58), a majority displayed a delay in the early stages of mitosis (through metaphase). An example of the time-lapse imaging can be found in Supplemental Video 4.
Nup153 Does Not Localize to the Kinetochore or the Midbody
In other cases in which mitotic functions have been discovered for a nucleoporin, this has mostly corresponded to targeting of the nucleoporin to kinetochores and involvement in proper attachment of the kinetochore to the microtubule spindle. We therefore assessed the localization of Nup153 during mitosis. DNA morphology was used to track the stage of cell division and costaining for CENP-E provided a reference point for various mitotic structures (Figure 6). Nup153 was not detected at kinetochores, distinguishing it from members of the Nup107 complex and Nup358 (Joseph et al., 2002; Salina et al., 2002; Loiodice et al., 2004). Furthermore, despite the delays observed in midbody resolution when Nup153 levels are reduced, this nucleoporin was not detected at midbodies (Figure 6). Although we did not find evidence for kinetochore or midbody targeting of Nup153, it is known to be very dynamically associated with the NPC (Daigle et al., 2001; Griffis et al., 2004) and could likewise dynamically associate with these mitotic structures, perhaps escaping detection.
Figure 6.
Nup153 is not detected at kinetochores or the midbody at mitosis in HeLa cells. Nup153 localization was assessed using the anti-Nup153-Z antibody. Antibody directed against CENP-E was used to detect kinetochores (prophase through anaphase), the cleavage furrow (telophase) and the midbody (cytokinesis). Bar, 5 μm.
Two Roles for Nup153 in Mitosis
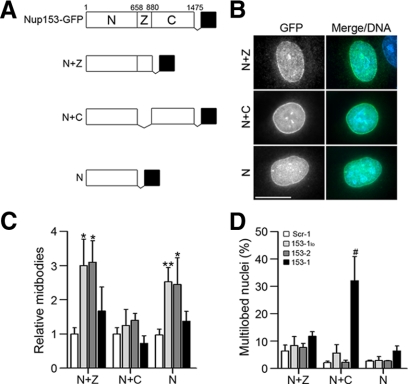
To determine whether the phenotypes observed here upon Nup153 depletion are different manifestations of the same defect, with the severity dependent on the extent of depletion achieved, or whether mitotic roles for Nup153 can be uncoupled, we capitalized on the rescue strategy. We first tested variants of Nup153 with deletions of the C-terminal region, the zinc-finger domain, or both (N+Z, N+C, and N, respectively). These constructs all localize to the nuclear rim at interphase (Figure 7B; Enarson et al., 1998). At these expression levels, which did not exceed the endogenous amount of Nup153, the N+Z, N+C, and N constructs alone did not dominantly affect the number of midbodies or multilobed nuclei (Figure 7, C and D; data not shown). Upon reduction of endogenous Nup153 in HeLaN+Z and HeLaN cells, an increase in midbodies similar to that seen when Nup153 was reduced in HeLaGFP cells was observed (compare Figure 7C with Figure 2C). In contrast, an increase in midbodies was not observed after reducing endogenous Nup153 in HeLaN+C cells (Figure 7C), indicating that this late mitotic phenotype is complemented only if the rescue construct includes the Nup153 C-terminal region.
Figure 7.
Rescue analysis indicates two separable roles for Nup153 in mitosis. (A) Schematic representation of Nup153 and panel of domain constructs. The N-terminal domain [N], zinc finger domain [Z], and C-terminal region [C] are indicated. GFP is indicated as a black box. (B) Indirect immunofluorescence with an anti-GFP antibody was used to detect expression of Nup153 derivatives in HeLa cells stably transfected with the respective expression cassettes. (C) HeLaN+Z, HeLaN+C, and HeLaN were transfected with siRNA as described in Figure 1. The number of midbodies relative to control (Scr-1) treatment was quantified for each cell line. (D) Percentage of cells displaying multilobed nuclei after the indicated siRNA treatments in each cell line. Error bars indicate the SD of four independent experiments in which >900 cells were counted for each treatment. p values were calculated and reported as in Figure 1: *p < 0.003, **p < 0.001, and #p < 0.005. Bar, 15 μm.
We observed a distinctly different pattern of complementation for the multilobed nuclei phenotype that results from further Nup153 depletion. This phenotype was not rescued by expression of Nup153 with an internal deletion of the zinc-finger domain (HeLaN+C cells; Figure 7D), consistent with previous reports that implicate the zinc-finger domain in efficient nuclear envelope breakdown (Liu et al., 2003; Prunuske et al., 2006). However, expression of the N-terminal domain of Nup153 was found to rescue the multilobed nuclei phenotype (Figure 7D). This pattern of complementation indicates that robust depletion of Nup153 results in mitotic defects that are more complex than can be resolved by our current strategy, either involving interplay between domains and/or a balance of more than one Nup153 activity. Yet, a clear point revealed by this rescue strategy is that the phenotypes resulting from graded Nup153 depletion are complemented by different regions of this protein and thus reflect at least two separable roles for Nup153 in mitosis.
Domain Analysis of Nup153 Function
A role for the Nup153 zinc-finger domain in mitotic events is underscored by the observation that ectopic expression of this domain in the HeLaZ cell line results in a significant presence (∼10%) of multinucleated cells (Figure 8), which likely indicates failure to initiate or complete cytokinesis. Interestingly, the zinc finger domain itself was also found to be sufficient to rescue the multilobed nuclei phenotype (Figure 9B). Further work will be needed to investigate how these activities are related.
Figure 8.
Expression of the zinc finger domain from Nup153 results in multinucleated cells. Indirect immunofluorescence of GFP and FG-nucleoporins (detected by mAb414), along with DNA detection, was used to track nuclei in HeLa cells stably transfected with plasmids expressing either GFP or a fusion of the Nup153 zinc finger domain with GFP. Bar, 50 μm.
Figure 9.
Analysis of domain requirements for Nup153 function. (A) HeLaz and HeLac were transfected with siRNA as described in Figure 1. Midbodies were counted and quantified relative to control (Scr-1) treatment for each cell line. (B) The percentage of cells displaying multilobed nuclei was tracked after the indicated siRNA treatments. (C) Indirect immunofluorescence with an anti-GFP antibody was used to detect stable expression of the N1-339+C in HeLa cells. (D) Western blot analysis indicates similar expression levels of the N+C and N1-339+C constructs (detected by α-GFP) after depletion of endogenous Nup153. (E) Quantification of the number of unresolved midbodies compared with control treatment. Note that although the N1-339+C construct was able to rescue the midbody phenotype, the residual levels of midbodies remained somewhat higher than in control treatment. Error bars indicate the SD of at least three independent experiments in which >900 cells were counted for each treatment. p values were calculated and reported as described in Figure 1: *p < 0.0003, **p < 0.002, and #p <0.005.
We next turned to the requirement for the C-terminal domain of Nup153 in late mitosis. Although this FG-rich region is linked to a general role in transport as a docking site for importin-β and other transport receptors (Shah et al., 1998; Ben-Efraim and Gerace, 2001; Ullman and Higa, 2008), our results indicate that protein import and export are not significantly perturbed under these conditions of depletion (Figure 3). Because importin-β is also an important mitotic regulator (Harel and Forbes, 2004), we considered whether the Nup153 FG-domain exerts its effects during late mitosis by controlling the availability or activity of either importin-β or other potential mitotic partners. If so, one would predict that the presence of the FG-domain itself would supply the essential function of Nup153. Midbodies remained significantly increased, however, in HeLaC cells after endogenous Nup153 reduction (Figure 9A). Thus, an activity associated with the N-terminal domain works in conjunction with the C-terminal region of Nup153 to contribute to mitotic exit.
One major difference between the Nup153 C construct versus N+C is subcellular localization, because the NPC targeting region is located within the N-terminal domain (amino acids 39-339; Enarson et al., 1998). To determine whether delivery to the NPC is the critical function conferred by the Nup153 N-terminal domain in the context of this late mitotic role, we assessed whether the midbody phenotype could be rescued by expression of an N1-339+C construct. As expected, this construct localized to the nuclear rim in stably transfected cells and was expressed in similar amounts as the N+C construct (Figure 9, C and D). When endogenous Nup153 was depleted to levels that give rise to an increase in midbodies (153-1lo and 153-2), expression of the N1-339+C construct rescued this phenotype somewhat more robustly than the C-terminal domain alone (Figure 9D), consistent with a role for NPC targeting. However, the number of midbodies in Nup153 depleted HeLaN1-339+C remained higher than in the control (Scr-1) treatment of this same cell line (Figure 9D). The reproducible persistence of a late mitotic defect suggests that NPC targeting is not the sole function conferred by the N-terminal domain.
DISCUSSION
We have shown here that reduction of Nup153 levels results in an increase in cells with unresolved midbodies, a signature of late cytokinesis. Although at face value, this could actually reflect an increase in proliferation with a proportional increase in cells at this stage of division, live imaging analysis clearly demonstrated a specific delay in the postmetaphase stages of mitosis. Rescue experiments confirm that the increase in unresolved midbodies is due to decreased Nup153 expression. Given the role for Nup153 in nucleocytoplasmic trafficking, this aspect of function was considered first. We found that under the depletion conditions of this study, global pore transport function remained robust and that Nup62 and Nup133, members of different core subcomplexes of the NPC, remain at the nuclear rim. This latter observation is consistent with a previous report in which localization of other members of the Nup107-160 complex were unperturbed by Nup153 depletion (Hase and Cordes, 2003). It is notable that even in the case where Nup153 depletion was found to result in deficient NLS-mediated import in Drosophila S2 cells, the reduction measured was significant, but not acute (∼25%) (Sabri et al., 2007). The exact effect on transport may depend on the extent of Nup153 reduction or the method by which its activity is impaired. Overall, transport is a very robust process that can proceed with relative efficiency even when changes in pore composition are introduced (Stavru et al., 2006; Dawlaty et al., 2008).
Although our results do not link the mitotic functions of Nup153 to a role in bulk transport of protein or mRNA, certain observations raise the question of whether a more specific alteration in nucleocytoplasmic trafficking underlies the accumulation of unresolved midbodies when Nup153 levels are reduced. The C-terminal region of Nup153 implicated in our rescue analysis of this phenotype houses a highly repetitive motif consisting of (FX)FG-repeats known to aide in mediating transport (Terry et al., 2007). Furthermore, the finding that the N-terminal region of Nup153 is needed in conjunction with C-terminal sequences indicates that this FG-rich domain does not carry out a mitotic role simply by sequestering and down-regulating a mitotic factor as cells progress to G1. This suggests that the N-terminal domain may be required to direct the C-terminal domain of Nup153 to the pore for this FG-rich region to participate in the transport of cargo particularly important to mitotic exit and/or cell cycle control. Such a role was recently identified for Nup96 (Chakraborty et al., 2008). To test this, the pore targeting region (amino acids 1-339) of Nup153 was linked directly to its C-terminal domain. In comparison to the C-terminal domain alone, however, this construct provided only a modest improvement in complementing the late mitotic phenotype. Although expression of N+C resulted in midbody numbers that were not significantly different in control versus Nup153 depletion, expression of N1-339+C did not fully attenuate the increase in midbodies that arise during Nup153 depletion. This indicates that pore targeting is not the sole function of the N-terminal domain and suggests that a transport role—at least one directed by the FG-rich domain—does not account for the activity of Nup153 in late mitosis. These results also implicate additional contributions made by the distal half of the N-terminal domain of Nup153.
While this manuscript was in preparation, a study was published in which Tpr, another component of the NPC, was found to bind directly to both Mad1 and Mad2 and to play a role in proper spindle checkpoint activation (Lee et al., 2008). The NPC localization of Tpr is known to be directed by its interaction with Nup153 (specifically amino acids 228-439 of the Nup153 N-terminal domain; Hase and Cordes, 2003). Tpr is an excellent candidate for collaborating with Nup153 in mitotic function, but it is important to note that loss of Tpr from the NPC, per se, would not account for the phenotypes observed upon Nup153 depletion. First, Tpr depletion was not found to have an effect on mitotic timing, whereas Nup153 levels were found critical for timely progression through both early and late mitosis. Second, although expression of a Nup153 construct lacking the C terminus has been shown previously to be sufficient to restore Tpr localization to the NPC (Sabri et al., 2007), here we found that expression of this construct does not rescue the increase in midbodies seen after Nup153 depletion. Although the phenotypes and rescue patterns observed in this study point toward contributions of Nup153 that are independent of Tpr, these two components of the nuclear pore basket are likely to participate in concert to provide a newly-emerging layer in mitotic regulation. The pore basket may act as a scaffold on which mitotic factors are sequestered and regulated. In addition to Mad1 and Mad2, the SUMO protease Ulp1p, required for late mitosis in yeast, is localized to the nuclear pore during interphase (Makhnevych et al., 2007). It is interesting to note that the mammalian homologue of Ulp1p, SENP2, also localizes to the nuclear pore and has been found to associate with the C-terminal domain of Nup153 (Hang and Dasso, 2002; Zhang et al., 2002).
Additional observations indicate that a subset of nucleoporins converge in a regulatory pathway during late mitosis and/or mitotic exit. Depletion of either Nup133, a member of the Nup107-160 complex, or ELYS, a protein associated with this complex, also results in accumulation of unresolved midbodies (Rasala et al., 2006). However, Nup153 is the only FG-containing nucleoporin so far identified to impinge on this stage of the cell cycle. Our finding that the FG-domain is required for rescue of midbody accumulation suggests that Nup153 contributes in a unique manner to this regulatory network.
When Nup153 levels are further depleted, many cells display abnormal, multilobed nuclei. This phenotype could reflect a direct role for Nup153 in forming nuclear structure. Alternatively, disruption of proper chromosome dynamics during mitosis is known to result in a similar nuclear morphology and indeed has been the basis of a screen for mitotic regulators (Draviam et al., 2007; Stegmeier et al., 2007; Ohsugi et al., 2008). We found in live imaging analysis that significant delays occur early in mitosis under these conditions of Nup153 depletion. This supports a model in which Nup153 function impacts progression out of prophase and/or chromosome segregation itself, and the miscoordination that follows results in aberrant nuclear morphology. More extensive imaging will be needed to distinguish where the defect in early mitosis lies more precisely. Further investigation into whether Nup153 has a separable role in assembly of proper nuclear architecture is also warranted.
In summary, we have found that two distinct phenotypes result from depleting Nup153 to different levels and, further, that rescue of these phenotypes requires distinct regions within Nup153. This graded response to disruption of Nup153 expression likely reflects different thresholds at which Nup153 becomes limiting for one or more processes. Taken together, our results indicate that Nup153 has at least two functions important for proper mammalian cell division and that the FG-rich region of Nup153 plays an important role at mitosis. Given the complicated multifunctional nature of nucleoporins, the strategy we have developed here, combining graded depletion and domain rescue, will aid in further elucidating how nucleoporins contribute to mitosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tracey Zundel and Vishesh Agrawal for assistance with cloning; Mary Dasso, Maureen Powers, Brian Burke, and Douglass Forbes for reagents; Chris Rodesch and Keith Carney at the University of Utah Fluorescence Microscopy Core Facility for assistance with time-lapse imaging and image processing; and Jody Rosenblatt for critical review of the manuscript. D.R.M. was supported by the American Cancer Society–Michael Schmidt Postdoctoral Fellowship PF-07-103-01-CSM. Core facilities were supported in part by P30 CA042014 awarded to the Huntsman Cancer Institute. This work was funded by the National Institutes of Health grant R01 GM-61275, the Leukemia and Lymphoma Society, and the Huntsman Cancer Foundation (to K.S.U.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0883) on January 21, 2009.
REFERENCES
- Antonin W., Ellenberg J., Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Bastos R., Lin A., Enarson M., Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I., Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 2001;152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Ellenberg J. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- Chakraborty P., et al. Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev. Cell. 2008;15:657–667. doi: 10.1016/j.devcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M. M., Malureanu L., Jeganathan K. B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J. M. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V. M., Stegmeier F., Nalepa G., Sowa M. E., Chen J., Liang A., Hannon G. J., Sorger P. K., Harper J. W., Elledge S. J. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat. Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- Enarson P., Enarson M., Bastos R., Burke B. Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma. 1998;107:228–236. doi: 10.1007/s004120050301. [DOI] [PubMed] [Google Scholar]
- Griffis E. R., Craige B., Dimaano C., Ullman K. S., Powers M. A. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol. Biol. Cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang J., Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Harborth J., Elbashir S. M., Bechert K., Tuschl T., Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Harel A., Forbes D. J. Importin beta: conducting a much larger cellular symphony. Mol. Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Hase M. E., Cordes V. C. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell. 2003;14:1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M. W., Walther T. C., Mattaj I. W. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Jeganathan K. B., Malureanu L., van Deursen J. M. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- Joseph J., Liu S. T., Jablonski S. A., Yen T. J., Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Joseph J., Tan S. H., Karpova T. S., McNally J. G., Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Sterling H., Burlingame A., McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P., Rabut G., Daigle N., Hand A. R., Terasaki M., Ellenberg J. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J. Cell Biol. 2003;160:1055–1068. doi: 10.1083/jcb.200211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Prunuske A. J., Fager A. M., Ullman K. S. The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev. Cell. 2003;5:487–498. doi: 10.1016/s1534-5807(03)00262-4. [DOI] [PubMed] [Google Scholar]
- Loiodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J. B., Doye V. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love D. C., Sweitzer T. D., Hanover J. A. Reconstitution of HIV-1 rev nuclear export: independent requirements for nuclear import and export. Proc. Natl. Acad. Sci. USA. 1998;95:10608–10613. doi: 10.1073/pnas.95.18.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T., Ptak C., Lusk C. P., Aitchison J. D., Wozniak R. W. The role of karyopherins in the regulated sumoylation of septins. J. Cell Biol. 2007;177:39–49. doi: 10.1083/jcb.200608066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V. M., Sorger P. K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Ohsugi M., Adachi K., Horai R., Kakuta S., Sudo K., Kotaki H., Tokai-Nishizumi N., Sagara H., Iwakura Y., Yamamoto T. Kid-mediated chromosome compaction ensures proper nuclear envelope formation. Cell. 2008;132:771–782. doi: 10.1016/j.cell.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Orjalo A. V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S. G., Fontoura B., Briggs S., Dasso M., Forbes D. J. The Nup107–160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunuske A. J., Liu J., Elgort S., Joseph J., Dasso M., Ullman K. S. Nuclear envelope breakdown is coordinated by both Nup358/RanBP2 and Nup153, two nucleoporins with zinc finger modules. Mol. Biol. Cell. 2006;17:760–769. doi: 10.1091/mbc.E05-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunuske A. J., Ullman K. S. The nuclear envelope: form and reformation. Curr. Opin. Cell Biol. 2006;18:108–116. doi: 10.1016/j.ceb.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B. A., Orjalo A. V., Shen Z., Briggs S., Forbes D. J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rines D. R., et al. Whole genome functional analysis identifies novel components required for mitotic spindle integrity in human cells. Genome Biol. 2008;9:R44. doi: 10.1186/gb-2008-9-2-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri N., Roth P., Xylourgidis N., Sadeghifar F., Adler J., Samakovlis C. Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J. Cell Biol. 2007;178:557–565. doi: 10.1083/jcb.200612135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D., Bodoor K., Eckley D. M., Schroer T. A., Rattner J. B., Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Salina D., Enarson P., Rattner J. B., Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Forbes D. J. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- Shah S., Tugendreich S., Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F., Hulsmann B. B., Spang A., Hartmann E., Cordes V. C., Gorlich D. NDC 1, a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 2006;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Terry L. J., Shows E. B., Wente S. R. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Ullman K. S., Higa M. Nup153. UCSD–Nature Molecule Pages. 2008 doi: 10.1038/mp.a003702.01. [DOI]
- Walther T. C., Fornerod M., Pickersgill H., Goldberg M., Allen T. D., Mattaj I. W. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Saitoh H., Matunis M. J. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M., et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.