Abstract
We used antibody microinjection and genetic manipulations to dissect the various roles of the homotetrameric kinesin-5, KLP61F, in astral, centrosome-controlled Drosophila embryo spindles and to test the hypothesis that it slides apart interpolar (ip) microtubules (MT), thereby controlling poleward flux and spindle length. In wild-type and Ncd null mutant embryos, anti-KLP61F dissociated the motor from spindles, producing a spatial gradient in the KLP61F content of different spindles, which was visible in KLP61F-GFP transgenic embryos. The resulting mitotic defects, supported by gene dosage experiments and time-lapse microscopy of living klp61f mutants, reveal that, after NEB, KLP61F drives persistent MT bundling and the outward sliding of antiparallel MTs, thereby contributing to several processes that all appear insensitive to cortical disruption. KLP61F activity contributes to the poleward flux of both ipMTs and kinetochore MTs and to the length of the metaphase spindle. KLP61F activity maintains the prometaphase spindle by antagonizing Ncd and another unknown force-generator and drives anaphase B, although the rate of spindle elongation is relatively insensitive to the motor's concentration. Finally, KLP61F activity contributes to normal chromosome congression, kinetochore spacing, and anaphase A rates. Thus, a KLP61F-driven sliding filament mechanism contributes to multiple aspects of mitosis in this system.
INTRODUCTION
Mitosis, the process by which identical copies of the replicated genome are distributed to the products of each cell division, involves a highly dynamic sequence of coordinated motility events, mediated by a bipolar protein machine, the mitotic spindle (Karsenti and Vernos, 2001; Mitchison and Salmon, 2001; Gadde and Heald, 2004; Wadsworth and Khodjakov, 2004; Mogilner et al., 2006; Brust-Mascher and Scholey, 2007; Walczak and Heald, 2008). These motility events are driven by molecular-scale forces generated by mitotic kinesins and dyneins, together with dynamic microtubules (MTs), whose activities are controlled by a network of regulatory proteins, e.g., mitotic kinases, phosphatases, and proteolytic enzymes (Sharp et al., 2000c; Bettencourt-Dias et al., 2004; Maiato and Sunkel, 2004; Rogers et al., 2005; Goshima et al., 2007). Among these mitotic proteins, the kinesin-5 motor is thought to play a key role (Cottingham et al., 1999; Valentine et al., 2006a; Civelekoglu-Scholey and Scholey, 2007).
Purified kinesin-5 is a slow, modestly processive, plus-end–directed bipolar homotetramer capable of cross-linking adjacent MTs and sliding apart antiparallel MTs in motility assays (Sawin et al., 1992; Cole et al., 1994; Kashina et al., 1996a; Kapitein et al., 2005; Tao et al., 2006; Valentine et al., 2006b; Krzysiak et al., 2008; Van den Wildenberg et al., 2008). In yeast cells the homotetrameric structure of kinesin-5 appears to be essential for mitosis (Hildebrandt et al., 2006), and in Drosophila embryos KLP61F displays dynamic properties consistent with an association with spindle MTs (Cheerambathur et al., 2008) and forms presumptive MT–MT cross-bridges (Sharp et al., 1999a). These results suggest that ensembles of multiple kinesin-5 motors could serve as dynamic cross-links that organize spindle MTs into bundles and drive a sliding filament mechanism that pushes apart antiparallel spindle MTs (Sharp et al., 1999a; Brust-Mascher et al., 2004), although alternative mechanisms of action for kinesin-5 motors have also been proposed (Kapoor and Mitchison, 2001; Tsai et al., 2006; Johansen and Johansen, 2007; Gardner et al., 2008).
The most obvious and frequently observed consequence of loss of activity of kinesin-5 motors, induced by loss-of-function mutation, antibody inhibition, small molecule inhibition, or RNA interference, is the formation of abnormal monoastral spindles (Enos and Morris, 1990; Saunders and Hoyt, 1992; Sawin et al., 1992; Heck et al., 1993; Saunders et al., 1997; Cottingham et al., 1999; Mayer et al., 1999; Sharp et al., 1999b; Sharp et al., 2000a; Goshima and Vale, 2003). This suggests that kinesin-5 may normally contribute to spindle bipolarity by sliding apart interpolar (ip) MTs to drive spindle pole separation during mitotic spindle assembly, elongation and function. (Saunders and Hoyt, 1992; Sawin et al., 1992; Heck et al., 1993; Saunders et al., 1997; Straight et al., 1998; Walczak et al., 1998; Sharp et al., 2000a; Brust-Mascher and Scholey, 2002; Brust-Mascher et al., 2004; Cheerambathur et al., 2007). It is clear that this activity is deployed differently in different systems, however. For example, kinesin-5 activity is required for initial spindle assembly in yeast but not in Drosophila embryos, whereas it is required for spindle maintenance in both systems (Saunders and Hoyt, 1992; Sharp et al., 1999b).
Inhibition of kinesin-5 in different systems has also resulted in defects in poleward flux (Miyamoto et al., 2004; Cameron et al., 2006) and spindle pole organization (Gaglio et al., 1996). However, in Drosophila S2 cells, perturbation of kinesin-5 levels did not interfere with metaphase spindle length (Goshima et al., 2005b). In Caenorhabditis elegans, kinesin-5 was also found to exert a braking effect on the spindle midzone that governs the rate of spindle elongation mediated by cortical motors (Saunders et al., 2007). It is unclear if these findings are relevant to all spindles and if they all reflect the antiparallel MT sliding activity of kinesin-5, or if kinesin-5 functions differently in different systems as implied by the proposal that kinesin-5 functions as a MT depolymerase rather than a sliding motor to control chromosome congression in Saccharomyces cerevisiae (Gardner et al., 2008). To distinguish these possibilities, therefore, systematic studies of kinesin-5 function throughout mitosis are needed, but this is difficult because the most frequently observed effect of perturbing kinesin-5 function, spindle collapse, obscures later effects in mitosis.
In Drosophila embryos, where the centrosomes dictate the assembly of remarkably dynamic, fast-acting spindles (Megraw et al., 2001; Cheerambathur et al., 2007), inhibiting the function of the kinesin-5, KLP61F produced collapsed monoastral spindles at a specific stage during prometaphase, and this was rescued by loss of function of the kinesin-14, Ncd (Sharp et al., 1999b, 2000a). Biochemical evidence supports the idea that the prometaphase spindle is maintained by a KLP61F-Ncd force balance (Tao et al., 2006), but it is unclear if the monoastral spindles form by the drawing together of focused poles due to Ncd-induced inward MT sliding or by the disorganization of spindle MTs combined with the fusion of microtubule organizing centers (Goshima and Vale, 2003). We previously proposed that the KLP61F-driven outward sliding of ipMTs drives pre-anaphase B poleward flux and anaphase B spindle elongation (Brust-Mascher and Scholey, 2002; Brust-Mascher et al., 2004). However, reductionist modeling (Brust-Mascher et al., 2004) and system level modeling (Wollman et al., 2008) surprisingly predicted that the rate of anaphase B pole–pole separation is insensitive to the KLP61F concentration and is controlled instead by the unloaded KLP61F MT sliding velocity together with a pole-associated MT depolymerase. Although experimental evidence supports a requirement for KLP61F activity in anaphase B (Sharp et al., 2000a), we had not tested the effect of changing its concentration on the rate of anaphase B or its contribution to poleward flux and metaphase spindle length. Furthermore, it is unclear if some of these activities also require the activity of the overlying cortex, e.g., via cortical motors pulling astral MTs outward.
In the current study we addressed some of these uncertainties by undertaking a systematic investigation of the spectrum of mitotic functions of kinesin-5, through antibody-induced dissociation of KLP61F from Drosophila syncytial blastoderm-stage embryo spindles, using an experimental system where the extent of dissociation can be directly observed. This was complemented by studies of the effects of varying the KLP61F gene dosage in embryos and by the investigation of mutant spindles after depletion of the maternal load of KLP61F. We find that kinesin-5 contributes to various aspects of mitosis in this system, some of which are novel and perhaps unexpected.
MATERIALS AND METHODS
Drosophila Stocks
Flies were maintained and embryos were collected as described (Sharp et al., 1999). Experiments were performed on embryos expressing green fluorescent protein (GFP)::tubulin or GFP::CNN (provided by Dr. Thomas Kaufman, Indiana University Bloomington) or GFP::CID, the Drosophila CENPA homolog (Henikoff et al., 2000), which is a stable and specific kinetochore marker (provided by Dr. Steven Henikoff, Fred Hutchinson Cancer Research), or KLP61F::GFP (Cheerambathur et al., 2008), as well as on Claret nondisjunctional (cand) and klp61f mutant embryos.
Generation of klp61f Mutant Fly Lines Expressing GFP-Tubulin
Standard genetic techniques were used to generate flies with GFP::tubulin on the second chromosome and these were then crossed into different alleles of klp61f mutants for genetic analyses. Briefly, we first crossed y[1] w[*]; P{w[+mC] = UASp-GFPS65C-alphaTub84B}14-6-II/P{w[+mC] = UASp-GFPS65C-alphaTub84B}14-6-II with yw;P{Act5C-Gal4}/CyO,y+ or w*; P{w[+mC] = matalpha4-GAL-VP16}V2H. We selected P{w[+mC] = UASp-GFPS65C-alphaTub84B}14-6-II/P{Act5C-Gal4}or P{w[+mC] = UASp-GFPS65C-alphaTub84B}14-6-II/P{w[+mC] = matalpha4-GAL-VP16}V2H females and crossed them with w; ln(2LR)Gla,wgGla-1Bc1/CyO males. We selected progeny in which recombination between the UAS-GFP-tubulin and the Gal4 driver had occurred by picking fluorescent larvae and used individual flies to make balanced stocks. All stocks were from the Bloomington Stock Center. To cross these balanced lines expressing GFP-tubulin under the Gal 4 driver with klp61f mutants (provided by Margarete Heck, University of Edinburgh; Andrea Pereira, University of Massachusetts; and Patricia Wilson, Georgia State University), we first crossed both lines with w; Sp/CyORoi; Ly/TM3Sbe flies (provided by Dr Jeanette Naetzle, University of California Davis), in which we had replaced the TM3 balancer with KrGFPTM3 or TM6 to distinguish homozygous from heterozygous embryos or larvae, respectively. The klp61f alleles used (7012 and 7415) are null or severe hypomorphic, late larval lethals, obtained by P-element insertion in the untranslated region of KLP61F (Heck et al., 1993).
Antibody Generation
KLP61F antibody was generated against amino acids 900-1066, which includes the BimC box containing a mutation in which the putative phosphorylation site, Threonine 933, was mutated to aspartic acid. To generate this mutant, two sets of primers were used in three sequential PCR steps: two anchor primers, flanking the region of interest and containing BamHI and EcoRI restriction sites, and two mutagenic primers containing the T933D mutation. To produce KLP61F antibody, PCR-amplified KLP61F (nt 2698-3201) containing the T933D mutation was subcloned into pGEX-JDK and the expressed protein was purified using glutathione-agarose affinity chromatography. Two rabbit polyclonal antibodies were generated using standard procedures and affinity-purified on GST-KLP61F columns. The antibody was acid eluted, neutralized in Tris buffer, dialyzed into PBS buffer, and concentrated. For embryo injection, the concentration in the needle was 7–15 mg/ml.
Time-Lapse and Fluorescent Speckle Microscopy of Embryos
Embryos were injected with rhodamine labeled tubulin (Cytoskeleton, Denver, CO). Time-lapse images were acquired with an Olympus (Melville, NY) microscope equipped with an Ultra-View spinning disk confocal head (Perkin Elmer-Cetus, Boston, MA) and a 100× 1.35 NA objective at time intervals of 1–3 s. Images were analyzed with Metamorph Imaging software (Universal Imaging, West Chester, PA). No neighbors deconvolution or sharpen high filters followed by a low-pass filter were applied to all images. Pole-to-pole distance as a function of time was measured from the position of the poles in each image. Kymography was used to quantify speckle movement. Calculations and statistical analyses were done on Microsoft Excel (Redmond, WA). To quantify the amount of motor on the spindle we measured the intensity of GFP and that of rhodamine on the spindle and subtracted background intensity and ratioed these two values. This allowed us to compare the amount of motor remaining on spindles within single embryos, but we cannot control the amount of injected rhodamine tubulin precisely enough to permit quantitative comparisons between different embryos.
Time-Lapse Analysis of Mitosis in Late Embryos and Larval Brains
Embryos were collected for 1 h and imaged after at least 7 h. Brains were dissected from wild-type larvae expressing G147-GFP (Bloomington Stock Center) or GFP-tubulin and from klp61f mutant second and third instar larvae expressing GFP-tubulin in D-22 (pH 6.7) insect medium (U.S. Biological, Swampscott, MA) supplemented with 7.5% fetal bovine serum and 0.5 mM ascorbic acid (Sigma, St. Louis, MO). The explanted brain was then moved carefully in a drop of medium onto a No. 1 coverslip placed on a special stainless steel slide with an indentation to allow observation of the sample. The brain was pushed carefully into direct contact with the coverslip, and an additional coverslip was placed on top of the stainless steel slide to keep the medium in place and prevent dehydration of the sample. Late embryos and brains were imaged as above except that stacks of 14–18 confocal planes spaced 0.5 μm apart were taken at time intervals of 30 s. Images were analyzed as above.
Online Supplemental Material
Supplemental Video 1 shows a time-lapse movie of an embryo expressing KLP61F-GFP and injected with rhodamine tubulin and anti-KLP61F (see Figure 1). Supplemental Video 2 shows a time-lapse movie of an embryo injected with rhodamine tubulin and anti-KLP61F (see Figure 2, A and C). Supplemental Video 3 shows a time-lapse movie of an embryo expressing GFP-CNN and injected with rhodamine tubulin and anti-KLP61F (see Figure 2, E–G).
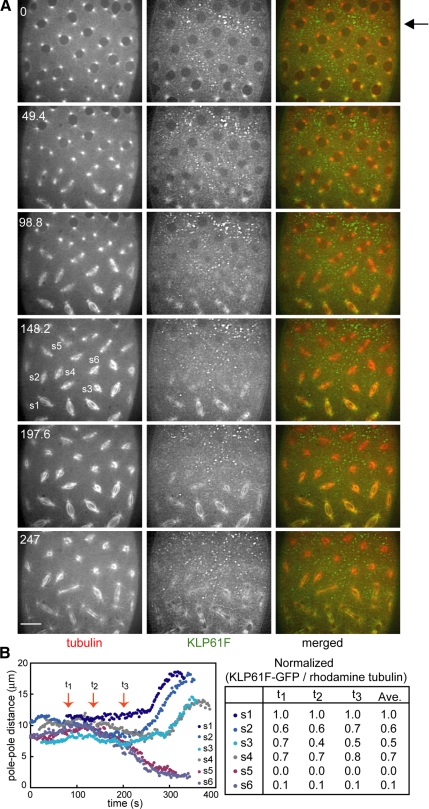
Figure 1.
(A) Microinjection of an anti-KLP61F antibody (designed to dissociate KLP61F from spindles) results in a gradient of antibody concentration and produces a gradient in the KLP61F content of different spindles. Images from time-lapse movie of an embryo expressing KLP61F-GFP and injected with rhodamine tubulin and anti-KLP61F (see Supplemental Video 1). Time in each frame is given in seconds from NEB. Bar, 10 μm. The injection site was close to the top of the embryo (arrow), KLP61F-GFP forms immunoprecipitates, and most KLP61F-GFP is depleted from the spindles. These spindles collapse, as seen at 247 s. Toward the bottom of the embryo, KLP61F-GFP is still present on the spindles and consequently these spindles assemble, though they may exhibit defects. (B) Graph of pole–pole distance as a function of time (left) and quantification of KLP61F remaining on these spindles (right). The normalized ratio of KLP61F-GFP to rhodamine tubulin is used to compare the amount of motor remaining on each spindle at different time points. Spindles that collapse have practically no motor on them, whereas spindles that do not collapse or recover from partial collapse have at least 40% remaining.
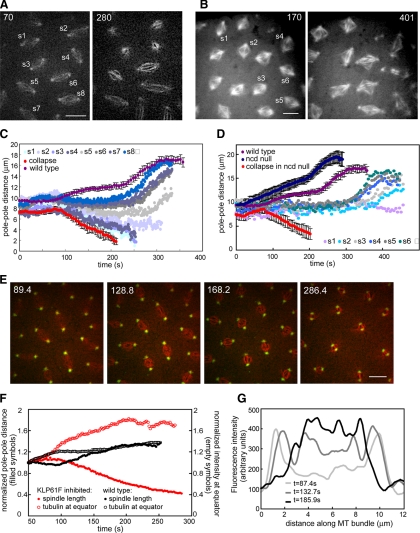
Figure 2.
(A) Two time points from a wild-type embryo injected with anti-KLP61F antibody showing a gradient of defects (see Supplemental Video 2). Bar, 10 μm. (B) Two time points from an Ncd null embryo injected with anti-KLP61F antibody showing more disorganized spindles due to the loss of two motors. Bar, 10 μm. (C) Graph of pole–pole distance as a function of time for the embryo shown in A. Close to the injection site spindles collapse (see s1 and s3), and other spindles shorten but remain small (see s2). Further away from the injection site, spindles do not elongate during prometaphase (s5, and the most distal spindles exhibit anaphase elongation (s6, s7, and s8). For comparison, we show graphs from a wild-type embryo (purple) and most severe effect (an embryo injected with enough antibody to collapse all spindles within the field of view; red). (D) Spindle length as a function of time for the Ncd null embryo shown in B. For comparison graphs of wild-type embryos (purple), Ncd null embryos (dark blue), and Ncd null embryos injected with a high concentration of anti-KLP61F (red) are shown. The gradient of concentration of antibody leads to a gradient of spindle pole lengths. A high inhibition of KLP61F leads to spindle collapse; lower concentrations lead to less severe defects. At a certain concentration the spindle maintains a steady length (light purple). (E) Anti-KLP61F antibody causes spindle collapse at prometaphase without apparent disruption of centrosomes or spindle poles. Images from time-lapse movie of an embryo expressing GFP-CNN (green) and injected with rhodamine tubulin (red) and anti-KLP61F (see Supplemental Video 3). Time in each frame is given in seconds. Bar, 10 μm. Note how the centrosomes move toward each other, but do not appear disorganized. (F) Tubulin fluorescence in the central spindle increases as the spindle collapses. Normalized spindle length (filled symbols) and fluorescent tubulin intensity at the equator (empty symbols) as a function of time for wild-type (black) and KLP61F inhibited spindles (red). After KLP61F inhibition spindles collapse with a correlated increase in tubulin intensity, suggesting that MTs are sliding inward and not depolymerizing. The increase is much higher than that observed in wild-type spindles, indicating that it is not due to a normal increase in tubulin. (G) Linescans of fluorescent tubulin intensity along a single ipMT bundle at different time points during the collapse. The intensity increases as the spindle collapses.
RESULTS
Injection of Anti-KLP61F Antibody into the Drosophila Syncytial Embryo Causes a Gradient of Mitotic Defects
We raised new rabbit affinity-purified polyclonal antibodies to the C-terminal tail domain of KLP61F. Studies using transient transfection with Eg5 mutants in cultured Xenopus cells (Sawin and Mitchison, 1995), with KLP61F mutants in live Drosophila embryos (Cheerambathur et al., 2008) and using antibodies raised against phospho- and unphospho-epitopes in Drosophila embryos (Sharp et al., 1999a) suggest that the cyclin-dependent kinase-mediated phosphorylation of a conserved Thr-933 residue in the bimC box of kinesin-5 is critical for its localization to the spindle. We reasoned that antibodies to the phosphorylated form of this domain would competitively inhibit the binding of native phosphorylated KLP61F to the spindle and, by displacing the motor from its site of action, would inhibit its function. Accordingly antibodies were raised against a tail domain fragment in which the Thr-933 residue was changed to an aspartate phospho-mimic by site-directed mutagenesis (Materials and Methods). These antibodies react with a single polypeptide of appropriate molecular weight on immunoblots of embryonic extracts and have been shown to stain mitotic spindles by immunofluorescence (our unpublished results and Silverman-Gavrila and Wilde, 2006), consistent with our previous results (Sharp et al., 1999a; Cheerambathur et al., 2008). As reported previously, the microinjection of affinity-purified antibody into the syncytial blastoderm embryo blocked mitosis and produced monoastral arrays of microtubules (Sharp et al., 1999b, 2000a) similar to those seen in fixed loss-of-function klp61f mutants after depletion of the maternal load in larvae (Heck et al., 1993; see below).
To directly visualize the extent of the anti-KLP61F induced dissociation of KLP61F from mitotic spindles in the injected embryos, we coinjected fluorescent tubulin and anti-KLP61F into stable transgenic strains of Drosophila that express functional KLP61F-GFP (Cheerambathur et al., 2008; Figure 1; Supplemental Video 1). The antibody rapidly displaced KLP61F from spindles and formed visible, fluorescent immunoprecipitates in the adjacent cytoplasm (Figure 1, arrow). Moreover this experiment provided clear visual evidence of the formation of a spatial gradient of inhibition of the target antigen throughout the syncytial blastoderm, as had been inferred indirectly from the effects of inhibitory antibody microinjection in previous studies (Sharp et al., 2000b; Blower and Karpen, 2001; Kwon et al., 2004). Proximal to the antibody microinjection site, where the anti-KLP61F concentration is highest, KLP61F is completely dissociated from spindles and consequently spindles form monoasters. The injected antibody diffuses through the cytoplasm, setting up a concentration gradient, highest at the injection site and decreasing at increasingly distal sites. Accordingly, moving away from the injection site, the extent of depletion of KLP61F from spindles decreases, and increasingly higher concentrations of fluorescent motor are retained on spindles (Figure 1). Thus at distal sites, spindles do not form monoasters but display less severe defects, producing a range of “phenotypes” analogous to an allelic series of genetic mutants (Figure 1). We quantified the amount of KLP61F present on spindles at different sites within this gradient by measuring the fluorescence intensity ratio of KLP61F-GFP to injected rhodamine tubulin, which allows us to compare spindles within a single embryo, but not between multiple embryos, due to small variations in the amount of injected tubulin. As shown in Figure 1B, the severity of spindle pole separation defects correlates reasonably well with the extent of depletion of KLP61F from the spindle, so that spindles containing maximal levels of KLP61F-GFP (arbitrarily assigned a value 1.0) displayed spindle pole separation profiles similar to uninjected controls, spindles containing only 0–10% residual KLP61F-GFP collapsed into monoasters, and those containing intermediate levels of the motor (50–70%) displayed spindle length defects but completed mitosis nonetheless.
Spindle Pole Separation Defects Produced by Varying Levels of KLP61F Depletion in Wild-Type Versus Ncd Null Mutant Embryos
In previous studies, we reported that prometaphase spindles collapse into monoasters after KLP61F inhibition and are “rescued” in Ncd null mutants, but the behavior of spindles containing intermediate levels of KLP61F was not systematically examined. Results presented in the previous section now permit us to do so. In wild-type embryos, we observed that, at increasing distances from the injection site, the extent of perturbation of spindle length, as characterized by changes in pole–pole spacing versus time, decreases in parallel with the decrease in extent of depletion of KLP61F from spindles (Figures 1B and 2C). The most severe effect, prometaphase spindle collapse, occurred at a rate of 0.06 μm/s (Figure 1B, spindles s5 and s6), although some spindles close by retained sufficient levels of KLP61F to slow down the rate of collapse (Figure 2C, spindles s1–s4). Further away, spindles were observed to shorten somewhat, but did not collapse and instead maintained a short prometaphase length, and then elongated at anaphase B (Figures 1B, s3 and s4, and 2C, s5). Even further away, spindles did not elongate during prometaphase, or did so only slightly, but then elongated at approximately wild-type rates during anaphase B sometimes after a slight delay (Figures 1B, s1 and s2, and 2C, s6–s8). Thus, particularly noteworthy features of the spindles that do not collapse after partial loss of KLP61F are their short lengths during metaphase and anaphase A and their relatively normal rates of anaphase B spindle elongation.
The latter result was intriguing because we had previously proposed that KLP61F-driven ipMT sliding drives anaphase B, yet in studying the gradient of intermediate phenotypes (Figures 1B and 2C), we did not observe spindles that circumvented the prometaphase collapse but then failed to elongate normally during subsequent anaphase B. This is, in fact, consistent with mathematical modeling predicting that the rate of anaphase spindle elongation is independent of the number of motors, at least above a low threshold number (Brust-Mascher et al., 2004; Wollman et al., 2008). We hypothesized that spindles containing sufficient KLP61F motor to circumvent collapse and progress through prometaphase might also contain enough motor to elongate the spindle at normal rates during anaphase B. We further reasoned that, because Ncd generates an inward force capable of antagonizing KLP61F (Sharp et al., 2000a; Tao et al., 2006), in the absence of Ncd function, we might improve our chances of encountering spindles with sufficient KLP61F to proceed through prometaphase but below the threshold level of KLP61F needed to drive normal rates of anaphase B. In such spindles, a decreased rate of anaphase B spindle elongation should be observed.
Accordingly, we inhibited KLP61F in Ncd null mutant embryos, which totally lack kinesin-14 function, in order to circumvent prometaphase spindle collapse and to study the role of KLP61F during spindle elongation. In uninjected mutants, pole–pole separation occurred prematurely from prometaphase onward, supporting our hypothesis that Ncd serves as a “brake” that exerts an inward force on spindle poles, antagonistic to KLP61F (Figure 2D, dark blue). Pole–pole separation in Ncd null embryos occurs shortly after nuclear envelope breakdown (NEB), earlier than in wild-type embryos, suggesting that normally Ncd's braking activity is turned on throughout prometaphase. Anti-KLP61F injection into Ncd null mutant embryos produced a gradient of defects (Figure 2D), where the most severe effect was spindle collapse. This was not observed in our previous studies (Sharp et al., 1999b, 2000a) because more antibody is needed to collapse the spindle in an Ncd null background than in a wild-type background. This result suggests that, when KLP61F and Ncd function are both missing, another inward force-generator must drive pole–pole collapse. In addition, spindles lacking the function of both motors are disorganized (Figure 2B), suggesting that these motors contribute to spindle organization by cross-linking MTs throughout the spindle, in agreement with their localization in these cells. Significantly, at more distal sites where spindles retain partial KLP61F function but Ncd function is missing, the spindles do not collapse and either do not elongate at all (Figure 2D, s1) or elongate after a significant delay (Figure 2D, s2–s6). This failure to elongate was not observed in anti-KLP61F–injected wild-type embryos, and it supports the hypothesis that KLP61F is necessary for anaphase B.
The Collapse Observed after Strong Inhibition of KLP61F Is Due to the Drawing Together of Intact, Organized Spindle Poles by Inward MT Sliding
We wanted to better understand if the spindle length defects we observe after loss of kinesin-5 function reflect defects in spindle pole separation (Saunders et al., 1997; Sharp et al., 2000a) or in spindle pole focusing (Gaglio et al., 1996; Goshima and Vale, 2003). To simultaneously observe the behavior of MTs and spindle poles and see if anti-KLP61F has any detectable effect on pole organization, we microinjected anti-KLP61F with rhodamine-labeled tubulin into embryos expressing GFP-centrosomin (CNN), an integral component of the mitotic centrosome (Megraw et al., 2002; Figure 2E; Supplemental Video 3). During collapse, intact spindle poles steadily moved toward one another at a rate of ∼0.06 μm/s, and the appearance of focused, radial sets of fluorescent MTs projecting from discrete centrosomes persisted throughout the collapse (Figure 2E). There was no obvious disorganization of the spindle poles, although some MT bundles appeared to “splay” laterally, projecting from the pole into the adjacent cytoplasm. Injection of a high concentration of antibody caused all observed spindles to collapse, and this was accompanied by a concomitant increase in the fluorescent intensity of tubulin in the central spindle (Figure 2F) as seen in linescans along the pole–pole axis of the collapsing spindles (Figure 2G). This indicates that ipMT bundles become thicker during the collapse, consistent with spindle shortening being due to an inward sliding of adjacent, antiparallel ipMTs, although serial section electron microscopy (EM) done at time intervals during the collapse, which is probably not feasible, would be required to establish this.
Genetic Evidence for a Role of KLP61F in Driving Spindle Pole Separation
Loss-of-function mutants provide a complementary method to antibody inhibition for probing motor protein function (Scholey, 1998; Figure 3). Homozygous klp61f mutants undergo mitosis in embryos due to the maternal load of KLP61F but display late larval lethality due to mitotic defects that arise when the maternal load of KLP61F is depleted (Heck et al., 1993). Fixed larval neuroblasts were previously found to contain abnormal monoastral spindles, consistent with our antibody inhibition results (Heck et al., 1993). However, in the absence of real-time studies, it is unclear if these monoasters arise from an initial failure in spindle pole separation, from a collapse of assembled bipolar spindles, or from the fixation of transiently collapsing structures. To study this problem, we created klp61f mutant flies (Heck et al., 1993) expressing GFP-tubulin using laborious genetic crosses (Materials and Methods).
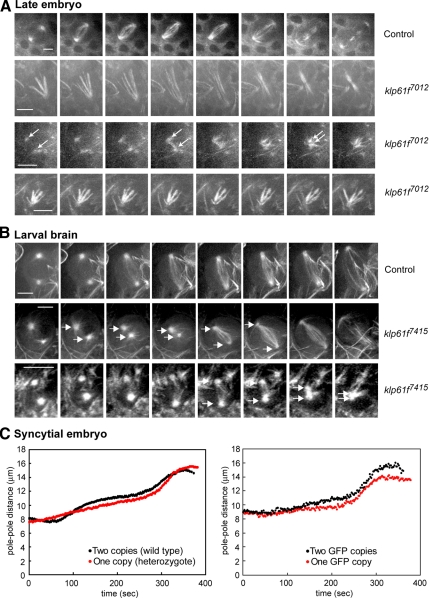
Figure 3.
(A) Still images from real-time recordings of spindle pole separation dynamics in klp61f7012 mutant embryos expressing GFP-tubulin. Top row, spindle in wild-type embryo. Second row, a monoastral spindle that becomes bipolar. Third row, a spindle that collapses to form a permanent monoaster. Arrows mark the poles. Fourth row, monoaster that does not change during observation time. Bar, 5 μm. (B) Still images from real-time recordings of spindle pole separation dynamics in wild-type and klp61f7415 mutants expressing GFP-tubulin. Top row, a wild-type neuroblast. Second row, a mutant neuroblast with separated spindle poles that transiently collapse, then recover and separate fully. Third row, a mutant larval spindle that collapses to form a permanent monoaster. Bar, 5 μm. (C) Differences in spindle pole separation due to varying KLP61F gene dosage. Left, embryos from heterozygous mothers have a shorter metaphase length than wild-type embryos. Right, klp61f mutant embryos rescued with two copies of a KLP61F-GFP transgene have a longer metaphase spindle length than those rescued with one copy.
In one allele (klp61f7012), larvae have smaller brains than wild types and spindles are rare. In these mutants, spindles start collapsing in mitotic domains of the late embryo (Figure 3A), where we observed that some spindles collapse into stable monoasters that fail to complete mitosis. Other monoasters seem to form monoastral bipolar spindles, which may assemble via a chromosome-directed pathway in which KLP61F activity is required to focus the anastral spindle pole as proposed for S2 cells (Goshima and Vale, 2003). Such monoastral bipolar spindles were not observed in our antibody inhibition experiments in syncytial embryos, but they have previously been observed in fixed larval brains and gonial cells of klp61f mutants (Wilson et al., 1997). These observations are consistent with the idea that KLP61F is required for the separation of spindle poles in the rapid, centrosome-dominant mitoses of the syncytial embryo, but as development proceeds, mitosis slows down (see below) and centrosomes become less dominant, so that KLP61F plays increasingly important roles in anastral spindle pole focusing.
In a different allele (klp61f7415), homozygous larvae had brains of normal size, and we could follow the progression of living mutant cells through mitosis (Figure 3B). We observed collapsing spindles with similar pole–pole separation defects to those seen in anti-KLP61F–injected embryos. In wild-type brains we never observed such collapsing or shortening spindles. Figure 3B shows the dynamics of spindle pole separation in a wild-type neuroblast spindle (Figure 3B, top row), a mutant spindle that initially separates its poles then collapses into a stable monoaster (Figure 3B, bottom), and a mutant spindle that transiently collapses and then recovers (Figure 3B, center). Careful 3D observations and measurements confirm that we were indeed visualizing spindle pole separation dynamics and not simply changes in the orientation of the spindle. The results are consistent with a complete and partial depletion of the maternal load of KLP61F giving rise to permanent and transient spindle collapse, respectively.
Because the mutants examined are recessive lethal (Heck et al., 1993), this result makes it extremely unlikely that the collapse of prometaphase spindles seen after severe antibody inhibition is due to dominant effects of the antibody. Interestingly, the rate of spindle pole separation in late embryos and neuroblasts is slower than in syncytial embryos; the time from NEB to telophase takes 10–15 min in late embryos and ∼20 min in neuroblasts compared with 4–6 min in the syncytial embryo. However, whether this difference in time reflects differences in the balance of forces operating in the spindles of different cell types or some other unknown factors is unclear.
Further evidence in support of the idea that KLP61F exerts outward forces on spindle poles was obtained by examining embryos carrying varying “doses” of the KLP61F gene (Figure 3C). For example, during prometaphase wild types harboring two copies of the KLP61F gene separated their spindle poles further than klp61f mutant heterozygotes containing a single copy, suggesting that the predicted increase in the concentration of the motor increases the outward force that pushes apart the poles (Figure 3C, left). In addition, the viability of klp61f mutants could be rescued with one or two copies of a KLP61F-GFP transgene that express the expected amount of protein based on immunoblotting (Cheerambathur et al., 2008). Measurement of pole–pole distances in these embryos also showed that two copies of the GFP transgene supported faster prometaphase spindle elongation and the formation of longer metaphase spindles than did a single copy of the transgene (Figure 3C, right), again consistent with the gradient observed after antibody inhibition.
KLP61F Is Required for Poleward Flux and Normal Metaphase Spindle Length
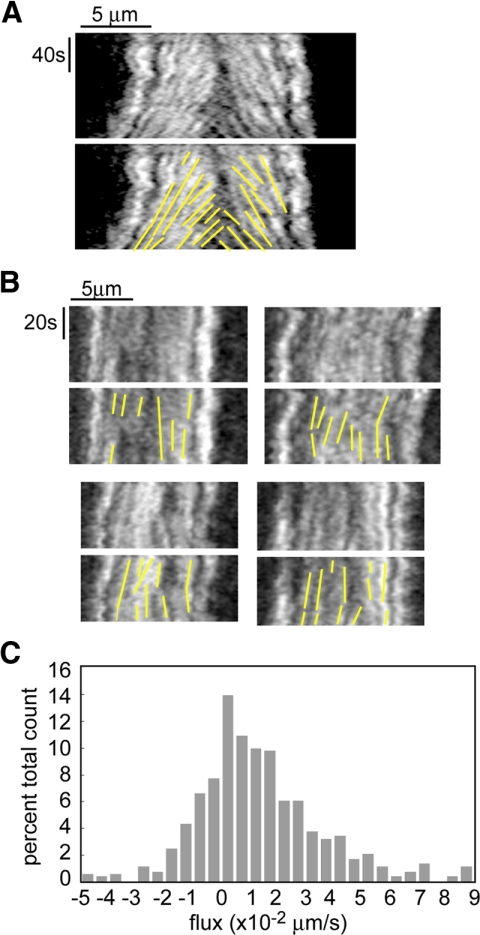
The gradient of defects elicited by antibody inhibition experiments revealed additional roles for KLP61F in embryos. On the basis of comparisons of the rates of poleward flux within ipMT bundles and the rates of MT sliding induced by purified KLP61F, as well as the observation that poleward flux stops at the onset of anaphase B, we previously proposed that KLP61F drives poleward flux in Drosophila embryos, but this had not been tested in vivo (Cole et al., 1994; Brust-Mascher and Scholey, 2002; Brust-Mascher et al., 2004; Tao et al., 2006). If we classify the gradient induced by KLP61F antibody injection by analyzing spindle pole length, we find as stated above that the most severe effect is spindle collapse. The next most severe effect is partial spindle shortening followed by a steady-state length that is smaller than the pole spacing at NEB. Other spindles maintained the characteristic NEB pole spacing until anaphase B or displayed only a small prometaphase elongation. In these spindles we measured the steady-state length and the rate of poleward flux during these periods of steady spindle length maintenance (i.e., metaphase and anaphase A). The average metaphase length decreased from 11.8 ± 0.08 to 8.8 ± 1.4 μm (Table 1), and poleward flux was significantly inhibited both within ipMTs and kinetochore (k) MTs (Figure 4). The average rate of flux was 0.01 ± 0.02 μm/s in KLP61F inhibited embryos compared with 0.05 ± 0.02 μm/s in control embryos (Figure 4; Table 1). This indicates that KLP61F-driven MT sliding is necessary to achieve the wild-type metaphase spindle length and that it also contributes to poleward flux in both ipMTs and kMTs. Interestingly, 17% of our flux measurements were negative (Figure 4C). A possible source of negative flux numbers, especially close to zero, is noise in the measurements. A more interesting possibility is that they represent speckles on ipMTs associated with one pole that slide inward as the spindle transiently shortens, bringing them closer to the pole in the opposite half spindle. Such a speckle would flux toward the first pole but away from the second, now closer, pole, and this would be measured as negative flux.
Table 1.
Partial inhibition of KLP61F decreases the rates of flux and anaphase A chromosome to pole movement as well as the metaphase spindle length and kinetochore-kinetochore distance
| Wild type | Partial inhibition of KLP61F | |
|---|---|---|
| Rate of flux during steady length (μm/s) | 0.05 ± 0.02 (15/45/822) | 0.01 ± 0.02 (3/21/577) |
| Metaphase spindle length (μm) | 11.8 ± 0.08 (3/15) | 8.8 ± 1.4 (6/59) |
| Interkinetochore distance (μm) | 1.31 ± 0.22 (3/15/33) | 0.77 ± 0.20 (6/59/143) |
| Rate of anaphase A (μm/s) | 0.11 ± 0.02 (6/27/106) | 0.06 ± 0.02 (6/37/144) |
The number of 1) embryos, 2) spindles and 3) speckles, or kinetochore pairs or kinetochores analyzed are indicated in parentheses.
Figure 4.
Poleward flux requires the function of KLP61F. (A) In wild-type embryos kymographs show that tubulin speckles flux toward the poles during the metaphase–anaphase A steady state, but move at the same rate as the poles during anaphase B. (Brust-Mascher and Scholey, 2002). (B) In embryos injected with anti-KLP61F antibody, kymographs obtained from spindles that did not collapse but maintained a steady length show that tubulin speckles do not flux toward the pole, indicating that the function of KLP61F is required for this movement (see Supplemental Video 2 for speckled movie). (C) Histogram of all the flux data obtained on spindles that did not collapse after KLP61F inhibition.
Congression Is Defective, Kinetochore Spacing and the Rate of Anaphase A Are Reduced, and the Relative Timing of Anaphase A and B Is Perturbed in Spindles Partially Depleted of KLP61F
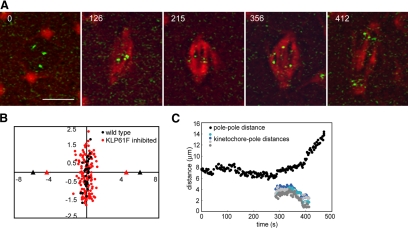
In those spindles that retain sufficient KLP61F to maintain pole–pole spacing and resist prometaphase spindle collapse, we were also able to study congression and measure the subsequent rate of anaphase A in transgenic embryos expressing GFP-CID that were injected with rhodamine tubulin and anti-KLP61F (Figure 5). In spindles that shorten or do not undergo prometaphase-to-metaphase elongation, but then proceed through anaphase, we find that congression is defective. Kinetochores occupy a larger area around the equator compared with wild-type spindles, revealing subtle defects in chromosome positioning on the metaphase plate (Figure 5B). Interestingly, kinetochore–kinetochore distance is reduced from 1.31 ± 0.22 μm in wild type to 0.77 ± 0.20 μm in KLP61F-inhibited spindles (Table 1), indicating a reduction in the magnitude of poleward forces acting on the kinetochores. Subsequently, the average rate of kinetochore-to-pole movement is 0.06 ± 0.02 μm/s (144 kinetochores in 37 spindles) compared with 0.11 ± 0.02 μm/s for wild-type anaphase A movement (Figure 5; Table 1). Some spindles that maintained a reduced but steady pole–pole spacing displayed no kinetochore-to-pole movement and a single nucleus reformed around the unsegregated parental chromosomes. Such spindles were excluded from our calculations of the average rate.
Figure 5.
Chromosome congression and anaphase A are perturbed by partial KLP61F inhibition. (A) Time-lapse images of one spindle in an embryo expressing GFP-CID injected with rhodamine tubulin and anti-KLP61F antibody. The spindle shortens slightly (compare 215 with 126) but does not collapse after KLP61F inhibition. Kinetochores move to the poles albeit slower than in control spindles. Time in each frame is given in seconds from NEB. Bar, 5 μm. (B) Positions of the center of kinetochore pairs in wild-type (black) and KLP61F-inhibited (red) spindles. In anti-KLP61F–inhibited spindles, kinetochores do not congress as tightly as in controls, and they exhibit a larger displacement around the equator. Triangles mark the average positions of the poles. (C) Pole–pole distance and kinetochore-to-pole distances for the spindle shown in A (averages in Table 1).
Significantly, the lower rate of 0.06 μm/s parallels the observed lack of poleward flux (previous section) and supports the idea that anaphase A is driven by a combined “flux-pacman” mechanism in Drosophila embryos (Brust-Mascher and Scholey, 2002; Maddox et al., 2002; Rogers et al., 2004). Interestingly, we noticed that in many of these spindles, kinetochore-to-pole movement occurs at the same time as pole–pole separation, whereas in wild-type spindles, anaphase A always precedes anaphase B. How loss of kinesin-5 function could lead to changes in the temporal relationship between chromosome-to-pole motility and spindle elongation during anaphase is unclear. As predicted (Sharp et al., 2000a; Brust-Mascher et al., 2004) the rate of anaphase B is relatively insensitive to KLP61F concentration to a certain point: spindles that exhibited a partial collapse and had a metaphase length of <7 μm elongated at a slower rate, but spindles that maintained at least the NEB length elongated at approximately wild-type rates.
Influence of Cortical Organization on KLP61F-dependent Mitotic Events
Cortical pulling forces could complement the outward sliding of ipMTs to control pole–pole spacing. To examine this, we measured spindle pole dynamics and MT speckle movements in two mutants that have severely disrupted actin cortices surrounding the nucleus. In the Drosophila syncytial embryo, actin caps are normally formed over each nucleus and a furrow is formed around the spindle during metaphase (Cytrynbaum et al., 2005). The cortex-defective Scrambled (sced; Stevenson et al., 2001) and Sponge (spg; Postner et al., 1992) mutants have highly reduced actin caps and no metaphase furrows (Supplemental Figure S1). If cortical dynein or cortical MT depolymerases play a significant role in pulling spindle poles apart by sliding astral MTs outward relative to the actin cortex, this activity should be reduced or eliminated due to the depletion of actin caps and furrows in these mutants. Plots of pole–pole separation dynamics (Supplemental Figure S1) show that the rate of separation during prometaphase is slower in these mutants, but during anaphase B there is no significant difference. Using fluorescence speckle microscopy, we examined the rate of movement of tubulin speckles away from the equator during metaphase and anaphase. This could reflect the sliding apart of MTs either by outward ipMT sliding by KLP61F or, during anaphase B, by outward pulling by cortical forces. In wild-type spindles, speckles move at a rate of 0.05 ± 0.02 μm/s, in spg mutants they move at 0.05 ± 0.02 μm/s, and in sced mutants at 0.06 ± 0.02 μm/s (Supplemental Figure S1), indicating that outward pulling by cortical forces is not significant throughout metaphase and anaphase. Together these results are consistent with the hypothesis that cortical forces augment ipMT sliding to contribute to spindle elongation during the prometaphase-to-metaphase transition, for example, via cortical dynein activity (Sharp et al., 2000a). However, cortical pulling forces apparently do not play a significant role after metaphase onset, whereupon KLP61F becomes the major force generator for the outward sliding of ipMTs that underlies poleward flux and anaphase spindle elongation.
DISCUSSION
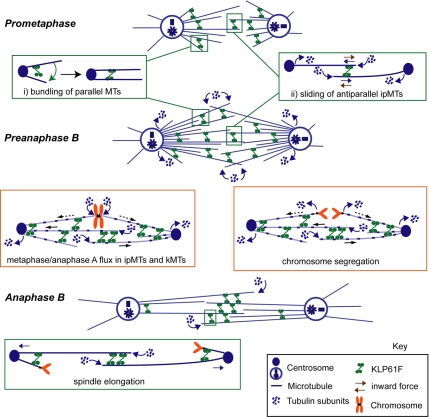
This study exploits our ability to create and directly visualize the KLP61F concentration gradient produced by the microinjection of an antibody to the motor's spindle targeting domain in wild-type and Ncd null mutant Drosophila syncytial blastoderm embryos in order to examine the spectrum of functions of kinesin-5. The work suggests that MT–MT crosslinking and sliding by KLP61F contributes to 1) the maintenance of prometaphase spindles as chromosomes are captured; 2) poleward flux in ipMTs and kMTs; 3) the bundling and organization of MTs throughout the spindle; 4) the control of pre-anaphase B spindle length; 5) tight congression, kinetochore–kinetochore spacing and the rate of anaphase A chromatid-to-pole motility; and 6) the elongation of the anaphase B spindle in a manner that is relatively insensitive to the KLP61F concentration (Figure 6). The hypothesis that KLP61F drives MT sliding from NEB through anaphase B to push apart spindle poles was also supported by gene dosage experiments in embryos and real-time fluorescence microscopy of transgenic late embryonic/larval klp61f mutants expressing GFP-tubulin. The latter studies also revealed monoastral bipolar spindles that were never seen in syncytial embryos, suggesting that chromosome-directed spindle assembly becomes more evident as embryonic and larval development proceeds and mitosis slows down.
Figure 6.
Model for the multiple roles of KLP61F in Drosophila syncytial embryo mitosis. We propose that kinesin-5 motors function as ensembles of dynamic, transient, MT crosslinkers throughout the spindle (Sharp et al., 1999a; Cheerambathur et al., 2008), organizing parallel MTs into bundles (inset i) and sliding apart antiparallel MTs (inset ii), in accordance with the biochemical properties of KLP61F (e.g., Cole et al., 1994; Kashina et al., 1996a; Tao et al., 2006; Van den Wildenberg et al., 2008). The results suggest that KLP61F-driven MT-MT sliding is required to oppose an inward force (ii, brown arrows) and maintain the spindle during early prometaphase, as well as to elongate it to the metaphase steady state length. In preanaphase B (i.e. metaphase and anaphase A) spindles, KLP61F driven ipMT outward sliding (blue solid arrows) is required for poleward flux in both kMT and ipMT bundles and for anaphase A chromosome movement. The kMTs may be slid poleward (dashed blue arrows) via crosslinks to the actively sliding antiparallel ipMTs. Finally, KLP61F drives spindle elongation during anaphase B. This model is proposed only for Drosophila embryos (see Discussion).
Experimental Strategy
Several methods are currently available for inhibiting target proteins in cells, including mutants, injection of dominant negative protein fragments, chemical inhibitors, RNA interference, and inhibitory antibody microinjection. In the current study, microinjected antibody to the spindle-targeting phospho-tail domain on kinesin-5 (Sawin and Mitchison, 1995; Sharp et al., 1999a; Cheerambathur et al., 2008) rapidly dissociated KP61F from mitotic spindles. By utilizing stable transgenic embryos that express GFP-tagged KLP61F, we were able to directly visualize the extent of dissociation of the target antigen from spindles in different regions of the syncytium relative to the injection site. This in turn allowed us to correlate the extent of KLP61F depletion with the mitotic perturbation observed, which permitted an exploration of the range of mitotic functions of KLP61F, in addition to the unseparated/collapsed spindle pole phenotype seen after severe inhibition or loss of function (Heck et al., 1993; Sharp et al., 1999b). Our results with respect to spindle pole spacing were confirmed by the observation of similar defects in klp61f mutants, including spindle collapse into monoasters after severe loss of KLP61F function and the observation that spindles depleted of ∼30–50% of KLP61F by antibody injection, like those in embryos carrying half the normal dose of the corresponding gene, are shorter than normal at metaphase, but complete anaphase successfully (Figures 1 and 3).
Relation to Previous Experimental Studies of Kinesin-5 in Drosophila Syncytial Embryos
The results support previous studies suggesting that KLP61F is dispensable for the assembly of astral syncytial embryo spindles but is essential for prometaphase spindle maintenance where it is antagonized by Ncd (Sharp et al., 1999b; Sharp et al., 2000a; Tao et al., 2006), and it further implicates the action of an additional, unknown, inward motor, possibly the MT depolymerase, KLP10A. Previously, we had predicted that KLP61F has a role in driving poleward flux and that the rate of anaphase B is insensitive to changes in the concentration of KLP61F (Brust-Mascher et al., 2004; Wollman et al., 2008). Here, we confirmed these predictions experimentally and also obtained evidence for new roles in controlling the length of the metaphase spindle, the tightness of chromosome congression, the distance between sister kinetochores, the rate of anaphase A chromatid-to-pole motility, and the relative timing of anaphase A and B, none of which have been reported previously.
For example, in spindles lacking Ncd function, spindle collapse driven by the inward force-generators can be circumvented and the results, specifically the failure or delay of spindle elongation, then reveal a role for KLP61F-driven sliding in anaphase B. In the spindles that fail to elongate, we propose that there is enough KLP61F to prevent spindle collapse but not enough to drive anaphase B (Figure 2). An outward, KLP61F-driven ipMT sliding, coupled to MT depolymerization at the poles, could also underlie KLP61F's contribution to poleward flux as predicted, but not previously tested (Brust-Mascher and Scholey, 2002). Perhaps surprisingly, after partial KLP61F inhibition, flux is slowed down in all MT bundles, suggesting that all bundles are interconnected. We propose that the sliding apart of ipMTs by KLP61F would also push kMTs poleward, if the kMTs are mechanically coupled to the sliding ipMTs by KLP61F (or other) MT–MT cross-linkers (Figure 6; Sharp et al., 1999a; Cheerambathur et al., 2008). Depolymerization of ipMTs and kMTs at the poles would then create KLP61F-dependent poleward flux. The slowing down of anaphase A chromatid-to-pole motility resulting from KLP61F inhibition could be explained by the loss of flux because it has been shown that in Drosophila embryos, anaphase A is driven by a combined flux-pacman mechanism (Rogers et al., 2004). Surprisingly, we also observed a loss of tension between sister kinetochores and a loss of congression. This may also be due to the loss of flux; i.e., normally as MTs slide outward, they generate a pulling force on the kinetochores. This force maintains the tension between sister kinetochores so that the loss of this outward sliding leads to a decrease in the distance between sisters.
Relation to Experimental Studies of Kinesin-5 Function in Other Systems
A comprehensive review of the diverse roles of kinesin-5 in different systems is beyond the scope of this article (see Valentine et al., 2006a; Civelekoglu-Scholey and Scholey, 2007), but a brief comparison between Drosophila syncytial embryos and other systems is appropriate. Live imaging of klp61f mutant late embryos and larvae (Figure 3) revealed that bipolar spindles collapse into monoasters, as observed in syncytial embryos, and also revealed the formation of functional bipolar, monoastral spindles, as observed previously in studies of fixed larval brains and testis (Wilson et al., 1997) but not in anti-KLP61F–injected embryos. It is plausible that this reflects the formation of bipolar monoastral spindles by chromosome-directed, KLP61F-mediated acentrosomal pole focusing as proposed for Drosophila S2 cells, based on RNA interference (RNAi) experiments (Goshima and Vale, 2003). We hypothesize that the syncytial embryo spindles that we study are adapted for rapid mitoses (Civelekoglu-Scholey et al., 2006) and consequently their assembly and function is dominated by the centrosome-directed pathway so that, unlike in larvae, S2 cells and many other cells, the chromosome-directed spindle assembly pathway plays a lesser role (Heald et al., 1997; Megraw et al., 2001). The relative importance of centrosome- versus chromosome-directed spindle organization may be a key factor influencing how kinesin-5 is deployed in different spindles, e.g., in spindle pole focusing versus spindle pole separation, though it is obviously not the only factor.
Our results showing that KLP61F appears to contribute to the length of pre-anaphase B spindles in living embryos differ from the results obtained in fixed S2 cells where, above the critical concentration required for bipolar spindle assembly, increasing concentrations of KLP61F did not influence metaphase spindle length (Goshima et al., 2005b). Although we cannot rule out an influence of technical differences between the two studies, we suspect that they reflect genuine system-specific differences, akin to the different consequences of loss of Ncd function in embryos versus cultured cells of Drosophila (Sharp et al., 2000a; Goshima et al., 2005a; Morales-Mulia and Scholey, 2005).
The Drosophila embryo spindle differs from that of S. cerevisiae in using only one, rather than two kinesin-5 motors. In both these systems, kinesin-5 function is required to prevent kinesin-14–induced prometaphase spindle collapse, but only in yeast is it essential for de novo spindle assembly (Saunders and Hoyt, 1992). As in Drosophila embryos, the kinesin-5 motors appear to contribute to metaphase spindle length (Saunders et al., 1997) and to anaphase spindle elongation (Saunders et al., 1995; Straight et al., 1998). In some respects, therefore, the manner in which kinesin-5 is deployed in Drosophila embryos resembles yeast more closely than S2 cells, again possibly reflecting the relative dominance of centrosomes and spindle pole bodies. In other respects the two systems may differ significantly; for instance, although kinesin-5 contributes to congression in Drosophila embryos and yeast, the proposed underlying mechanisms are very different (Figure 6 and Gardner et al., 2008).
The kinesin-5 inhibitor, monastrol, unfortunately does not interfere with purified kinesin-5 motility or mitosis in Drosophila (our unpublished observations), but it has been used extensively to probe the diverse functions of vertebrate kinesin-5. This work, together with complementary approaches, has revealed multiple roles for kinesin-5 in early spindle assembly, in spindle maintenance (in Xenopus but not cultured cells), in the sorting out of antiparallel chromosome nucleated MTs, and in the proper biorientation of chromosomes (Gaglio et al., 1996; Walczak et al., 1998; Kapoor et al., 2000; Uteng et al., 2008). Roles for kinesin-5 in driving poleward flux and controlling metaphase spindle length were proposed previously for Xenopus extract spindles (Miyamoto et al., 2004; Shirasu-Hiza et al., 2004) as we report here for Drosophila embryo spindles. In vertebrate cultured cells, in contrast, the inhibition of Eg5 function led to only a 25% decrease in the rate of poleward flux while spindle length remained normal, suggesting only minor contributions from kinesin-5 (Cameron et al., 2006). The latter authors also reported a small decrease in kinetochore–kinetochore spacing after Eg5 inhibition, supporting the notion that kinesin-5 may contribute to the exertion of poleward tension on kinetochores in some systems, including the Drosophila embryo (Figure 5).
Thus kinesin-5 motors have diverse, often essential mitotic functions in many systems including the Drosophila embryo. However, in Dictyostelium amoebae and C. elegans embryos, kinesin-5 motors play relatively minor mitotic roles, and they appear to be dispensable for mitosis (Saunders et al., 2007; Tikhonenko et al., 2008). Thus, mitotic motors can be deployed to carry out very different functions in different types of spindles, presenting a challenge for the formulation of general, integrated models of kinesin-5 function.
Relation to Studies of the Mechanism of Kinesin-5 Action
The multiple mitotic functions of KLP61F in Drosophila syncytial embryos can be explained by a model in which KLP61F cross-links adjacent MTs throughout the spindle (Figure 6), in accordance with the biochemical properties of the purified native embryonic or recombinant baculovirus-expressed motor (Cole et al., 1994; Kashina et al., 1996a; Kashina et al., 1996b; Tao et al., 2006; Van den Wildenberg et al., 2008), its immunolocalization (Sharp et al., 1999a), and its localization and dynamics in living cells (Cheerambathur et al., 2008). By cross-linking parallel MTs, KLP61F would “zip” together adjacent MTs to bundle and organize spindle MTs but would generate no force on them. By cross-linking and moving toward the plus ends of antiparallel ipMTs, however, it would persistently slide them apart to drive poleward flux when ipMT sliding is balanced by ipMT depolymerization at spindle poles, or to exert outward forces on spindles poles when ipMT depolymerization is turned off (McIntosh et al., 1969; Sharp et al., 1999a; Van den Wildenberg et al., 2008).
In the current study, this KLP61F-driven, outward ipMT sliding was directly visualized by monitoring fluorescent tubulin speckle behavior (Brust-Mascher and Scholey, 2002; Brust-Mascher et al., 2004), and it appeared to be antagonized by the inward sliding of ipMTs during prometaphase. In support of this, the decrease in pole–pole separation after KLP61F inhibition occurs without spindle pole disorganization, as documented in embryos expressing GFP-CNN, and is accompanied by an increase in microtubule density, as revealed by an increase in fluorescent tubulin intensity at the central spindle, consistent with the proposed sliding filament mechanism (Figure 2). The results obtained with sponge and scrambled mutants, which have severely disrupted cortices, suggest that cortical organization, and by inference, the pulling activity of cortical force generators, do not play a significant role in spindle pole separation after NEB. This is consistent with the hypothesis that KLP61F-driven outward sliding of ipMTs, augmented at specific stages by other ipMT-bound proteins such as KLP3A (Kwon et al., 2004), plays a central role.
We do not want to extrapolate our sliding filament model for kinesin-5 function in Drosophila syncytial embryos (Figure 6) to other systems where the molecular mechanism of action of kinesin-5 may be different (e.g., Kapoor and Mitchison, 2001; Gardner et al., 2008). The slow, plus-end–directed motility of kinesin-5 was first observed using recombinant motor domain subfragments of Eg5 (Sawin et al., 1992), and subsequently, in elegant, pioneering work, full-length recombinant Eg5 was shown to cross-link and slide adjacent MTs (Kapitein et al., 2005). The dynamic behavior of Eg5 in spindles is also consistent with it driving MT–MT sliding (Uteng et al., 2008), but to our knowledge, the ultrastructure of Eg5 and the properties of the purified native protein have not been studied. The yeast kinesin-5, Kip1, has a bipolar ultrastructure, like the Drosophila embryo protein, but its motility properties have not been described (Gordon and Roof, 1999). Elsewhere, many kinesin-5 motors have not been purified and studied biochemically, so it is possible that some kinesin-5 motors may act differently, e.g., serving as MT depolymerases (Gardner et al., 2008) or sliding MTs relative to a spindle matrix (Kapoor and Mitchison, 2001). Purified kinesin-5 motors have been shown to slide MTs over inert surfaces but not to depolymerize MTs (Sawin et al., 1992; Tao et al., 2006). Further biochemical studies on the mechanism of action of purified kinesin-5 from multiple systems would obviously be fruitful.
Relation to Quantitative Force-Balance Models for Mitosis
System level and reductionist models have been used to describe spindle pole separation after NEB in Drosophila (Brust-Mascher et al., 2004; Goshima et al., 2005b; Wollman et al., 2008; Civelekoglu-Scholey, Tao, Wollman, Brust-Mascher, and Scholey, unpublished data). In Wollman et al. (2008) a system level search was undertaken to find the integration of force generator activity that best explains spindle pole separation versus time in embryos, using experimental data to constrain the number of plausible models. Although ∼1000 models emerged from the search, these models fall into only six groups, which fit the experimental data, including most of the data on spindle pole separation presented here, remarkably well. Some discrepancies remain, e.g., the timing of activation of Ncd, but this is perhaps not surprising, given the inherent assumptions in such a broad theoretical approach.
Reductionist models were developed for Drosophila S2 cells (Goshima et al., 2005b) and for embryos (Brust-Mascher et al., 2004; Civelekoglu-Scholey, G., Tao, L., Wollman, R., Brust-Mascher, I., and Scholey, J. M., unpublished data). In the former model, which complemented novel high throughput microscopy and RNAi experiments, coupling of outward ipMT sliding to MT depolymerization at the poles was introduced to account for the lack of effect of KLP61F concentration on spindle length. We now understand that this model contains a physically unrealistic assumption in which an inward motion of the poles through increased depolymerization, coupled to decreased sliding, occurs in the absence of an associated force to drive this movement. Of course, this imperfection would remain undetected if the same mechanism had been described only qualitatively, but it shows that this model requires further refinement to explain the KLP61F concentration versus spindle length data.
The latter models were based closely on experimental data, including the dynamics, physical properties, and geometry of embryo spindles as well as the biochemical properties of MTs and mitotic motors. The Anaphase B (Brust-Mascher et al., 2004) model made several useful predictions, for example, that although KLP61F acts on dynamically unstable ipMTs to drive steady spindle elongation, the actual rate of anaphase B is insensitive to KLP61F concentration, in agreement with the experimental data presented here. It also predicts a spatial gradient of MT catastrophe events at anaphase B onset, which would be difficult to formulate using purely qualitative arguments and which is now being tested (Cheerambathur et al., 2007). In the prometaphase model, the proposed KLP61F and Ncd force balance was subjected to a focused reductionist analysis of individual spindles and purified proteins (Civelekoglu-Scholey, Tao, Brust-Mascher, Wollman, and Scholey, unpublished data). This work supports the idea that the KLP61F/Ncd force balance engages immediately after NEB as dynamic ipMT bundles start to form, whereas the subsequent elongation phase requires additional outward acting force-generators, for example, KLP3A on chromosome arms and cortical dynein (Sharp et al., 2000a; Kwon et al., 2004). Although all modeling approaches are useful and merit further refinement and exploration, reductionist models that pay close attention to both experimental evidence and physical chemical principles provide the most useful and realistic descriptions of mitosis.
Our own force-balance models for spindle assembly and function are based on the premise that antagonistic motors mediate the relative sliding (e.g., kinesin-5 and -14) or depolymerization (e.g., kinesin-13) of ipMTs, but other scenarios are also possible. For example, a new “slide-and-cluster” model for anastral spindle assembly proposes that kinesin-5 transports MTs that assemble around chromosomes to the spindle poles where they are focused by minus-end–directed motors (Burbank et al., 2007). This model apparently generates a force-balance capable of maintaining a robust steady-state spindle length, and it provides a plausible description of chromosome-directed, anastral spindle assembly in Xenopus extracts and elsewhere, possibly including Drosophila late embryos/larvae and S2 cells. However, in Drosophila syncytial embryo spindles, where MT assembly is nucleated predominantly at centrosomes, MTs turn over very quickly (half-time, 5 s), and it seems unlikely that kinesin-5 would have sufficient time to transport MTs to the spindle poles for focusing there (Cheerambathur et al., 2007). This suggestion again implies that kinesin-5 is deployed in different ways in different systems.
Summary
This comprehensive study has revealed the spectrum of mitotic functions of the kinesin-5 motor, KLP61F, in a single system, the Drosophila embryo. We conclude that KLP61F has multiple mitotic functions, all of which are consistent with its MT–MT cross-linking and antiparallel MT sliding activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Sandra Morales-Mulia for help in preparing the antibody. We thank Drs. Heck, Henikoff, Kaufmann, Naetzle, Pereira, and Wilson for fly stocks. We thank Drs. Civelekoglu-Scholey, Mogilner, McNally, and Wollman for comments. This work was supported by National Institutes of Health Grant GM55507 to J.M.S. P.S. was partially supported by Grant GM068952 to J.M.S. and A.M. (and subsequently to A.M. alone).
Glossary
Abbreviations used:
- ipMT
interpolar MT
- kMT
kinetochore MT
- MT
microtubule
- NEB
nuclear envelope breakdown.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-10-1033) on January 21, 2009.
REFERENCES
- Bettencourt-Dias M., et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I., Civelekoglu-Scholey G., Kwon M., Mogilner A., Scholey J. M. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA. 2004;101:15938–15943. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I., Scholey J. M. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol. Biol. Cell. 2002;13:3967–3975. doi: 10.1091/mbc.02-05-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I., Scholey J. M. Mitotic spindle dynamics in Drosophila. Int. Rev. Cytol. 2007;259:139–172. doi: 10.1016/S0074-7696(06)59004-7. [DOI] [PubMed] [Google Scholar]
- Burbank K. S., Mitchison T. J., Fisher D. S. Slide-and-cluster models for spindle assembly. Curr. Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Cameron L. A., Yang G., Cimini D., Canman J. C., Kisurina-Evgenieva O., Khodjakov A., Danuser G., Salmon E. D. Kinesin 5-independent poleward flux of kinetochore microtubules in PtK1 cells. J. Cell Biol. 2006;173:173–179. doi: 10.1083/jcb.200601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D. K., Brust-Mascher I., Civelekoglu-Scholey G., Scholey J. M. Dynamic partitioning of mitotic kinesin-5 crosslinkers between microtubule-bound and freely diffusing states. J. Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D. K., Civelekoglu-Scholey G., Brust-Mascher I., Sommi P., Mogilner A., Scholey J. M. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J. Cell Biol. 2007;177:995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Scholey J. M. Mitotic motors: kinesin-5 takes a brake. Curr. Biol. 2007;17:R544–R547. doi: 10.1016/j.cub.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Sharp D. J., Mogilner A., Scholey J. M. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys. J. 2006;90:3966–3982. doi: 10.1529/biophysj.105.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D. G., Saxton W. M., Sheehan K. B., Scholey J. M. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J. Biol. Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- Cottingham F. R., Gheber L., Miller D. L., Hoyt M. A. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 1999;147:335–350. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cytrynbaum E. N., Sommi P., Brust-Mascher I., Scholey J. M., Mogilner A. Early spindle assembly in Drosophila embryos: role of a force balance involving cytoskeletal dynamics and nuclear mechanics. Mol. Biol. Cell. 2005;16:4967–4981. doi: 10.1091/mbc.E05-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos A. P., Morris N. R. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Gadde S., Heald R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J. B., Hasbani M. J., Gill S. R., Schroer T. A., Compton D. A. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. K., et al. Chromosome congression by kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. M., Roof D. M. The kinesin-related protein Kip1p of Saccharomyces cerevisiae is bipolar. J. Biol. Chem. 1999;274:28779–28786. doi: 10.1074/jbc.274.40.28779. [DOI] [PubMed] [Google Scholar]
- Goshima G., Nedelec F., Vale R. D. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 2005a;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin N., Zhang J. M., Scholey J. M., Vale R. D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Kelso R. J., Stuurman N., Scholey J. M., Vale R. D. Length control of the metaphase spindle. Curr. Biol. 2005b;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Habermann A., Karsenti E., Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck M. M., Pereira A., Pesavento P., Yannoni Y., Spradling A. C., Goldstein L. S. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J. Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Platero J. S., van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt E. R., Gheber L., Kingsbury T., Hoyt M. A. Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J. Biol. Chem. 2006;281:26004–26013. doi: 10.1074/jbc.M604817200. [DOI] [PubMed] [Google Scholar]
- Johansen K. M., Johansen J. Cell and molecular biology of the spindle matrix. Int. Rev. Cytol. 2007;263:155–206. doi: 10.1016/S0074-7696(07)63004-6. [DOI] [PubMed] [Google Scholar]
- Kapitein L. C., Peterman E. J., Kwok B. H., Kim J. H., Kapoor T. M., Schmidt C. F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kapoor T. M., Mayer T. U., Coughlin M. L., Mitchison T. J. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T. M., Mitchison T. J. Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 2001;154:1125–1133. doi: 10.1083/jcb.200106011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Kashina A. S., Baskin R. J., Cole D. G., Wedaman K. P., Saxton W. M., Scholey J. M. A bipolar kinesin. Nature. 1996a;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina A. S., Scholey J. M., Leszyk J. D., Saxton W. M. An essential bipolar mitotic motor. Nature. 1996b;384:225. doi: 10.1038/384225a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak T. C., Grabe M., Gilbert S. P. Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. J. Biol. Chem. 2008;283:2078–2087. doi: 10.1074/jbc.M708354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Morales-Mulia S., Brust-Mascher I., Rogers G. C., Sharp D. J., Scholey J. M. The chromokinesin, KLP3A, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell. 2004;15:219–233. doi: 10.1091/mbc.E03-07-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P., Desai A., Oegema K., Mitchison T. J., Salmon E. D. Poleward microtubule flux is a major component of spindle dynamics and anaphase a in mitotic Drosophila embryos. Curr. Biol. 2002;12:1670–1674. doi: 10.1016/s0960-9822(02)01183-1. [DOI] [PubMed] [Google Scholar]
- Maiato H., Sunkel C. E. Kinetochore-microtubule interactions during cell division. Chromosome Res. 2004;12:585–597. doi: 10.1023/B:CHRO.0000036587.26566.81. [DOI] [PubMed] [Google Scholar]
- Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- McIntosh J. R., Hepler P., Van Wie D. G. Model for mitosis. Nature. 1969;224:659–663. [Google Scholar]
- Megraw T. L., Kao L. R., Kaufman T. C. Zygotic development without functional mitotic centrosomes. Curr. Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Kilaru S., Turner F. R., Kaufman T. C. The centrosome is a dynamic structure that ejects PCM flares. J. Cell Sci. 2002;115:4707–4718. doi: 10.1242/jcs.00134. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Salmon E. D. Mitosis: a history of division. Nat. Cell Biol. 2001;3:E17–21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- Miyamoto D. T., Perlman Z. E., Burbank K. S., Groen A. C., Mitchison T. J. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol. 2004;167:813–818. doi: 10.1083/jcb.200407126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A., Wollman R., Civelekoglu-Scholey G., Scholey J. M. Modeling Mitosis. Trends Cell Biol. 2006;16:88–96. doi: 10.1016/j.tcb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Morales-Mulia S., Scholey J. M. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postner M. A., Miller K. G., Wieschaus E. Maternal effect mutations of the sponge locus affect actin cytoskeletal rearrangement in Drosophila melanogaster embryos. J. Cell Biol. 1992;119:1205–1218. doi: 10.1083/jcb.119.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., Vale R. D., Scholey J. M., Sharp D. J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Sharp D. J. Spindle microtubules in flux. J. Cell Sci. 2005;118:1105–1116. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- Saunders A. M., Powers J., Strome S., Saxton W. M. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr. Biol. 2007;17:R453–454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M. A. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W. S., Hoyt M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders W. S., Koshland D., Eshel D., Gibbons I. R., Hoyt M. A. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K. E., LeGuellec K., Philippe M., Mitchison T. J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sawin K. E., Mitchison T. J. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J. M. Functions of motor proteins in echinoderm embryos: an argument in support of antibody inhibition experiments. Cell Motil. Cytoskelet. 1998;39:257–260. doi: 10.1002/(SICI)1097-0169(1998)39:4<257::AID-CM1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Brown H. M., Kwon M., Rogers G. C., Holland G., Scholey J. M. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 2000a;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., McDonald K. L., Brown H. M., Matthies H. J., Walczak C., Vale R. D., Mitchison T. J., Scholey J. M. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 1999a;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000b;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. Microtubule motors in mitosis. Nature. 2000c;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Yu K. R., Sisson J. C., Sullivan W., Scholey J. M. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1999b;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Shirasu-Hiza M., Perlman Z. E., Wittmann T., Karsenti E., Mitchison T. J. Eg5 causes elongation of meiotic spindles when flux-associated microtubule depolymerization is blocked. Curr. Biol. 2004;14:1941–1945. doi: 10.1016/j.cub.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Silverman-Gavrila R. V., Wilde A. Ran is required before metaphase for spindle assembly and chromosome alignment and after metaphase for chromosome segregation and spindle midbody organization. Mol. Biol. Cell. 2006;17:2069–2080. doi: 10.1091/mbc.E05-10-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson V. A., Kramer J., Kuhn J., Theurkauf W. E. Centrosomes and the Scrambled protein coordinate microtubule-independent actin reorganization. Nat. Cell Biol. 2001;3:68–75. doi: 10.1038/35050579. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Sedat J. W., Murray A. W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., Mogilner A., Civelekoglu-Scholey G., Wollman R., Evans J., Stahlberg H., Scholey J. M. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr. Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- Tikhonenko I., Nag D. K., Martin N., Koonce M. P. Kinesin-5 is not essential for mitotic spindle elongation in Dictyostelium. Cell Motil. Cytoskelet. 2008;65:853–862. doi: 10.1002/cm.20307. [DOI] [PubMed] [Google Scholar]
- Tsai M. Y., Wang S., Heidinger J. M., Shumaker D. K., Adam S. A., Goldman R. D., Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Uteng M., Hentrich C., Miura K., Bieling P., Surrey T. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J. Cell Biol. 2008;182:715–726. doi: 10.1083/jcb.200801125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine M.T., Fordyce P.M., Block S.M. Eg5 steps it up! Cell Div. 2006a;1:31–38. doi: 10.1186/1747-1028-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine M. T., Fordyce P. M., Krzysiak T. C., Gilbert S. P., Block S. M. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 2006b;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Wildenberg S., Tao L, Kapitein L, Schmidt C. F., Scholey J. M., Peterman E.J.G. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr. Biol. 2008;18:1860–1864. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P., Khodjakov A. E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol. 2004;14:413–419. doi: 10.1016/j.tcb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Heald R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Vernos I., Mitchison T. J., Karsenti E., Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Wilson P. G., Fuller M. T., Borisy G. G. Monastral bipolar spindles: implications for dynamic centrosome organization. J. Cell Sci. 1997;110(Pt 4):451–464. doi: 10.1242/jcs.110.4.451. [DOI] [PubMed] [Google Scholar]
- Wollman R., Civelekoglu-Scholey G., Scholey J. M., Mogilner A. Reverse engineering of force integration during mitosis in the Drosophila embryo. Mol. Syst. Biol. 2008;4:195. doi: 10.1038/msb.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.