Abstract
Within the mitotic spindle, there are multiple populations of microtubules with different turnover dynamics, but how these different dynamics are maintained is not fully understood. MCAK is a member of the kinesin-13 family of microtubule-destabilizing enzymes that is required for proper establishment and maintenance of the spindle. Using quantitative immunofluorescence and fluorescence recovery after photobleaching, we compared the differences in spindle organization caused by global suppression of microtubule dynamics, by treating cells with low levels of paclitaxel, versus specific perturbation of spindle microtubule subsets by MCAK inhibition. Paclitaxel treatment caused a disruption in spindle microtubule organization marked by a significant increase in microtubules near the poles and a reduction in K-fiber fluorescence intensity. This was correlated with a faster t1/2 of both spindle and K-fiber microtubules. In contrast, MCAK inhibition caused a dramatic reorganization of spindle microtubules with a significant increase in astral microtubules and reduction in K-fiber fluorescence intensity, which correlated with a slower t1/2 of K-fibers but no change in the t1/2 of spindle microtubules. Our data support the model that MCAK perturbs spindle organization by acting preferentially on a subset of microtubules, and they support the overall hypothesis that microtubule dynamics is differentially regulated in the spindle.
INTRODUCTION
The cytoskeleton must undergo dramatic reorganization as cells transition from interphase to mitosis to assemble the mitotic spindle. The spindle is a macromolecular structure composed of a dynamic array of microtubules (MTs) and their associated proteins that is necessary for proper chromosome segregation (Compton, 2000; Walczak and Heald, 2008). At the onset of mitosis, there is an increase in MT turnover that allows for breakdown of the interphase array and assembly of the mitotic spindle (Saxton et al., 1984; Kline-Smith and Walczak, 2004). This change is correlated with an increased catastrophe frequency (transition from growth to shrinkage) and a decreased rescue frequency (transition from shrinkage to growth) (Belmont et al., 1990; Gliksman et al., 1992; Verde et al., 1992; Rusan et al., 2001). The changes in MT dynamics are also correlated with a change in MT polymer levels during mitosis (Zhai et al., 1996). At nuclear envelope breakdown, there is a dramatic decrease in MT polymer levels, which may allow the cell to rapidly assemble a mitotic spindle because the released tubulin dimers can polymerize into new spindle MTs. As the cell progresses from mitosis to the next interphase, there is no change in the levels of MT polymer (Zhai et al., 1996), suggesting that MT polymer gets reorganized to reform the interphase MT cytoskeleton, without a significant change in the dynamics of the MTs. These studies highlight the need for the cell to possess a mechanism to temporally regulate MT dynamics.
In addition to the temporal changes in dynamics throughout the cell cycle, the dynamics of MTs within mitosis are also highly regulated. The spindle MTs or nonkinetochore MTs (nonkt-MTs) represent the bulk of the MT polymer in the spindle and are highly dynamic, with a half-time for recovery (t1/2) of ∼18 s (Saxton et al., 1984; Cimini et al., 2006). The kinetochore MTs (K-fibers) are organized into bundles of MTs that mediate the attachment of the chromosomes to the spindle (Rieder, 1982; McDonald et al., 1992; McIntosh et al., 2002). K-fibers are overall more stable, with a t1/2 of ∼5 min (Gorbsky et al., 1988; Zhai et al., 1995; Cimini et al., 2006), but locally they are highly dynamic as the polymerization/depolymerization cycles at their plus ends are coupled to chromosome movement (McIntosh et al., 2002). Astral MTs extend from the spindle pole toward the cell cortex and may be important in spindle positioning (Rosenblatt, 2005). They have similar turnover dynamics as spindle MTs (Zhai et al., 1996). All classes of MTs within the spindle as well as astral MTs exhibit dynamic instability; however, only spindle MTs and kinetochore MTs exhibit poleward MT sliding and MT flux (Wadsworth and Salmon, 1986; Gorbsky et al., 1988; Mitchison, 1989; Mitchison and Salmon, 1992; Waterman-Storer et al., 1998). MT flux is a specialized MT behavior wherein tubulin subunits undergo net addition at the plus end and net loss at the minus end, creating a poleward translocation of tubulin subunits through the spindle (Mitchison, 1989; Kwok and Kapoor, 2007). The overall dynamics within the spindle are clearly highly complex and require a diverse set of cellular factors to control their intricate regulation. A major question becomes: to what extent can specific cellular factors influence the spatial and temporal coordination of MT dynamics?
Several families of proteins alter MT turnover in cells, including both MT-stabilizing and MT-destabilizing proteins (Wittmann et al., 2001; Scholey et al., 2003). Of particular interest are members of the kinesin superfamily that regulate the polymerization dynamics of MTs (Wordeman, 2005; Moores and Milligan, 2006; Howard and Hyman, 2007). These include members of the kinesin-13 family (Kif2A, Kif2B, and MCAK/Kif2C), which seem to be strictly MT-depolymerizing enzymes (Desai et al., 1999; Hunter et al., 2003; Helenius et al., 2006), as well as members of the Kinesin-8 family, which are MT plus end-directed motors and plus end-specific depolymerases (Gupta et al., 2006; Howard and Hyman, 2007; Mayr et al., 2007; Varga et al., 2008). MCAK has been shown to be a critical regulator of MT dynamics and organization in vitro, in Xenopus egg extracts and in mammalian cells (Walczak et al., 1996; Desai et al., 1999; Maney et al., 2001; Kline-Smith and Walczak, 2002; Cassimeris and Morabito, 2004; Zhu et al., 2005; Stout et al., 2006; Manning et al., 2007; Ohi et al., 2007; Wordeman et al., 2007; Hedrick et al., 2008). In addition, the action of MCAK is critical at the kinetochore in which it may promote error correction (Kline-Smith et al., 2004) by facilitating kinetochore MT turnover and the coordination of chromosome movement (Wordeman et al., 2007). The action of Kif2A has been more controversial. Some studies implicate the protein in controlling MT flux (Gaetz and Kapoor, 2004; Ganem et al., 2005), whereas other studies clearly show that it is not required for MT flux (Cameron et al., 2006; Ohi et al., 2007). More recently, it has been demonstrated that although both MCAK and Kif2A can depolymerize MTs with nearly equal efficiency, they cannot substitute for each other functionally during spindle assembly in Xenopus egg extracts (Ohi et al., 2007). In addition, knockdown of Kif2A results in monopolar spindles whose bipolarity can be restored by codepletion of MCAK (Ganem et al., 2005), suggesting that individual members of the kinesin-13 family may act at different ends of the MTs to contribute to spindle organization. Finally, it has been demonstrated that each of the members of the kinesin-13 family plays a distinct role during mitosis (Rogers et al., 2004; Buster et al., 2007; Manning et al., 2007).
The above-mentioned studies support a model in which different members of the kinesin-13 family affect the dynamics of individual populations of MTs during mitosis to coordinate spindle organization. However, there has not been a systematic test of what would happen to spindle MT dynamics if a single kinesin-13 were perturbed and how that would compare with an overall suppression of spindle MT dynamics. In this study, we looked at both spindle MT dynamics and organization under conditions in which MT dynamics were perturbed by treatment with paclitaxel, which should act as a global inhibitor of MT dynamics, or by MCAK knockdown, which may be spatially controlled. Our data are consistent with the model that MCAK perturbs spindle organization by acting preferentially on a subset of spindle MTs, and they support the overall hypothesis that MT dynamics are differentially regulated in the spindle.
MATERIALS AND METHODS
Cell Culture and Drug Treatments
PtK2 cells and PtK-T-cells (PtK2 cells that stably express green fluorescent protein [GFP]-α-tubulin) (Khodjakov et al., 2003) were maintained in complete Opti-MEM (Invitrogen, Carlsbad, CA) supplemented with 1% penicillin/streptomycin (Invitrogen), 1% GlutaMAX (Invitrogen), and 10% fetal bovine serum (Invitrogen) at 37°C, 5% CO2. For immunofluorescence analysis or for microinjection, cells were plated on 12-mm glass coverslips at one third to one sixth dilutions and grown for 3–6 d. For fluorescence recovery after photobleaching (FRAP) experiments, cells were plated on 22- × 22-mm glass coverslips at one third to one sixth dilutions and grown for 3–6 d. All drugs were made as concentrated stocks in tissue culture grade dimethyl sulfoxide (DMSO). Cells were treated with paclitaxel (Sigma-Aldrich, St. Louis, MO) at 20, 30, or 40 nM for 24 h, or at 10 μM for 2 h. In each case, the drugs were diluted to a stock concentration in DMSO and then diluted in Opti-MEM such that the final amount of DMSO added was 0.1% for all treatments. Control cells were incubated in 0.1% DMSO in complete Opti-MEM. For experiments involving monoastral spindles, monastrol was added to cells at 100 μM final concentration for 2–3 h. For measurements of tubulin diffusion rates, cells were incubated in 2 μM nocodazole for 3 h.
Small Interfering RNA (siRNA) Transfections
RNA interference (RNAi) experiments in PtK2 cells were performed as described in Stout et al. (2006) and Stout et al. (2009) by using a 21-base pair siRNA (Dharmacon TNA Technologies, Chicago, IL) to PtK-MCAK (UGACUUUCUUUGAGAUCUA[dT][dT]), or control nontargeting siRNA to luciferase.
Microinjection
Microinjection was carried out as described in Kline-Smith and Walczak (2002), by using either control nonimmune immunoglobulin G (IgG), anti-Xenopus MCAK (Walczak et al., 1996), or anti-PtK MCAK antibodies, each at needle concentration of 2 mg/ml. Antibodies were stored in antibody dialysis buffer (10 mM HEPES and 100 mM KCl, pH 7.2). Anti-PtK MCAK antibodies were raised against the N terminus of PtK MCAK and affinity-purified before use. All antibodies were spun at 90K rpm for 5 min in a TLA100 rotor to get rid of any aggregates before injection. Injection of either anti-MCAK antibody led to indistinguishable morphologies in PtK2 cells. Before microinjection, each coverslip was transferred to a 35-mm tissue culture dish containing 2 ml of Complete Opti-MEM media supplemented with 20 mM K-HEPES, pH 7.2, prewarmed to 37°C. Cells on each coverslip were injected over a period of 5 min. For analysis of bipolar spindles, cells were injected at prophase before nuclear envelope breakdown. After injection, cells were placed on a 37°C warming tray for 30 min before they were fixed and processed for immunofluorescence. For analysis of monoastral spindles, cells were treated with 100 μM monastrol for 2–3 h, and then cells were injected at either prophase or prometaphase and placed on a 37°C warming tray. Twenty-five minutes after injection, cells were fixed and stained to examine the MT array.
Immunofluorescence
Cells were rinsed with phosphate-buffered saline (PBS) (12 mM PO42−, 137 mM NaCl, and 3 mM KCl, pH 7.4) and then fixed for 20 min with 4% formaldehyde and 0.1% glutaraldehyde in PHEM [60 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 25 mM HEPES, 10 mM EGTA, and 2 mM MgCl2, pH 6.9] for MT staining. For MCAK staining, 2% formaldehyde in PHEM was used. Fixed cells were rinsed with Tris-buffered saline/Triton X-100 (TBS-TX: 20 mM Tris, 150 mM NaCl, pH 7.5, + 0.1% Triton X-100) and blocked in AbDil (2% bovine serum albumin and 0.1% NaN3 in TBS-TX) for 30 min. All subsequent rinses between antibody incubations were performed using TBS-TX; all antibodies were diluted in AbDil, and all antibody incubations were 30 min. The following antibodies were used: for MTs, DM1-α was used at 1/500; for kinetochores, anti-centromere antibodies (ACA) (Antibodies, Davis, CA) were used at 1/10; and for MCAK, anti-PtK-MCAK antibodies were used at 5 μg/ml. Cells were then incubated with fluorescein isothiocyanate, Texas Red, or Alexa-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA; or Invitrogen). Cells were incubated in 2 μg/ml Hoechst for 5 min to stain DNA. Coverslips were mounted onto microscope slides with 0.5% p-phenylenediamine and 20 mM Tris, pH 8.8, in 90% glycerol.
Microscopy, Image Acquisition, and FRAP
Wide-field epifluorescence images were captured with a CoolSNAP HQ charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) mounted on a 90i Eclipse microscope (Nikon, Melville, NY) with either a 60× (1.45 numerical aperture [NA]) Plan Apo VC or a 100× (1.4 NA) Plan Apo objective. The microscope, filters, and shutters were controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). Z-series stacks of images at 0.5-μm intervals through each cell were collected and deconvolved with fixed settings using AutoDeblur 9.3 software (Media Cybernetics, Bethesda, MD). For quantification, all images were acquired at identical acquisition settings. For each slide, late prometaphase cells were randomly selected and analyzed for fluorescence intensity as described below. For identification of injected cells on the slides, cells were stained for the injectate (IgG or anti-MCAK).
For FRAP imaging, PtK-T-cells were grown on 22- × 22-mm coverslips, mounted on a microscope slide in Opti-MEM containing 20 mM HEPES and 1/100 Oxyrase (Oxyrase, Mansfield, OH), and sealed with VALAP (equal volumes of Vaseline, lanolin, and paraffin). During imaging, temperature was maintained at 37°C by using an airstream incubator (NEVTEK, Williamsville, VA). Data were collected using a spinning disk confocal microscope. A Nikon TE2000U inverted microscope equipped with a Yokogawa CSU-10 spinning disk confocal head (PerkinElmer Life and Analytical Sciences, Boston, MA) and illuminated via a 10-mW 488-nm laser controlled by MetaMorph was used. Images were collected on a Cascade-II EMCCD camera (Photometrics). For the majority of the paclitaxel-treated cells in which FRAP was performed, the imaging and bleaching were carried out on a similar spinning disk system, equipped with a Hamamatsu (Bridgewater, NJ) C9100-50 EM CCD camera and a 50-mW, 488-nm laser and controlled by Volocity software (Improvision, Waltham, MA). Similar experiments performed on both systems gave indistinguishable results. Photobleaching for FRAP was carried out using a 300-mW, 488-nm laser controlled by Mosaic software (Photonics Instruments, St. Charles, IL) by emitting a high-intensity laser for 500 ms at a 3- × 10-μm masked region on the spindle placed between the pole and the spindle equator. Imaging and photobleaching were carried out with the 60× or 100× 1.4 NA oil objectives. Images were captured at an average of 300-ms prebleach. Postbleach images were collected every 2 s for at least 90 s by using the same settings as the prebleach imaging.
Immunofluorescence Quantification and Data Collection
Fluorescence quantification of MT polymer distribution was performed on deconvolved three-dimensional (3-D) reconstructed z-series stacks of mitotic cells. A comparison of raw versus deconvolved images of fluorescently labeled beads showed that deconvolution did not affect the linearity of the average fluorescence intensity measured on z-series images. Using MetaMorph, the spindles were rotated so that the pole-to-pole axis was horizontal. Fixed size boxes were drawn on the 3-D reconstructed images at distinct regions to define each MT polymer subclass or on a region outside the cell for background measurements. To measure astral-cortical MTs, four 30 × 150 pixel boxes (20 μm2 each or 80 μm2 total area) were positioned outside of the spindle (2 boxes vertically positioned outside of the spindle poles, and 2 boxes horizontally positioned above and below the spindle region (Figure 1A); to measure pole intensity, two 30 × 100 pixel boxes (13.4 μm2 each or 26.8 μm2 total area) were placed over the centrosome region; and to measure spindle intensity, two 100 × 150 pixel boxes (67 μm2 each or 134 μm2 total area) were placed on the spindle region equidistant between the spindle pole and the spindle equator. The K-fiber fluorescence was measured by first locating the kinetochore by using the kinetochore marker ACA, drawing a 10 × 10 pixel (0.5-μm2) box that surrounded the kinetochore and then making an adjacent 10 × 10 pixel box on the K-fiber (Figure 1B). A background box was subtracted from the image by placing a 10 × 10 pixel box on a region of the spindle adjacent to the kinetochore but that does not contain any kinetochore MTs. All fluorescence values collected were in average pixel fluorescence in deconvolved images and were collected from at least 15 cells in each of three independent experiments. Spindle length was measured from late prometaphase spindles by drawing a line between the centers of the poles using the 3-D reconstructed images. The interkinetochore distance was measured using the centromere marker ACA. Distance was determined by drawing a line from the outer kinetochore extending to the outer edge of its sister kinetochore using the z-stacks. The length of the line was converted from pixels to microns, and the distance was calculated using the Pythagorean Theorem to account for the z-step(s) distance (Hoffman et al., 2001). The interkinetochore distances were measured from three to five kinetochores/cell and then averaged per cell. At least 15 cells were averaged in each of three independent experiments for a total of >100 kinetochore pairs in 45 cells.
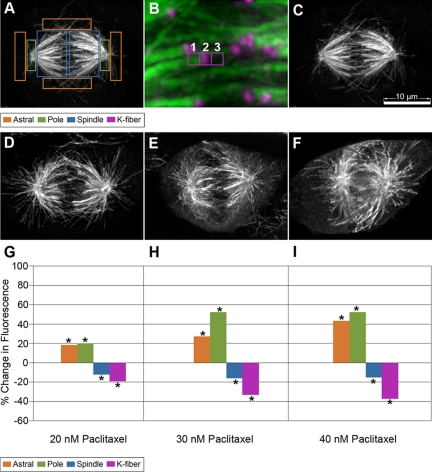
Figure 1.
Paclitaxel addition alters MT polymer distribution in the mitotic spindle. (A) Fluorescence quantification of MTs was carried out by drawing fixed size boxes on distinct regions of the spindle, astral-cortical MTs (orange); pole region (green); and spindle MTs (blue). (B) For K-fiber quantification, three equivalent boxes were drawn at the end of a K-fiber. Box 1 marks the end of the K-fiber, box 2 marks the centromere, and box 3 is drawn as a background measurement. The fluorescence intensity of the K-fiber is determined by measuring the average fluorescence intensity in boxes 1–3. Cells were either incubated in media containing 0.1% DMSO (C), 20 nM (D), 30 nM (E), or 40 nM (F) paclitaxel for 24 h and then fixed and stained for MTs, the kinetochore marker ACA, and DNA (only MTs shown). Fluorescence quantification of MT polymer fluorescence intensity is plotted for each paclitaxel concentration as a percentage of change in comparison with the DMSO controls (G–I). Average fluorescence values were collected at the specified regions from the deconvolved 3-D reconstructed images. Asterisks at each bar reflect a statistically significant change (p < 0.05). Bar, 10 μm.
For monastrol-treated cells, K-fiber length, astral MT length, and area were all collected from 3-D reconstructed images. The length of K-fibers was determined by drawing lines that connect each single kinetochore to a specified point at the center of the monoastral structure. The average length of all the lines drawn was calculated as the mean distance of the kinetochores to the centrosome region reflecting the average length of a K-fiber. For measurements of astral MT length, the same central point specified was used to extend at least eight lines from that point to the tip of astral MTs near the cortex. The average of the lines was calculated to estimate the mean length of an astral MT. The area of the monoastral structure was measured by drawing a freehand object around the structure in the MT channel. Lengths of lines and area measurements were converted from pixel and pixel2 to micrometers and square micrometers, respectively. Total and average fluorescence measurements were collected for each given area for each structure where the total fluorescence represents the integrated fluorescence of the measured region and the average fluorescence = the integrated fluorescence/area, which is important for distinguishing between treatments that change the overall size of the structure versus those that affect the amount of MT polymer in that region. A background measurement outside the structure was collected and subtracted from each measurement.
FRAP Statistical Analysis
FRAP data analysis were performed generally as described previously in Maddox et al. (2000). Data collection was carried out using either MetaMorph or Volocity in the same manner: to measure the FRAP recovery rate, a 2-μm2 box was placed on either the “bleached” region or a “background” region outside the cell (for subtraction of background noise). To measure the fluorescence dissipation due to photobleaching that occurs during the imaging, measurements of an “unbleached” region within the spindle were collected by placing a free hand drawn shape of variable size surrounding the unbleached opposite spindle half. The average fluorescence for each region (bleached, unbleached, or background) was collected for each image in the time-lapse series, and then the background value was subtracted from either the “bleached” or the “unbleached” region. To calculate fluorescence recovery, we needed to take into account the amount of photobleaching that occurs due to the imaging laser. The rate of photobleaching due to imaging was corrected for by normalizing to values obtained from the unbleached spindle half as follows: for each experimental condition (e.g., control RNAi, MCAK RNAi, DMSO, and paclitaxel), we calculated the mean of the average fluorescence intensity at each time point. We then plotted the mean average fluorescence intensity versus time and fit it to the power regression model (y = axb), which gave the highest r2 value of multiple models tested. The equation representing the curve from the power regression model was then subtracted from each fluorescence recovery curve to generate the normalized FRAP recovery data. Normalized fluorescence recovery data were then individually plotted for each cell and fit to a single or double exponential recovery curve using Prism (GraphPad Software, San Diego, CA). The double exponential curve, Y = Ymax1 × (1 − exp(−K1 × X)) + Ymax2 × (1 − exp(−K2 × X)), was favored over the single exponential curve as judged by having a higher r2 value. Any recovery curve with an r2 value <0.8 was not used in the subsequent data analysis. To calculate the t1/2 values, we used the equation t1/2 = ln 2/k, where k = K1 or K2 was obtained from the double exponential recovery equation. We then calculated the average ± SEM for each t1/2 for the fast or slow phase for each experimental condition. To determine the amount of fluorescence representing each phase, we calculated: Ymax1 = Ymax1/(Ymax1 + Ymax2) × 100, and Ymax2 = Ymax2/(Ymax1 + Ymax2) × 100, respectively. We then calculated the average ± SEM for Ymax1 and for Ymax2 for each experimental condition. For all data analysis, we used a two-tailed t test in Excel (Microsoft, Redmond, WA). A significant difference was considered when p ≤ 0.05.
RESULTS
Paclitaxel Treatment and MCAK Perturbation Alter Spindle Structure Differently
We wanted to understand how MCAK contributes to mitotic spindle organization and function. Prior localization and perturbation studies suggested that MCAK might act to differentially affect the dynamics of individual subpopulation of MTs during mitosis. To test this idea, we needed a way to determine what effect a general inhibitor of MT dynamics would have on spindle structure. We hypothesized that treating cells with relatively low concentrations of paclitaxel would act as a global regulator of MT dynamics by reducing the catastrophe frequency of all MTs without preference for any subpopulation of spindle MTs. We treated PtK2 cells with paclitaxel for 24 h, chemically fixed the cells, and performed immunofluorescence labeling of tubulin and centromeres. At paclitaxel concentrations of ≤40 nM, nearly all mitotic spindles were bipolar, and cells progressed through mitosis. Higher paclitaxel concentrations resulted in a significant fraction of monoastral, monopolar, and multipolar spindles. We therefore elected to use 20, 30, and 40 nM paclitaxel treatments to study the progression of effects on spindle structure.
To evaluate spindle organization, we quantified the distribution of immunofluorescence labeling of MTs in different regions of the mitotic spindle for control cells and for cells treated with each of the specified paclitaxel concentrations. Fixed size boxes were drawn on the indicated regions of the spindle, and the average fluorescence intensities were collected for each region as detailed in Materials and Methods (Figure 1, A and B). Increasing concentrations of paclitaxel resulted in progressively larger changes in spindle morphology (Figure 1, C–F). The largest relative change for paclitaxel-treated cells was an increase in fluorescence intensity near the spindle poles. The average fluorescence intensity increased 20–52% relative to the intensity measured for control cells (Figure 1, G–I; Supplemental Figure 1, A–C). Fluorescence intensity measured for the astral-cortical MT population also increased progressively from 18% more than controls in 20 nM paclitaxel to up to 43% more than controls in 40 nM paclitaxel (Figure 1, G–I; Supplemental Figure 1, A–C). The inner spindle regions distal to the poles showed decreased intensity (13–15%) relative to controls in part due to a loss of fluorescence intensity measured for K-fibers. The average K-fiber intensity in cells treated with 20–40 nM paclitaxel was 19–37% less than control cells, respectively (Figure 1, G–I, and Supplemental Figure 1). This decrease in K-fiber intensity near kinetochores did not correlate with a reduction in interkinetochore distance (Supplemental Figure 1D). Additionally, there was a gradual reduction in spindle length that ranged from 3 to 13% as paclitaxel concentrations were increased (Supplemental Figure 1D).
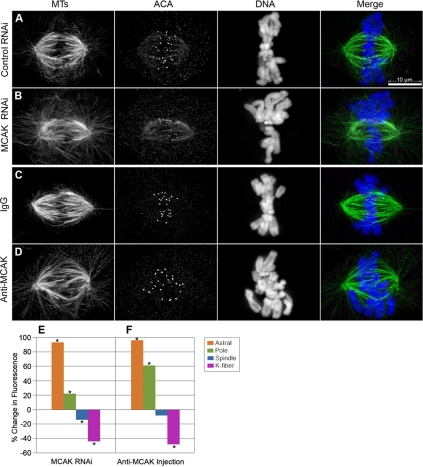
To ask whether perturbation of MCAK caused similar disruptions in spindle organization as that caused by treatment with low levels of paclitaxel, we inhibited MCAK function in PtK2 cells by using two separate methods: by siRNA-mediated knockdown of transcripts or by microinjection of anti-MCAK inhibitory antibodies. Cells were treated with the MCAK inhibitory reagent or with a mock (control) treatment, chemically fixed, and processed for tubulin immunofluorescence labeling in exactly the same manner as for paclitaxel-treated cells. MCAK perturbation by both RNAi knockdown and by antibody inhibition did not interfere with the completion of mitosis; however, it led to abnormal MT polymer distribution in mitotic cells (Figure 2, A–D). The MCAK-perturbed cells showed a large increase in the average fluorescence intensity for astral-cortical MTs compared with controls. For both the RNAi and the microinjection experiments, the astral-cortical regions were increased 90% compared with corresponding control cells (Figure 2, E and F; Supplemental Figure 2, A and B). The intensity of MTs at the pole region was also greater, 22 and 61% greater than controls in RNAi and microinjection experiments, respectively. The inner spindle region showed a small decrease in fluorescence intensity, 14% for RNAi and 8% for antibody injection, with the antibody injection not yielding a statistically significant change from controls (Figure 2, E and F, and Supplemental Figure 2). Measurements of K-fiber fluorescence showed substantially reduced intensity, 44 and 48% less than controls for RNAi and antibody injection, respectively, suggesting that loss of MCAK leads to a decrease in the amount of MT polymer per K-fiber. Unlike in paclitaxel-treated cells, with MCAK inhibition the decreased fluorescence intensity of K-fibers correlated with a decrease in the interkinetochore distance. There was a 15% decrease in interkinetochore distance upon MCAK knockdown and 28% decrease with antibody injection (Supplemental Figure 2, C and D). There was also a decrease in spindle length in both treatments; however, it was only statistically significant in RNAi experiments (Supplemental Figure 2, E and F).
Figure 2.
Altered MCAK levels disrupt MT polymer distribution in the bipolar spindle differently than paclitaxel addition. PtK2 cells were transfected with either control nonspecific (A) or MCAK specific siRNAs (B) and then fixed and stained for MTs (green), the kinetochore marker ACA (magenta), and DNA (blue). Alternatively, cells were injected with either control IgG (C), or inhibitory antibodies against MCAK (D), and then processed for immunofluorescence. Average fluorescence values were collected at the specified regions from the deconvolved 3-D reconstructed images and plotted for RNAi (E) and microinjection experiments (F) as a percentage of change in comparison with their corresponding control cells. Asterisks at each bar reflect a statistically significant change (p < 0.05). Bar, 10 μm.
MCAK Affects MT Polymer Distribution in a Monoastral Spindle
Our data indicate that MCAK perturbation results in a larger percentage of increase in fluorescence for the astral-cortical MTs than we would have predicted from observations of paclitaxel-treated cells (compare Figure 1, G–I with Figure 2, E and F). Because the total fluorescence signal from the astral-cortical MTs is small, and therefore less reliable for quantitative comparisons than the other spindle regions observed, we elected to compare astral MTs in a system enriched for astral-cortical polymers. We used the drug monastrol to inhibit the function of the kinesin-5 Eg5, which is required for spindle bipolarization (Mayer et al., 1999). Cells treated with paclitaxel and then monastrol were chemically fixed and processed for tubulin immunofluorescence. Treated cells had monoastral spindles that became progressively smaller as the concentration of paclitaxel was increased from 20 to 40 nM (Figure 3, A–D). To quantify these effects, we measured the area occupied by the monoasters, the total fluorescence within that area, the average fluorescence intensity, the length of the astral MTs, and the length of the K-fiber MTs.
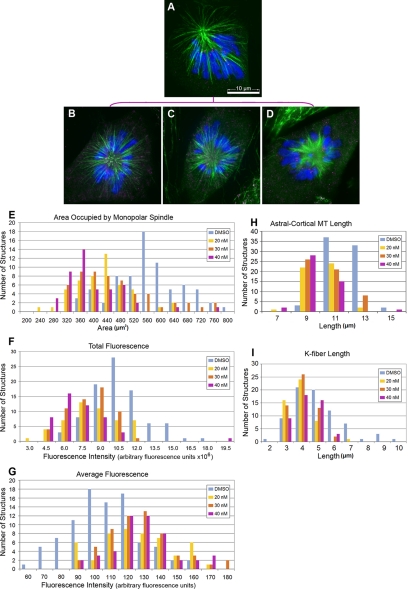
Figure 3.
Paclitaxel treatment of monoastral spindles decreases aster size. PtK2 cells were incubated in media containing 0.1% DMSO (A), 20 nM (B), 30 nM (C), or 40 nM (D) paclitaxel, and 24 h later incubated for 2–3 h in 100 μM monastrol. Cells were then fixed and stained for MTs (green), the kinetochore marker ACA (magenta), and DNA (blue). Measurements of area occupied by the monoastral spindle (E), total fluorescence (F), average fluorescence (G), the distance from the pole to the tip of astral-cortical MTs (H), and the distance from the pole to the kinetochores (I) were measured from 3-D reconstructed images and plotted as histograms for control DMSO (blue), 20 nM (yellow), 30 nM (orange), or 40 nM (magenta) paclitaxel. Each drug treatment resulted in a change in all measurements that were statistically different from the DMSO control with p < 0.001. Bar, 10 μm.
Paclitaxel significantly decreased the overall area occupied by the structures (Figure 3E), which correlated with a decrease in the total fluorescence of that structure (Figure 3F). This was not surprising because paclitaxel treatments resulted in a reduction in the size of the monoasters, thus reducing the number of pixels containing fluorescence signal. In contrast to the total fluorescence measurements, the average fluorescence gradually increased for each of the paclitaxel treatments, suggesting that there is a redistribution of the MT polymer with increasing paclitaxel concentration. To determine whether the length of MTs was consistent with these results, we measured the mean lengths of all MTs at all three drug concentrations, which showed a significant decrease consistent with the area and total fluorescence measurements (Figure 3, H and I).
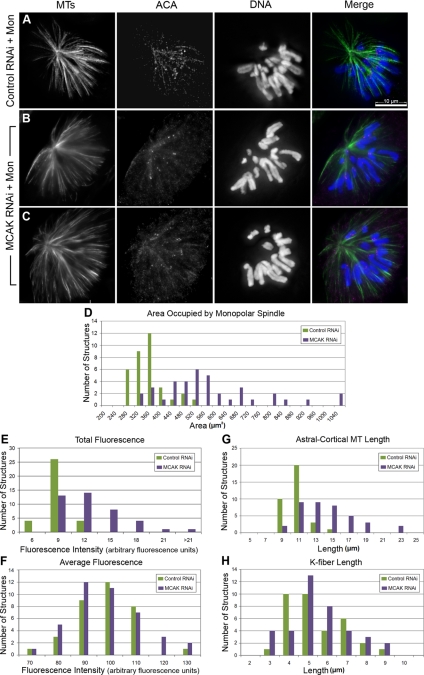
We next asked what happened to monoastral spindle organization in cells in which MCAK was knocked down by RNAi. Monastrol treatment after MCAK RNAi gave rise to a mixture of monoastral spindle morphologies in which some cells had spindle morphologies that were indistinguishable from control RNAi monastrol-treated cells, and other cells had asters with very long MTs that extended past chromosomes (Figure 4, A–C). To quantify these defects we measured the area, MT fluorescence intensity, and the length of MTs in a random selection of the mixture of morphological structures as we did for paclitaxel-treated cells. The area occupied by the monoastral structure in MCAK RNAi cells was significantly larger (65%) than in control cells (Figure 4D) as was the total fluorescence intensity (Figure 4E). Notably, the peak of area for the MCAK RNAi cells was broad and had a non-Gaussian distribution with a skew toward larger areas, supporting our qualitative assessment that the monoastral spindles after MCAK RNAi had a mixture of morphologies. However, measurements of average fluorescence showed no difference (Figure 4F), suggesting that MCAK knockdown does not affect the density of MT polymer in average per area. The mean pole to tip distance from astral MTs was longer (37%) than in controls (Figure 4G) and also had a broad distribution of lengths. However, K-fiber length remained unchanged from controls in the MCAK-depleted cells (Figure 4H). We infer that the longer MTs observed in MCAK RNAi-treated cells after monastrol treatment are astral-cortical MTs, consistent with our earlier observations from bipolar spindles.
Figure 4.
RNAi of MCAK in a monoastral spindle gives rise to a mixed population of spindle morphologies. PtK2 cells were transfected with either control nonspecific siRNAs (A) or MCAK specific-siRNAs (B and C), and 72 h posttransfection, cells were incubated for 2–3 h in 100 μM monastrol. Cells were then fixed and stained for MTs (green), the kinetochore marker ACA (magenta), and DNA (blue). Measurements of area occupied by the monoastral spindle (D), total fluorescence (E), average fluorescence (F), the distance from the pole to the tip of astral-cortical MTs (G), and the distance from the pole to the kinetochores (H) were measured from z-series 3-D reconstructed images of 34 control and 38 MCAK RNAi monoastral spindles and plotted as histograms for control RNAi (green) and MCAK RNAi (blue). The increase in area and astral MT length measurements were statistically significant each showing p < 0.001; however, there was no significant change in the K-fiber length (p = 0.6). Bar, 10 μm.
MCAK and Paclitaxel Have Opposite Effects on Spindle MT Dynamics
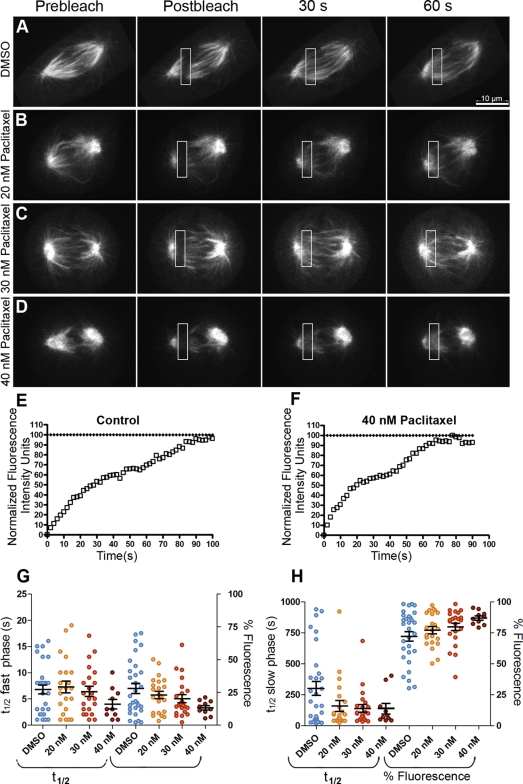
Our data support the idea that MCAK perturbation and paclitaxel addition give rise to distinct organizations of the mitotic spindle, suggesting that they do not act equivalently on all populations of spindle MTs. To ask whether these changes in spindle organization correlated with changes in spindle MT dynamics, we used FRAP to measure changes in MT dynamics in the spindle. PtK2 cells stably expressing GFP-α-tubulin (PtK-T) were mock treated or treated with low concentrations of paclitaxel and allowed to progress into late prometaphase or metaphase. A pattern generator and laser were used to rapidly photobleach a rectangular region within the mitotic spindle, whereas time-lapsed images were collected at 2-s intervals (Figure 5, A–D, and Supplemental Videos 1–4). FRAP was measured from these experiments and fit to models for exponential recovery (see Materials and Methods). In all cases, we determined that our fluorescence recovery measurements were better fit by a double exponential function than by a single exponential, indicating that minimally there is a biphasic recovery (Figure 5, E and F). For control cells, we calculated a fast phase recovery of t1/2 = 7 s and a slow phase recovery of t1/2 = 307 s (Table 1). At 20 nM paclitaxel, the fast phase was not significantly different from controls (p = 0.72). However, increasing drug concentrations resulted in a corresponding decrease in the t1/2 values for the fast phase of 14 and 43% at 30 and 40 nM paclitaxel, respectively, which was not statistically significant at 30 nM paclitaxel (p = 0.77) but was at 40 nM paclitaxel (p = 0.04; Figure 5G and Table 1). In contrast to the fast recovery phase, paclitaxel significantly altered the slow phase recovery at all three concentrations. At 20 nM paclitaxel, there was a 48% decrease in the t1/2 value and at 30 and 40 nM paclitaxel, a 55% decrease was observed (p = 0.05 for 20 nM, 0.02 for 30 nM, and 0.03 for 40 nM; Figure 5H; Table 1). There was a slight shift in the percentage of fluorescence from the fast phase to the slow phase as the paclitaxel concentration was increased (Figure 5, G and H). In addition, at 40 nM paclitaxel there was a 38% reduction in the total percentage of fluorescence recovery, suggesting the existence of a hyperstabilized population of MTs (Supplemental Figure 3A).
Figure 5.
Paclitaxel addition increases MT turnover. PtK-T-cells were incubated in media containing 0.1% DMSO (A), 20 nM (B), 30 nM (C), or 40 nM (D) paclitaxel for 24 h. Late prometaphase cells were photobleached by specifying a 3- × 10-μm box in the spindle region placed between the spindle equator and the pole, and then the spindle was imaged with an average of 300-ms exposures at 2-s intervals for ≥90 s. The data fit a biphasic recovery curve with a fast and a slow t1/2. An example of a fluorescence recovery curve for a single FRAP experiment for a control cell (E) or 40 nM placitaxel-treated cell (F) is shown. The t1/2 values and percent fluorescence in the fast phase (G) or slow phase (H) are summarized as dot plots with the mean and SEM indicated by the large and small horizontal bar(s), respectively. Note that there are only statistically significant differences in percent fluorescence for the fast and the slow phases at 40 nM: 20 nM (p = 0.51), 30 nM (p = 0.24), and 40 nM (p = 0.01). Bar, 10 μm.
Table 1.
Summary of MT turnover measurements
| t1/2 Fast phase (s) | t1/2 Slow phase (s) | R2 | n | |
|---|---|---|---|---|
| DMSO | 7 ± 3 | 307 ± 18 | 0.99 | 26 |
| 20 nM paclitaxel | 7 ± 3 | 159 ± 11 | 0.99 | 22 |
| 30 nM paclitaxel | 6 ± 3 | 139 ± 17 | 0.99 | 22 |
| 40 nM paclitaxel | 4 ± 2 | 137 ± 12 | 0.97 | 10 |
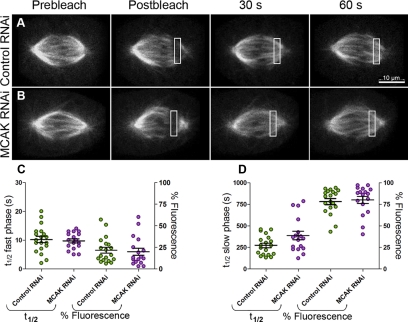
| Control RNAi | 10 ± 1 | 274 ± 26 | 0.97 | 19 |
| MCAK RNAi | 10 ± 0.7 | 387 ± 49 | 0.97 | 17 |
The decrease in t1/2 values in the presence of paclitaxel could suggest that MTs turn over more rapidly in the presence of paclitaxel, which is surprising given that paclitaxel normally stabilizes MTs (Schiff and Horwitz, 1981; Jordan et al., 1993). Increasing the paclitaxel concentration to 10 μM for the FRAP experiments eliminated observable fluorescence recovery (our unpublished data), indicating that higher doses of the drug stabilize MTs as predicted.
To address whether MCAK perturbation caused a similar change in MT dynamics as did paclitaxel treatment, MCAK was knocked down in PtK-T-cells by using RNAi. FRAP experiments were performed on cells from the RNAi-treated population using the same protocol for late prometaphase and metaphase cells as described above (Figure 6, A and B, and Supplemental Videos 5 and 6). Although MCAK knockdown resulted in no change from controls for the fast recovery phase (p = 0.65; Figure 6C; Table 1), there was a measurable increase of 41% in the t1/2 value measured for the slow phase compared with mock-transfected controls (p = 0.05; Figure 6D; Table 1). The distribution of fluorescence attributable to each phase of the biphasic recovery (Figure 6, C and D) remained approximately constant between control and RNAi-treated cells, suggesting that the change in slow phase recovery is not due to a shift in polymer from the fast phase to the slow phase. The percentage of fluorescence recovery, when total spindle bleaching is accounted for, was nearly 100% for our experiments, which discounts the hypothesis that a small population of hyperstabilized MTs might be contributing to the slow phase recovery (Supplemental Figure 3B).
Figure 6.
MCAK loss alters MT turnover of K-fibers. PtK-T-cells were transfected with control nonspecific siRNAs (A) or MCAK-specific siRNAs (B). For each treatment, late prometaphase-metaphase cells were photobleached by specifying a 3- × 10-μm box in the spindle region placed between the spindle equator and the pole and then the spindle was imaged with an average of 300-ms exposures at 2-s intervals for ≥90 s. The t1/2 values and percentage of fluorescence in the fast phase (C), or slow phase (D) are summarized as dot plots with the mean and SE indicated by the large and small horizontal bar(s), respectively. Note that there is no statistically significant difference in percentage of fluorescence in either the fast or the slow phase (p = 0.7). Bar, 10 μm.
To test the hypothesis that the first phase of fluorescence recovery might represent diffusion of unincorporated GFP-tubulin subunits into the bleach zone, we repeated the FRAP experiments on cells treated with nocodazole to depolymerize the MTs (2 μM for 3 h). A t1/2 value of 1.3 s from a single exponential fit to the recovery data was determined for the free GFP-tubulin subunits, a five- to sevenfold faster t1/2 than the estimated first phase of recovery measured for intact spindles (data not shown).
DISCUSSION
We set out to test the hypothesis that the dynamics of spindle MTs are differentially controlled by MT regulatory proteins. We found that general suppression of MT dynamics by incubation with low levels of paclitaxel produced distinct changes in spindle MT organization and dynamics that were not mimicked by inhibition of the kinesin-13 MCAK. Paclitaxel treatment caused a disruption in spindle MT organization marked by an increase in MTs near the pole and a reduction in the fluorescence intensity of the K-fibers. In contrast, MCAK inhibition caused a dramatic reorganization of spindle MTs with a significant increase in astral MTs and a more modest reduction (41%) in the fluorescence intensity of the K-fibers. Our data support the model that MCAK perturbs spindle organization by acting preferentially on astral MTs and that it contributes to error correction by controlling MT dynamics in the K-fiber.
Previous studies measured the dynamics of spindle MTs by using either FRAP or photoactivation (Salmon et al., 1984; Saxton et al., 1984; Mitchison, 1989; Zhai et al., 1995; Waterman-Storer et al., 1998; Cimini et al., 2006). In most studies using FRAP, the data were fit to a single exponential that represented the turnover of the highly dynamic spindle MTs. Because of the increased sensitivity and speed of new imaging systems, we achieved sufficiently high temporal resolution that our data best fit a biphasic recovery model, allowing us to gain information about the t1/2 values for both fast and slow phases of recovery in a single experiment. Our measured t1/2 values for the slow phase of the recovery for prometaphase cells are similar to those reported previously for the K-fibers (Cimini et al., 2006). Our values for the fast phase, however, are faster than those reported by others (Saxton et al., 1984; Zhai et al., 1995; Cimini et al., 2006). One possibility is that our fast phase actually represents the diffusion of unincorporated GFP–tubulin subunits into the bleach zone. However, when cells were treated with nocodazole we measured a t1/2 of 1.3 s from a single exponential fit, which is approximately a sevenfold faster t1/2 than the estimated fast phase of recovery measured for intact spindles. We think the more likely explanation is that the differences come from a combination of the time interval for imaging (our data are collected at 2-s intervals compared with 20–60-s intervals of others), the differences in techniques (in several cases comparing photobleaching vs. photoactivation), as well as the different imaging system used. Based on our analysis, we expect that the fast t1/2 represents the recovery of the nonkt-spindle MTs and the slow t1/2 represents the recovery of the K-fiber MTs. As such, we discuss the rest of our results based on this biological interpretation.
It should be noted that although the measurements of the t1/2 values are similar to those reported previously, there is a dramatic difference in the distribution in MT fluorescence between the two phases. Previous work estimated that ∼70% of the MTs were in nonkt-spindle MTs and 30% were in K-fibers (Mitchison and Salmon, 1992; Cimini et al., 2006). However, in our study we observed that only 20% of the recovery comes from the nonkt-spindle MTs and 80% is from the K-fibers. This disparity likely represents differences in the methodology used. Many of the early FRAP studies used wide-field microscopy in which the information was collected from a larger z-area, whereas we used a spinning disk confocal microscope in which we collected information from a thin optical plane. Because we focused on the central region of the spindle, which is rich in K-fibers, we are likely sampling more data from the K-fibers. This may have contributed to our ability to extract the slower t1/2 values of the K-fiber MTs from photobleaching experiments. Because our t1/2 measurements are similar to those from previous studies, we do not believe that the differences in the percentage of data from either phase alter the interpretation of our findings.
What Is the Relationship between Spindle MT Dynamics and Organization?
Our data clearly establish that differential perturbation of MT dynamics gives rise to different spindle morphologies and different effects on dynamics. In previous studies, treatment of cells with low levels of paclitaxel suppressed the plus end dynamics of individual MTs in interphase cells (Yvon et al., 1999). The defects in spindle organization we find are consistent with this effect because we see a significant increase in short MTs near poles, which is likely due to stabilization of the plus ends of spindle MTs that grow toward the spindle equator. This type of biased MT growth toward the chromosomes has been demonstrated in studies tracking GFP-EB1 in the spindle (Piehl and Cassimeris, 2003) and may be due to stabilization of MTs near chromosomes based on the Ran-GTP gradient (Wollman et al., 2005; Bastiaens et al., 2006; Kalab et al., 2006; O'Connell and Khodjakov, 2007; Kalab and Heald, 2008). Alternatively, the increase in MTs near the poles could be a result of an increase in nucleation of MTs from the centrosome as paclitaxel has been shown to affect the MT nucleation rate (Schiff and Horwitz, 1981).
We were surprised to find that treatment of cells with paclitaxel resulted in a faster t1/2 than in control cells, suggesting that the MTs turned over more rapidly with paclitaxel treatment. It is important to remember that the fluorescence recovery in a FRAP experiment is a complicated measurement that takes into account not only the change in dynamics of the MTs in the bleach zone but also reflects fluorescence dissipation due to MT flux, as well as the contribution of any new MT ends that enter the bleach zone whether they be elongating MT plus ends or newly nucleated MTs at the centrosomes. Given that our photobleaching marks are placed between the pole and the chromosomes on a half spindle, we think the most likely explanation of the faster t1/2 values is that they arise from new MT growth into the bleach zone, consistent with the morphological changes in the spindle, which showed an increase in MT fluorescence intensity near the poles.
The differences we find in both spindle organization and regulation of MT dynamics between paclitaxel addition and MCAK inhibition strongly support the idea that MCAK does not suppress the dynamics of all MT ends equivalently. The large increase in astral MTs seen after knockdown of MCAK is consistent with earlier studies showing that MCAK and its orthologues have a dramatic effect on astral MT organization (Walczak et al., 1996; Grill et al., 2001; Maney et al., 2001; Kline-Smith and Walczak, 2002; Goshima and Vale, 2003; Rogers et al., 2004; Srayko et al., 2005). Unfortunately, we were unable to obtain FRAP recovery data on the astral MTs because of the difficulty in obtaining sufficient fluorescence signal to get reliable FRAP curves. It is possible that the increase in astral MTs is a direct result of the diminished MTs in the K-fibers after MCAK perturbation. We think this is unlikely because our estimation is that if half of the cellular tubulin present in K-fibers were released into the cytoplasm, it would not make a significant impact on the amount of free tubulin in the cytoplasm (∼10 μM in a mammalian cell) that could then preferentially polymerize onto the astral MTs. In addition, low levels of paclitaxel also caused a reduction in K-fiber fluorescence without causing such a substantial increase in astral MTs seen after MCAK inhibition. We favor the idea that the large increase in astral MTs is due to MCAK acting to preferentially depolymerize astral MTs, an idea that will need to be tested directly.
Control of Kinetochore MT Dynamics
We found that addition of low levels of paclitaxel caused a reduction in the fluorescence of K-fibers with no change in the interkinetochore distance as well as a faster t1/2. These results suggest that the dynamics of the MTs in the K-fibers are actually faster in the presence of paclitaxel. One possibility is that paclitaxel addition directly increases the dynamicity of kinetochore-MTs, which is not unprecedented as studies from interphase cells showed that treatment with low levels of paclitaxel (5 nM) results in an increase in MT plus end dynamics (Pasquier et al., 2005). The change in dynamics may alter the kinetochore–MT interaction in a way that stimulates detachment, resulting in the decreased fluorescence in the K-fiber. However we think this possibility is unlikely because we use paclitaxel at significantly higher concentrations (20, 30, and 40 nM) than those used by Pasquier et al. (2005). In addition, we would expect that stimulating MT detachments at the kinetochores would reduce tension across the sister kinetochores; however, there was no corresponding change in the interkinetochore distance under our tested conditions. Instead, we favor the idea that the recovery seems faster because of new MT growth, but the questions then become: where does that new MT growth come from and, what makes it distinct from the dynamics of MTs in the fast phase of the recovery? If the new MT growth comes from the centrosomes, the dynamics must be significantly different from those of the fast phase. Maybe the MTs that grow with a bias toward the chromosomes and become part of the K-fiber have intrinsically different MT dynamics (Carazo-Salas et al., 2001; Karsenti and Vernos, 2001; Caudron et al., 2005). Alternatively, there may be a population of MTs with a different distribution of catastrophe frequencies that form the K-fiber, and this distribution gets altered because of the suppression of dynamics caused by paclitaxel addition. A third possibility is that there are new MTs that grow from the chromosomes and become incorporated into the K-fiber bundles (Khodjakov et al., 2003). The MTs that grow from the chromosomes may quickly become coated with HURP or with TPX2, two specialized MT-associated proteins that have been implicated in chromosome-mediated spindle assembly (Gruss et al., 2002; Koffa et al., 2006; Sillje et al., 2006; Tulu et al., 2006; Wong and Fang, 2006; Casanova et al., 2008), such that their overall dynamics are more consistent with the stabilized MTs present in a K-fiber. This possibility is consistent with our observation in Figure 5, G and H, where as the concentration of paclitaxel is increased, the percentage of MTs in the slow phase is increased. It is possible that not all of the MT polymer that contributes to the slow phase is part of the K-fiber as our fixed analysis of individual K-fibers showed that there is a decrease in the intensity of K-fibers (Figure 1, G–I). Perhaps paclitaxel treatments perturb spindle MT dynamics and nucleation properties so that the short newly nucleated MTs form a more stable MT subset that exhibit dynamic properties more closely related to those of K-fibers and therefore contribute to the increase in percent fluorescence within the slow phase. The ability to distinguish between these models will require the ability to follow the dynamics of individual MTs within the spindle, a feat that has not yet been accomplished.
Unlike paclitaxel addition, MCAK inhibition causes a more dramatic reduction in the fluorescence intensity of the K-fiber that is correlated with a reduction in the interkinetochore distance and a slower t1/2. MCAK inhibition results in merotelic attachments of kinetochores (Kline-Smith et al., 2004), which may reduce the number of MTs per K-fiber either directly by error correction mechanisms contributing to the release of kinetochore-MTs, or by the aberrant binding sites caused by the merotelic attachments (Cimini et al., 2003; Salmon et al., 2005), which may limit the number of end-on binding sites for MTs. The reduction in interkinetochore distance is consistent with our previous findings (Kline-Smith et al., 2004), but different from that reported by others (Wordeman et al., 2007). It is most likely that their live imaging examined only a subset of kinetochores that were selected based on chromosome behavior, whereas we have sampled a broader distribution of chromosome attachments as well as included bioriented chromosomes in late prometaphase cells.
It is likely that MCAK is needed to help turn over the MTs in the K-fiber, thus loss of MCAK results in a slower t1/2 for the K-fiber. Our data are not consistent with those of Ganem et al. (2005), who found no change in the dynamics of the K-fiber in human U2OS cells. This is likely due to the increased temporal resolution in our studies as well as to the spatial resolution afforded by the spindle morphology in PtK2 cells. Our data are consistent with the recent findings of Wordeman et al. (2007), who propose an attractive idea that the role of MCAK is to loosen up MT ends to facilitate directional coordination of chromosome motility and MT release. Another interesting observation is that although it has been clearly established that MCAK is a substrate of Aurora B kinase (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Sampath et al., 2004; Zhang et al., 2007), the magnitude of the effects of MCAK inhibition versus Aurora B inhibition on the t1/2 value of the K-fiber strongly supports the idea that Aurora B causes release of aberrant MT attachments primarily through a mechanism that does not rely solely on MCAK (Cimini et al., 2006; DeLuca et al., 2006; Wordeman et al., 2007).
Overall our work supports a model where MCAK differentially affects the MT subclasses which coexist during mitosis, with specific effects on K-fibers and astral MTs. This is consistent with the complex localization of MCAK in the spindle and the highly complicated phosphorregulatory schemes that regulate its localization and activity at centromeres and at spindle poles (Andrews et al., 2004; Gorbsky, 2004; Lan et al., 2004; Ohi et al., 2004; Knowlton et al., 2006; Zhang et al., 2007, 2008). This suggests that MCAK is responsible for preferentially altering the dynamics individual MT subclasses and raises the important question of how other MT dynamics effectors contribute to the control of spindle MT subclasses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alexey Khodjakov for the PtK-T-cells. We are grateful to Tim Mitchison, Pat Wadsworth, and many members of the Walczak laboratory for thoughtful ideas and suggestions on this work. We are especially grateful to Bill Saxton for assistance in assembling the imaging setup as well as generous guidance on these studies and sharing of equipment, to Susan Strome for suggestions on examining monoastral spindles, and to Jason Cooper for measurements of fluorescence intensity of deconvolved versus nondeconvolved images. Jane Stout provided critical comments on the manuscript. This work was supported by National Institutes of Health grant GM-59618 (to C.E.W.) and an American Heart Association predoctoral fellowship (to R.S.R.). The imaging studies were performed in part at the Indiana University Light Microscopy Imaging Facility.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0985) on January 21, 2009.
REFERENCES
- Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J. R. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Bastiaens P., Caudron M., Niethammer P., Karsenti E. Gradients in the self-organization of the mitotic spindle. Trends Cell Biol. 2006;16:125–134. doi: 10.1016/j.tcb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Belmont L. D., Hyman A. A., Sawin K. E., Mitchison T. J. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Buster D. W., Zhang D., Sharp D. J. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol. Biol. Cell. 2007;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron L. A., Yang G., Cimini D., Canman J. C., Kisurina-Evgenieva O., Khodjakov A., Danuser G., Salmon E. D. Kinesin 5-independent poleward flux of kinetochore microtubules in PtK1 cells. J. Cell Biol. 2006;173:173–179. doi: 10.1083/jcb.200601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Gruss O. J., Mattaj I. W., Karsenti E. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- Casanova C. M., Rybina S., Yokoyama H., Karsenti E., Mattaj I. W. HURP is required for chromatin-induced microtubule assembly independently of TPX2. Mol. Biol. Cell. 2008;19:4900–4908. doi: 10.1091/mbc.E08-06-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization and bipolar spindle assembly. Mol. Biol. Cell. 2004;15:1580–1590. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Cimini D., Moree B., Canman J. C., Salmon E. D. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- Cimini D., Wan X., Hirel C. B., Salmon E. D. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Compton D. A. Spindle assembly in animal cells. Annu. Rev. Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., Salmon E. D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T. J., Walczak C. E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Gaetz J., Kapoor T. M. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Upton K., Compton D. A. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Gliksman N. R., Parsons S. F., Salmon E. D. Okadaic acid induces interphase to mitotic-like microtubule dynamic instability by inactivating rescue. J. Cell Biol. 1992;119:1271–1276. doi: 10.1083/jcb.119.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G. J. Mitosis: MCAK under the aura of Aurora B. Curr. Biol. 2004;14:R346–R348. doi: 10.1016/j.cub.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Gorbsky G. J., Sammak P. J., Borisy G. G. Microtubule dynamics and chromosome motion visualized in living anaphase cells. J. Cell Biol. 1988;106:1185–1192. doi: 10.1083/jcb.106.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S. W., Gonczy P., Stelzer E. H., Hyman A. A. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Gruss O. J., Wittmann M., Yokoyama H., Pepperkok R., Kufer T., Sillje H., Karsenti E., Mattaj I. W., Vernos I. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- Gupta M. L., Jr, Carvalho P., Roof D. M., Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hedrick D. G., Stout J. R., Walczak C. E. Effects of anti-microtubule agents on microtubule organization in cells lacking the kinesin-13 MCAK. Cell Cycle. 2008;7:2146–2156. doi: 10.4161/cc.7.14.6239. [DOI] [PubMed] [Google Scholar]
- Helenius J., Brouhard G., Kalaidzidis Y., Diez S., Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- Hoffman D. B., Pearson C. G., Yen T. J., Howell B. J., Salmon E. D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J., Hyman A. A. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hunter A. W., Caplow M., Coy D. L., Hancock W. O., Diez S., Wordeman L., Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. A., Toso R. J., Thrower D., Wilson L. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc. Natl. Acad. Sci. USA. 1993;90:9552–9556. doi: 10.1073/pnas.90.20.9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J. Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pralle A., Isacoff E. Y., Heald R., Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Copenagle L., Gordon M. B., Compton D. A., Kapoor T. M. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 2003;160:671–683. doi: 10.1083/jcb.200208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Khodjakov A., Hergert P., Walczak C. E. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol. Biol. Cell. 2002;13:2718–2731. doi: 10.1091/mbc.E01-12-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Knowlton A. L., Lan W., Stukenberg P. T. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- Koffa M. D., Casanova C. M., Santarella R., Kocher T., Wilm M., Mattaj I. W. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Kwok B. H., Kapoor T. M. Microtubule flux: drivers wanted. Curr. Opin. Cell Biol. 2007;19:36–42. doi: 10.1016/j.ceb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E., Stukenberg P. T. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Maddox P. S., Bloom K. S., Salmon E. D. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Wagenbach M., Wordeman L. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J. Biol. Chem. 2001;276:34753–34758. doi: 10.1074/jbc.M106626200. [DOI] [PubMed] [Google Scholar]
- Manning A. L., Ganem N. J., Bakhoum S. F., Wagenbach M., Wordeman L., Compton D. A. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Mayr M. I., Hummer S., Bormann J., Gruner T., Adio S., Woehlke G., Mayer T. U. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- McDonald K. L., O'Toole E. T., Mastronarde D. N., McIntosh J. R. Kinetochore microtubules in PTK cells. J. Cell Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., West R. R. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- Mitchison T. Poleward microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Salmon E. D. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 1992;119:569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores C. A., Milligan R. A. Lucky 13-microtubule depolymerisation by kinesin-13 motors. J. Cell Sci. 2006;119:3905–3913. doi: 10.1242/jcs.03224. [DOI] [PubMed] [Google Scholar]
- O'Connell C. B., Khodjakov A. L. Cooperative mechanisms of mitotic spindle formation. J. Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- Ohi R., Burbank K., Liu Q., Mitchison T. J. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr. Biol. 2007;17:953–959. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T. J. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier E., Honore S., Pourroy B., Jordan M. A., Lehmann M., Briand C., Braguer D. Antiangiogenic concentrations of paclitaxel induce an increase in microtubule dynamics in endothelial cells but not in cancer cells. Cancer Res. 2005;65:2433–2440. doi: 10.1158/0008-5472.CAN-04-2624. [DOI] [PubMed] [Google Scholar]
- Piehl M., Cassimeris L. Organization and dynamics of growing microtubule plus ends during early mitosis. Mol. Biol. Cell. 2003;14:916–925. doi: 10.1091/mbc.E02-09-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C. L. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., Vale R. D., Scholey J. M., Sharp D. J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J. Spindle assembly: asters part their separate ways. Nat. Cell Biol. 2005;7:219–222. doi: 10.1038/ncb0305-219. [DOI] [PubMed] [Google Scholar]
- Rusan N. M., Fagerstrom C. J., Yvon A. M., Wadsworth P. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell. 2001;12:971–980. doi: 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E. D., Cimini D., Cameron L. A., DeLuca J. G. Merotelic kinetochores in mammalian tissue cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:553–568. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E. D., McKeel M., Hays T. Rapid rate of tubulin dissociation from microtubules in the mitotic spindle in vivo measured by blocking polymerization with colchicine. J. Cell Biol. 1984;99:1066–1075. doi: 10.1083/jcb.99.3.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath S. C., Ohi R., Leismann O., Salic A., Pozniakovski A., Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Saxton W. M., Stemple D. L., Leslie R. J., Salmon E. D., Zavortink M., McIntosh J. R. Tubulin dynamics in cultured mammalian cells. J. Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff P. B., Horwitz S. B. Taxol assembles tubulin in the absence of exogenous guanosine 5′-triphosphate or microtubule-associated proteins. Biochemistry. 1981;20:3247–3252. doi: 10.1021/bi00514a041. [DOI] [PubMed] [Google Scholar]
- Scholey J. M., Brust-Mascher I., Mogilner A. Cell division. Nature. 2003;422:746–752. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- Sillje H. H., Nagel S., Korner R., Nigg E. A. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Srayko M., Kaya A., Stamford J., Hyman A. A. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Stout J. R., Rizk R. S., Kline S. L., Walczak C. E. Deciphering protein function during mitosis in PtK cells using RNAi. BMC Cell Biol. 2006;7:26. doi: 10.1186/1471-2121-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout J. R., Rizk R. S., Walczak C. E. Comparing protein function in tissue culture cells by antibody microinjection versus RNA-mediated interference. Methods Mol. Biol. 2009;518:77–97. doi: 10.1007/978-1-59745-202-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu U. S., Fagerstrom C., Ferenz N. P., Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A. A., Tanaka T. U., Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 2008;8:903–905. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Verde F., Dogterom M., Stelzer E., Karsenti E., Leibler S. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P., Salmon E. D. Microtubule dynamics in mitotic spindles of living cells. Ann. N Y Acad. Sci. 1986;466:580–592. doi: 10.1111/j.1749-6632.1986.tb38434.x. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Heald R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Mitchison T. J., Desai A. XKCM 1, a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C. M., Desai A., Bulinski J. C., Salmon E. D. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- Wittmann T., Hyman A., Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 2001;3:E28–E34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Wollman R., Cytrynbaum E. N., Jones J. T., Meyer T., Scholey J. M., Mogilner A. Efficient chromosome capture requires a bias in the ‘search and capture’ process during mitotic-spindle assembly. Curr. Biol. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Wong J., Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 2006;173:879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L. Microtubule-depolymerizing kinesins. Curr. Opin. Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wordeman L., Wagenbach M., von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon A. M., Wadsworth P., Jordan M. A. Taxol suppresses dynamics of individual microtubules in living human tumor cells. Mol. Biol. Cell. 1999;10:947–959. doi: 10.1091/mbc.10.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Kronebusch P. J., Borisy G. G. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Kronebusch P. J., Simon P. M., Borisy G. G. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J. Cell Biol. 1996;135:201–214. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ems-McClung S. C., Walczak C. E. Aurora A phosphorylates MCAK to control Ran-dependent spindle bipolarity. Mol. Biol. Cell. 2008;19:2752–2765. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lan W., Ems-McClung S. C., Stukenberg P. T., Walczak C. E. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol. Biol. Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J. B., Abraham R. T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.