Abstract
Actin plays an essential role in many eukaryotic cellular processes, including motility, generation of polarity, and membrane trafficking. Actin function in these roles is regulated by association with proteins that affect its polymerization state, dynamics, and organization. Numerous proteins have been shown to localize with cortical patches of yeast actin during endocytosis, but the role of many of these proteins remains poorly understood. Here, we reveal that the yeast protein Ysc84 represents a new class of actin-binding proteins, conserved from yeast to humans. It contains a novel N-terminal actin-binding domain termed Ysc84 actin binding (YAB), which can bind and bundle actin filaments. Intriguingly, full-length Ysc84 alone does not bind to actin, but binding can be activated by a specific motif within the polyproline region of the yeast WASP homologue Las17. We also identify a new monomeric actin-binding site on Las17. Together, the polyproline region of Las17 and Ysc84 can promote actin polymerization. Using live cell imaging, kinetics of assembly and disassembly of proteins at the endocytic site were analyzed and reveal that loss of Ysc84 and its homologue Lsb3 decrease inward movement of vesicles consistent with a role in actin polymerization during endocytosis.
INTRODUCTION
Endocytosis is the process through which the plasma membrane invaginates into the cell, resulting in the production of a vesicle that is then able to fuse with endosomes and enter the endolysosomal membrane system. Endocytosis is required for recycling of plasma membrane lipids and trafficking proteins, and for uptake or down-regulation of cell surface receptors (reviewed in Smythe and Ayscough, 2006). In yeast, a requirement for a dynamic actin cytoskeleton at sites of endocytosis has been recognized for many years (Kübler and Riezman, 1993; Raths et al., 1993). Several elegant studies using live imaging of fluorescently tagged proteins have allowed a clear idea of the main stages of the endocytic process to be elucidated (Kaksonen et al., 2003, 2005). Within endocytosis, actin plays a central role during later stages, that is, during vesicle invagination and post scission. Actin is not present at the early stages of endocytosis during the recruitment of the endocytic coat components. Interestingly, one of the key regulators of actin polymerization Las17, the yeast homologue of WASP, localizes to the endocytic sites relatively early but is proposed to be kept in an inactive state by association with certain endocytic coat components such as Sla1 and Bbc1 (Li, 1997; Rodal et al., 2003; Kaksonen et al., 2005). The mechanism of activation of Las17 is unclear. However, once activated it is thought that Las17 binds to and activates Arp2/3 that in turn initiates actin polymerization at the site (Winter et al., 1999; Rodal et al., 2005). The formation of branched and bundled actin structures at the patch have been shown to be critical for the invagination process (Galletta et al., 2008; Gheorghe et al., 2008). It is proposed that a strong actin framework allows force to be generated against this structure to allow the plasma membrane to be directed into the cell. After scission, it is also thought that actin plays a role in movement of the vesicle further into the cell (Huckaba et al., 2004; Kim et al., 2006).
The majority of actin-associated proteins localize only at the late stages of endocytosis. Many proteins in this group are actin-dependent components of endocytosis, because the addition of the actin monomer-sequestering drug latrunculin-A to cells leads to their dispersal (Ayscough et al., 1997). It is not clear, however, whether these proteins all bind directly to actin, or whether some of the association with cortical actin patches is mediated through other proteins. We have studied Ysc84, an actin-dependent endocytic patch, protein in detail to determine its function during endocytosis. Ysc84 is a member of a family of proteins that are found throughout fungi and also in vertebrates and plants. Most organisms seem to have a single protein showing homology with Ysc84. In mouse and humans, this protein is called Src homology 3 domain containing Ysc84 like-1 (SH3yl-1) (Aoki et al., 2000). Due to the whole genome duplication, Saccharomyces cerevisiae has two highly related genes, YSC84 and LSB3. Previous work showed that both Ysc84 (Lsb4) and Lsb3 have interactions with Las17 in two-hybrid assays (Madania et al., 1999). In our own work, we identified Ysc84 as an interactor of the endocytic adaptor protein Sla1 (Dewar et al., 2002). We also revealed that Ysc84 localizes to dynamic patches at the plasma membrane and that the mammalian homologue SH3yl-1 is able to affect the actin cytoskeleton in yeast (Dewar et al., 2002). In this study, we aimed to determine whether Ysc84 associates directly with actin, and if so, what the effect on F-actin dynamics or structure might be. We reveal that the N-terminal domain of Ysc84 is able to bind and bundle actin and as such represents a new class of actin-binding domain. We also show that the actin-binding activity is regulated by Las17/WASP. The functional importance of Ysc84 and its homologue are demonstrated in vivo during endocytosis.
MATERIALS AND METHODS
Plasmid and Strain Construction
Due to the presence of an intron, the YSC84 coding sequence was initially amplified by reverse transcription-polymerase chain reaction (RT-PCR) (Titan One Tube system; Roche Applied Science, Indianapolis, IN) from RNA isolated from wild-type yeast cells (KAY302) using a RNAeasy kit (QIAGEN, Dorking, Surrey, United Kingdom). The RT-PCR product was cloned into a pCR 4-TOPO-TA vector (Invitrogen, Paisley, United Kingdom) to generate pKA259. After sequencing, this plasmid was then used to amplify both full-length YSC84 and YSC84 fragments. Plasmids generated are listed in Table 1. Las17 two-hybrid construct pKA325 (Costa and Ayscough, 2005) was mutated using a QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Las17 GST construct was generated by cloning the appropriate fragment from pKA325 into pGEX6P1 (GE Healthcare, Buckinghamshire, United Kingdom). A diploid yeast strain carrying Ysc84-eGFP was a gift from Klaus Natter (University of Graz, Austria; KAY1308). This strain was sporulated and tetrads dissected using a Singer MSM micromanipulator. Haploid strains were then crossed with either Sac6mRFP or Las17mRFP to generate heterozygous diploid strains. It was noted that cells expressing the Ysc84GFP construct showed normal actin organization, but the timing of patch lifetimes for a number of proteins seemed shorter. For this reason, the timings given in Figure 5 (intensity and distance) are for the strains containing both green fluorescent protein (GFP) and red fluorescent protein (RFP) constructs to allow direct comparison of arrival and disassembly times for the proteins. These are not strictly comparable with timings when the Las17 and Sac6 are studied alone (e.g., in Figure 6A).
Table 1.
Plasmids used in this study
| pKA no. | Plasmid | Reference |
|---|---|---|
| 259 | TOPO TA-YSC84 | This study |
| 279 | pGEX4T1-YSC84Nt | This study |
| 539 | TOPO TrcHis YSC84Nt | This study |
| 540 | TOPO TrcHis YSC84 | This study |
| 411 | Untagged Ysc84 | This study |
| 412 | Untagged Ysc84Nt | This study |
| 558 | pGBDU-Ysc84 | This study |
| 325 | pGAD Las17 (292-536) | Costa and Ayscough, (2005) |
| 365 | pGAD Las17 (292-422) | Costa and Ayscough, (2005) |
| 366 | pGAD Las17 (292-422) | Costa and Ayscough, (2005) |
| 61 | pRS313 | Sikorski and Hieter, (1989) |
| 602 | pGAD Las17 P387A, P388A | This study |
| 566 | pGEX6P-Las17 (300-422) | This study |
| 88 | Abp1-GFP URA CEN | Warren et al. (2002) |
| pDA67 | pGBD-ACT1 | Amberg et al. (1995) |
| pGEX4T1-Ysc84-SH3 domain | Landgraf et al. (2004) |
Figure 5.
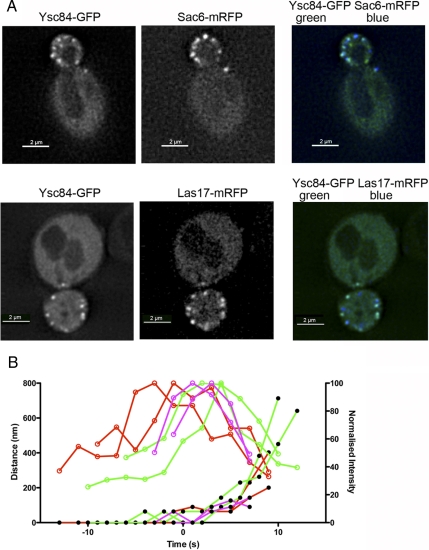
Ysc84 localization at endocytic patches. (A) Cells expressing Ysc84-eGFP and either Las17-mRFP (KAY1358) or Sac6-mRFP (KAY1357) were generated as described in Materials and Methods. GFP- and RFP-tagged proteins were visualized and images recorded as described in text. To facilitate visualization, RFP is shown in the blue channel (B) Using these cells, distance intensity measurements of the GFP and RFP spots were made. Movement of six or more spots was recorded for each marker, with distance representing distance from the original point of patch appearance, two are shown here for clarity on the graph. Intensity is normalized to a percentage to allow clearer comparison. Open circles denote intensity and closed circles, distance. Red, Las17; green, Ysc84; and purple, Sac6.
Figure 6.
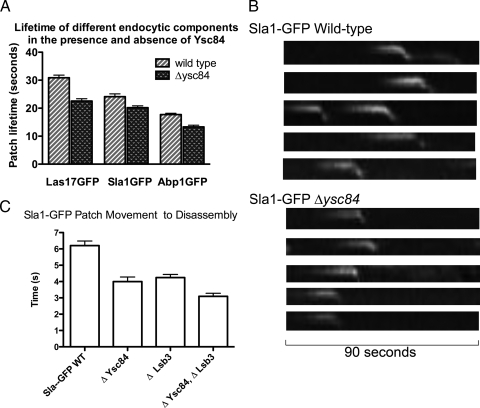
Deletion of Ysc84 affects the invagination stage of endocytosis. (A) Effect of deletion of ysc84 on markers of different stages of endocytosis was assessed. Lifetime of Sla1-GFP, Las17GFP, and Abp1GFP was assessed by measuring patch lifetime from movies recorded of cells expressing these markers. In all cases, there is a significant reduction in patch lifetime. More than 10 patches were assessed in wild-type and mutant strains. (B) The behavior of patches was also analyzed using kymographs with the marker Sla1GFP. As shown in the top panels in wild-type cells, Sla1GFP localizes to the plasma membrane and then shows a clear inward movement before disassembling. In the absence of ysc84 (bottom), the lifetime on the membrane and the extent of the inward movement is reduced. (C) To determine whether the Ysc84 homologue Lsb3 plays a distinct or overlapping role from Ysc84, the effect of deletion of lsb3 on the invagination step of endocytosis was also assessed. More than 15 patch lifetimes were measured for each strain. Differences between patch lifetimes in wild-type and Δysc84 and between Δysc84 and Δysc84Δlsb3 cells are significant (p value <0.001 and 0.0096, respectively).
Sequences encoding fluorescent protein tags were inserted into the genome by using an integrating polymerase chain reaction (PCR) strategy (Longtine et al., 1998). This was used to generate Las17-mRFP, Las17-GFP, Sla1GFP, and Abp1-GFP. All integrants were verified by colony-PCR amplification to confirm the replacement occurred at the expected locus. Functionality of strains was compared with untagged strains across a range of temperatures and for actin organization (using rhodamine phalloidin). Deletion strains (Δysc84 and Δlsb3) were also generated using the PCR integration approach. Plasmids and yeast strains used in this study are listed in Tables 1 and 2.
Table 2.
Yeast strains used in this study
| Yeast KAY no. | Genotype | Reference |
|---|---|---|
| 302 | MATa trp1-1, leu 2-3,112, lys2-801, his3-Δ200, ura 3-52 | Dewar et al. (2002) |
| 389 | MATatrp1-1, leu 2-3,112, lys2-801, his3-Δ200, ura 3-52 | Dewar et al. (2002) |
| 397 | KAY302 + integrated Sla1-GFP::HIS3 | Dewar et al. (2002) |
| 513 | KAY389 + Sla1GFP::TRP1 Δysc84::HIS3 | Dewar et al. (2002) |
| 510.1 | MATa, his3Δ200, leu2-3,112, ura3-52, trp1-1, lys2Δ, Δysc84::HIS3 | Dewar et al. (2002) |
| 997 | KAY389 + Δlsb3::LEU2 | This study |
| 934 | KAY510.1 + Δlsb3::LEU2 | This study |
| 446 | MATa, his3Δ1 leu2Δmet15Δura3Δ | Euroscarf |
| 473 | MATa Δlas17::KanMx his3Δ leu2Δ met15Δ ura3Δ | Euroscarf |
| 684 | MATa his3Δ1, leu2Δ, lys2Δ, ura3Δ Sac6mRFP::KanMx | Huh et al. (2003) |
| 1348 | KAY302 + Las17-7ala-mRFP::HIS3 | This study |
| 1308 | MATa/a ura3-52/ura3-52, trp1Δ63/TRP1, leu2Δ1/LEU2 + p416TEF1-YSC84-eGFP (FY1679) | Kals et al. (2005) |
| 1356 | MATaura3-52, trp1, leu2, + pTEF1-YSC84eGFP-(URA) | This study |
| 1357 | 684×1356 (Sac6mRFP/SAC6 + Ysc84-eGFP) | This study |
| 1358 | 1356×1348 (Las17mRFP/LAS17 + Ysc84-eGFP) | This study |
| pJ69-4a | MATatrp1-901, leu2-3,112, ura3-52, his3Δ200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, met2::GAL7-LacZ | James et al. (1996) |
Yeast Strains and Cell Growth
Cells were grown with rotary shaking at 30°C in liquid YPD medium (1% yeast extract, 2% Bacto-peptone, 2% glucose supplemented with 40 μg/ml adenine) or in synthetic medium (0.67% yeast nitrogen base and 2% glucose), with appropriate supplements. Transformations were performed using lithium acetate as described previously (Kaiser et al., 1994).
The yeast two-hybrid screen used bait and activation plasmids, and a yeast strain pJ69-4A as described previously (James et al., 1996). Matα strains carrying the binding domain constructs and MATa strains carrying the activation domain constructs were crossed, and diploids were selected on selective synthetic media (lacking tryptophan and leucine). Diploid strains were then grown overnight in selective liquid culture and assayed for β-galactosidase activity (Kaiser et al., 1994) or plated as serial (10×) dilutions onto plates lacking leucine, tryptophan, and histidine to allow combinations that trigger activation of reporter genes to be detected. Further stringency was made by adding 4 mM 3-aminotriazole to the plates lacking leucine, tryptophan, and histidine.
Protein Purification
Rabbit skeletal muscle actin was purified and gel filtered as described previously (Winder et al., 1995). Ysc84Nt and full length Ysc84 were cloned from pKA259 and modified by addition of a histidine-tag using a pTrcHis TOPO TA expression kit (Invitrogen). Plasmids pKA539 (Ysc84Nt) and 540 (Ysc84) were transformed into BL21 Escherichia coli, cells and they were grown to an OD600 = 0.6. Protein expression was induced by addition of isopropyl β-d-thiogalactoside to 1 mM for 3.5 h (37°C). Cell pellets were resuspended in 30 mM Imidazole, 1× phosphate buffer, and the protein was purified using HisTrap HP nickel columns (GE Healthcare) following the manufacturer's instructions. Ysc84 was then desalted into Ysc84 buffer (20 mM Tris, pH 8.0, 300 mM NaCl, and 0.1 mM dithiothreitol). pKA566 encoding GST-Las17 (300-422) was transformed into BL21 E. coli and induced as described above. The cell pellet was washed with phosphate-buffered saline (PBS), pH 7.0, and the cells lysed by sonication before being incubated with glutathione transferase (GST)-beads (GE Healthcare). GST was cleaved by incubation with Pre-Scission Protease (GE Healthcare) following the manufacturer's instructions. A plasmid expressing GST-Ysc84 SH3 domain was a gift from Gianni Cesareni (Rome, Italy). This plasmid was transformed into BL21 cells, and expression was induced as described above. Proteins were purified as for Las17 fragment, and the SH3 domain was cleaved from GST by using thrombin (Novagen, Madison, WI) overnight at room temperature.
Actin Assays
High- and low-speed pelleting assays and viscometry were performed as described previously (Winder et al., 1995). Fluorimetry assays were set up as for pelleting in 400-μl assays by using 5 μM actin. Pyrene actin was added to 5%. Polymerization salts-10× KME (500 mM KCl, 10 mM MgSO4, and 10 mM EGTA, pH 8.0) were added to 1× and mixed thoroughly before placing the assay in a fluorimetry cuvette. Polymerization as observed at settings excitation wavelength, 366 nm; emission, 384 nm, and slit width, 5 nm (RF-5301PC; Shimadzu Europe, Duisberg, Germany).
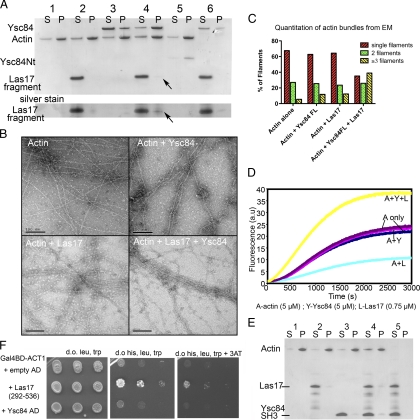
Electron microscopy was performed using assays set up as for pelleting. Samples were adsorbed on glow discharged carbon-coated copper grids and stained with 0.75% uranyl formate. Electron micrographs were recorded on a Philips CM100 electron microscope using a Gatan MultiScan 794 charge-coupled device camera. Quantification of electron micrographs was carried out using a stereologic approach. One-centimeter grids were placed over micrographs and every filament crossing a grid line was scored as to whether it was single, or associated with one or more other filaments (i.e., bundled). More than 200 filaments were scored for each condition.
Fluorescence Microscopy
Fluorescence microscopy was performed on an Olympus IX-81 inverted microscope with a DeltaVision RT Restoration Microscopy System running SoftWoRx image analysis and model-building application (Applied Precision Instruments, Seattle, WA). For live cell imaging, cells expressing tagged proteins were visualized after growing to early log phase in synthetic medium with appropriate supplements.
Time-lapse live cell imaging of GFP-tagged proteins was performed with 1-s time lapse. All image data sets were deconvolved; the distance of moving fluorescence spots was measured; and the arbitrary profile of intensity values, image coordinates, and tracking of patch movements were established using the SoftWoRx application. Images were exported as TIFF files, and image size was adjusted to 300 dpi and assembled using Adobe Photoshop CS2 (Adobe Systems, Mountain View, CA). Kymographs were assembled using ImageJ software (National Institutes of Health, Bethesda, MD). Statistical analysis of lifetimes was performed using GraphPad Prism software (GraphPad Software, San Diego, CA). Rhodamine phalloidin staining of actin structures was performed as described previously (Hagan and Ayscough, 2000).
RESULTS
Ysc84 N-Terminal Homology Domain Binds and Bundles Actin
Comparison of several Ysc84 homologues shows a distinct region with a high level of homology at the N terminus (Dewar et al., 2002). Between S. cerevisiae Ysc84 and human SH3yl-1 the identity in the region is 48%. This domain, or a fragment of it, has also been listed on databases as COG2930 and DUF 500. Based on the function that we demonstrate, we refer to this domain as the YAB domain, for Ysc84 Actin Binding. Interestingly, although we can identify clear homologues in vertebrate cells, no homologue has been found in any invertebrate, including Drosophila melanogaster, Caenorhabditis elegans, or Apis mellifera. In addition, a Ysc84 N-terminal homologous domain is found in plants, including Oryza sativa and Arabidopsis thaliana. However, in these organisms the domain seems to be localized toward the C-terminal region of the protein and is found in conjunction with a FYVE (Fab1, YOTB, Vac1, and EEA1) domain. An alignment and a phylogenetic tree of several vertebrate, fungal, and plant YAB domains is shown in Supplemental Figure 1, A and B. The second definable domain in Ysc84 family proteins is an SH3 domain at the extreme C terminus. Given the known interacting partners for many SH3 domains, it seemed less likely that the SH3 domain was involved in actin binding; so, constructs expressing the N-terminal region alone were generated. Ysc84Nt represents the first 218 amino acids of the sequence encompassing the YAB domain conserved across species. This protein was generated both as a His-tagged fusion and also an untagged protein.
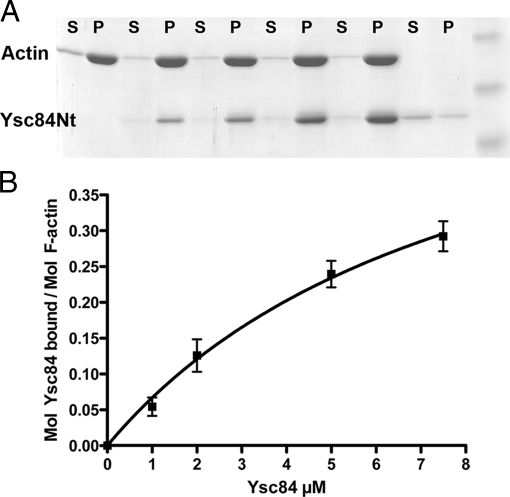
To determine whether Ysc84Nt is able to bind F-actin, high-speed pelleting assays were performed in which increasing concentrations of Ysc84Nt were incubated with actin. As shown in Figure 1A, Ysc84Nt is able to pellet in these conditions and therefore Ysc84Nt binds directly to F-actin. A dissociation constant was calculated by assessing binding of increasing concentrations of Ysc84Nt to actin (as described in Winder et al., 1995; Figure 1B). The value for Kd was calculated in GraphPad Prism software by taking the value at 50% of Bmax extrapolated from a nonlinear fit of the data. Bmax = 0.623 ± 0.16 mol Ysc84/mol actin, Kd = 8.4 ± 3.4 μM Ysc84.
Figure 1.
Ysc84Nt binds to F-actin. (A) Ysc84Nt can bind actin in a high-speed pelleting assay. Actin (5 μM) was induced to polymerize by addition of salts (see Materials and Methods) in the presence of increasing concentration of Ysc84Nt (1, 2, 5, and 10 μM). Copelleting of Ysc84Nt with actin is clearly observed. S, supernatant; P, pellet. (B) Densitometry was used to assess proportion of Ysc84Nt pelleting compared with actin over three independent experiments. A dissociation constant, Kd, of 8.4 μM was calculated.
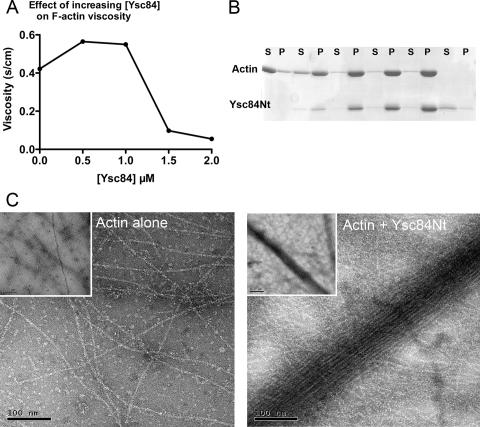
To address whether Ysc84 affected F-actin structure or dynamics, viscometry assays were performed. Ysc84Nt was copolymerized with actin and viscosity of the resulting solutions assessed in a falling ball assay. As shown (Figure 2A) the presence of Ysc84Nt initially increases viscosity, but at higher levels viscosity falls. This profile can be indicative of an actin-bundling activity, because at low levels of the protein, there is cross-linking that impedes movement of the ball, but as the levels of the binding protein increases, bundling is possible and these bundles produce channels in the fluid that permits faster passage of the ball. Further evidence for bundling was addressed in a low-speed pelleting assay. In this approach, only actin that has been bundled is able to sediment at these lower speeds (15,000 × g). As shown in Figure 2B, the presence of increasing concentrations of Ysc84 causes an increase in actin in the pellet. Finally, using electron microscopy, actin copolymerized with Ycs84Nt was analyzed. As shown in Figure 2C (right), Ysc84Nt is able to interact with, and bundle, actin filaments. Bundled filaments were not seen in the actin only control (Figure 2C, left). By gel filtration, Ysc84Nt runs as a monomer, indicating that the Ysc84Nt contains two actin-binding sites to facilitate bundling (data not shown).
Figure 2.
Ysc84Nt bundles F-actin. (A) Viscometry assays were performed in which Ysc84Nt was copolymerized with actin and viscosity of the resulting solutions was assessed in a falling ball assay as described previously (Winder et al., 1995). Shown is a representative assay of three independently conducted assays. (B) A low-speed pelleting assay at 15,000 × g with increasing concentration of Ysc84Nt with 5 μM actin shows that as Ysc84Nt levels increase, the proportion of actin in the pellet also increases. S, supernatant; P, pellet (C) Electron microscopy of negatively stained actin filaments polymerized alone or copolymerized with Ycs84NT demonstrates the presence of large, loose bundles of actin filaments in the presence of Ysc84Nt.
Given that these data used His-Ysc84Nt. We considered whether the His tag and linker region could play a part in actin binding and bundling. Purified untagged Ysc84Nt was generated and binding to F-actin is also observed (Figure 4A, lane 5).
Figure 4.
Las17 regulates Ysc84 actin binding. (A) Actin (5 μM) was polymerized and spun at high speed alone (1) with Las17 fragment (2), with Ysc84 full length alone (3), with both Las17 and Ysc84 (4), or with Ysc84Nt (5). Ysc84 and Las17 were also incubated and spun together to demonstrate that they did not interact to cause pelleting in the absence of actin (6). Lane 4 shows pelleting of actin and Ysc84 at about a 2:1 ratio, whereas Las17 fragment only pellets at substoichiometric levels. Pelleting is seen more clearly in the silver-stained section of the gel below. S, supernatant; P, pellet. (B) Electron microscopy of negatively stained actin filaments in the absence and presence of the Las17 fragment and full-length Ysc84. (C) A stereologic approach was applied to electron micrographs to quantify the effect of Ysc84 and Las17 on bundling. The number of filaments present in bundles of three or more filaments is shown to increase dramatically (>3-fold) only in the presence of both Ysc84 and Las17. (D) A pyrene actin assay was performed to assess the effect of Ysc84 and Las17 on actin polymerization. A, actin only controls; A+Y, actin + Ysc84; A+L, actin + Las17 fragment; and A+Y+L, actin + Ysc84 + Las17 (E) Actin was polymerized and spun at high speed alone (1), with Las17 fragment (2), with Ysc84 SH3 domain (3), and with both Las17 fragment and Ysc84 SH3 domain (4). Addition of Ysc84-SH3 domain to Las17 and actin does not increase actin polymerization. Las17 and Ysc84-SH3 domain in the absence of actin are only found in the supernatant (lane 5) (F) To confirm the possibility of the Las17 fragment binding to actin, a two-hybrid assay was performed with a Gal4Binding domain-ACT1 fusion.
Interestingly, during this biochemical analysis it was observed that Ysc84Nt does not interact with prepolymerized (filamentous) actin and only interacts with actin as polymerization initiates (Supplemental Figure 2). This finding indicates that Ysc84 is an atypical bundling protein, because the majority of known actin-bundling activities will bind and bundle preformed actin filaments.
Ysc84 SH3 Domain Interacts with a Specific Consensus Site in Las17
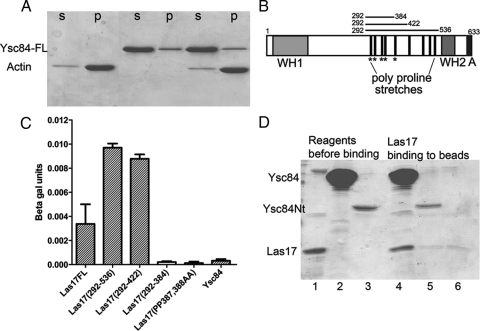
Having demonstrated an actin-binding activity for Ysc84Nt, it was then necessary to consider this function within the context of the full-length protein. Recombinant full-length Ysc84 was generated and incubated with actin to determine whether the presence of the SH3 domain and the linker region between these domains affected the activity of Ysc84 toward actin. Intriguingly, full length Ysc84 did not show any binding to actin in a high-speed, actin-pelleting assay (Figure 3A). This result suggests that the C-terminal region of Ysc84 may fold in such a way that it inhibits the actin-binding site. We then considered whether this potential autoinhibition of Ysc84 actin-binding could be relieved by an interaction with another protein. Such binding could alter the Ysc84 conformation to reveal its actin-binding site. Known interactors of Ysc84 were considered. Las17 has been reported to interact with Ysc84 C-terminal half (amino acids 270-468) in a two-hybrid screen (Madania et al., 1999). Further details of this interaction have not been reported. In addition, Sla1 had been demonstrated to bind to Ysc84 with the interaction mediated by the SH3 domain of Ysc84 to a specific site PAMPAR in Sla1 (amino acids 195-200) (Dewar et al., 2002; Gourlay et al., 2003). These data suggest that a possible site responsible for inhibition would be the SH3 domain. Because Las17 is known to be involved in actin polymerization, Las17 was selected for investigation (Li, 1997; Winter et al., 1999; Rodal et al., 2005).
Figure 3.
Ysc84 binds to a specific site in Las17. (A) Full-length ysc84 was expressed, purified, and incubated with actin. No binding was observed in a high speed pelleting assay. S, supernatant; P, pellet. (B) Schematic diagram of Las17 depicting recognized domains: WH1, WASP homology-1; WH2, WASP homology-2; and A, acidic. The central polyproline region contains both runs of prolines and type 1and 2 SH3-binding motifs. Motifs of the type PXXPXR are proposed to be the consensus binding site for the SH3 domain of Ysc84 (Landgraf et al., 2004; Beltrao and Serrano, 2005). Fragments used in the two-hybrid assay are shown. (C) Two hybrid assays were performed between Ysc84 and full-length Las17 or fragments of Las17. β-Galactosidase assays were used to assess the level of interaction. Deletion of the region 384-422 or mutation of the prolines P387 and P388 to alanines abrogated the interaction Errors are calculated from three independently conducted experiments. (D) An interaction between Las17 and Ysc84 was confirmed with recombinant proteins. His-tagged Ysc84 (Nt or full length) on nickel-agarose beads was incubated with GST-Las17. Lane 1, Las17; 2, Ysc84 full length; 3, Ysc84Nt; 4, Las17 bound to Ysc84 full length protein on beads; 5, Ysc84Nt beads with little Las17 bound; and 6, beads alone with background level of Las17 bound.
Two-hybrid analysis was performed with full-length Las17 or fragments of Las17 that carried its polyproline region or fragments thereof that contained reduced numbers of SH3-binding consensus sites (Figure 3B, schematic). As shown using β-galactosidase reporter assays, Ysc84 interacts with full-length Las17 and with the largest two Las17 fragments (amino acids 292-536 and 292-422) (Figure 3C). However, there was no interaction with the smaller fragment (amino acids 292-384). This interaction was confirmed with growth on plates containing appropriate selection (Supplemental Figure 3). Analysis of the sequence of this region revealed that the sequence truncated between 384 and 422 contained a single PXXPXR SH3 domain-binding consensus. This is the sequence motif that Ysc84 SH3 domain has been shown to bind to preferentially in phage display and other assays (Landgraf et al., 2004; Beltrao and Serrano, 2005). To determine whether this consensus marks a single Ysc84-binding site in Las17, it was mutated by in vitro mutagenesis from PPPPPR to AAPPPR (amino acids 387–392). This was then assessed and as shown (Figure 3C), this mutation abolished the Ysc84-Las17 two-hybrid binding interaction. As expected, deletion of Ysc84 SH3 domain also failed to interact with the Las17 fusion (Supplemental Figure 3).
To confirm that the Las17-Ysc84 interaction is direct, full-length Ysc84 was generated as a His-tagged fusion and bound to nickel agarose beads (Figure 3D, lane 2). The fragment of Las17 containing the majority of the sequence in the two-hybrid fusion was made as a GST fusion (amino acids 300-422) and then cleaved to yield a 12.7-kDa Las17 fragment. The controls for binding were beads carrying Ysc84Nt-6xHis, which has no SH3 domain (Figure 3D, lanes 3 and 5) and beads alone (Figure 3D, lane 6). As shown (Figure 3D), significant binding is only observed between full-length Ysc84 and the Las17 polyproline fragment (amino acids 300-422).
Yeast WASP Homologue Las17 Regulates Binding of Ysc84 to Actin
The effect of Las17 on the full-length Ysc84 interaction with actin was then assessed. If Las17 serves to regulate Ysc84 binding to actin, it is important to distinguish this from known effects of Las17 itself on actin dynamics. Rather than use full-length Las17, which has defined actin-binding sites at the WH2 domain close to its C terminus, the central polyproline region of Las17 that shows direct binding to Ysc84 was used (amino acids 300-422). This region has no reported interaction with actin.
High-speed pelleting assays were used to determine actin binding. These assays used recombinant untagged full-length Ysc84 and Ysc84Nt. As shown in Figure 4A, full-length Ysc84 alone does not interact with actin. However, addition of the Las17 fragment causes a marked shift of Ysc84 into the pellet demonstrating that Ysc84 is now able to interact with actin. The proportion of Ysc84 bound to actin in the presence of the Las17 fragment seems to be ∼1:2. Two other interesting observations can be made. First, that incubation of actin with the Las17 fragment alone causes a reproducible shift of actin from the pellet into the supernatant (Figure 4A, lane 2). This suggests that the Las17 fragment is binding to monomeric actin and preventing it from polymerizing in vitro, thereby potentially highlighting a novel actin-binding site in Las17. Second, in the presence of actin and Ysc84, whereas a small fraction of Las17 seems to enter the pellet, this proportion is clearly substoichiometric compared with Ysc84 and actin.
The effect of the Las17 fragment on the full-length Ysc84–actin interaction was also assessed by electron microscopy. As shown in Figure 4B, actin does not seem to be affected by Ysc84 or Las17 fragment alone, but when added together bundles can be observed. Quantitation of electron micrographs of actin filaments by using stereological approaches, in the presence and absence of Ysc84 and Las17 fragment, demonstrates a clear requirement for both proteins to be present to bundle actin filaments (Figure 4C). Thus, the Las17 fragment activates the actin-bundling activity of full-length Ysc84.
Las17 is known to be a nucleation-promoting factor that activates Arp2/3 to drive actin polymerization. We considered whether Las17 could also activate Ysc84 to have a similar activity. Using a pyrene actin assay, full-length Ysc84 and the Las17 fragment were added to actin and polymerization was initiated by addition of salts. As shown (Figure 4D), Ysc84 alone has no effect on actin polymerization. This is predicted as we had demonstrated (Figure 3A) that Ysc84 does not seem to bind to actin. Interestingly, the Las17 fragment alone inhibits actin polymerization kinetics providing further evidence for another actin interacting site on Las17 in addition to those previously identified. Combination of the Las17 fragment with Ysc84 caused a significant increase in the actin polymerization rate revealing a new mode of action of Las17 in stimulating actin filament formation.
To exclude the possibility that the Ysc84-SH3 domain is activating an unknown Las17 nucleation function, and to more conclusively demonstrate that the increased polymerization observed required the Ysc84 actin-binding region, a further control experiment was carried out. Ysc84-SH3 domain alone was expressed and incubated with actin alone, or with actin and Las17 fragment. As shown in Figure 4E, lane 2, incubation of actin with Las17 alone causes a shift of actin into the supernatant (S) from the pellet (P). In the presence of Ysc84-SH3 domain alone or with Las17 + Ysc84 SH3 domain, this shift of actin to the supernatant remains or in fact slightly increases (Figure 4E, lanes 3 and 4) demonstrating that the observed increase in actin polymerization requires the presence of the N-terminal Ysc84 actin-binding domain.
Given the possibility of the Las17 fragment interacting with monomeric actin a two-hybrid assay was performed with actin fused to the Gal4-binding domain and Las17 fragment to the Gal4-activation domain. The actin fusion has been used previously to reveal interactions with several monomer binding activities in yeast (Amberg et al., 1995). As shown (Figure 4F), the Las17 fragment (amino acids 292-536) but not full-length Ysc84 shows a clear interaction with actin.
Due to ease of purification, the studies thus far used rabbit muscle actin to investigate Ysc84 interaction. Rabbit muscle actin is widely used in actin-binding studies for proteins from a wide range of sources, including plants, fungi, and mammalian nonmuscle cells. However, to be certain that the interactions identified are genuine and physiological, we sought to confirm that these results by using yeast actin. Supplemental Figure 4 demonstrates that Ysc84Nt and full-length Ysc84 behave in the same way toward yeast actin as with rabbit muscle actin. Briefly, we show Ysc84Nt can bind and bundle yeast actin (Supplemental Figure 4, A and D) and that full-length Ysc84 can be induced to bind and bundle yeast actin in the presence of Las17(300-422) fragment (Supplemental Figure 4, B and C).
Function of Ysc84 in Endocytosis
The above-mentioned data demonstrate that a fragment of Las17 is able to bind full-length Ysc84 and activate its ability to regulate actin dynamics in vitro and the two-hybrid analysis indicates the interaction can occur within cells. Large-scale proteomic studies have also demonstrated interactions between Las17 and Ysc84, including isolation in complexes from cells (Gavin et al., 2002; Tong et al., 2002; Landgraf et al., 2004; Tarassov et al., 2008). However, to date, no role has been shown for Ysc84 in endocytosis. To gain an understanding of the stage at which Ysc84 is functioning in endocytosis, we investigated the localization of GFP-Ysc84 relative to both Las17-mRFP and Sac6-mRFP. We had previously visualized Ysc84 expressed under the Gal promoter but growth on galactose often causes morphological effects that we wished to avoid (Dewar et al., 2002). We were not able to visualize Ysc84 tagged at its C terminus in the genome possibly because of its levels of expression. We therefore used a C-terminally enhanced green fluorescent protein (eGFP)-tagged Ysc84 expressed under the TEF1 promoter (gift from Klaus Natter, University of Graz, Austria; Kals et al., 2005). Strains expressing Ysc84-eGFP were then crossed with those expressing either Sac6-RFP or Las17-RFP. In cells containing either Las17-mRFP or Sac6-mRFP with Ysc84-eGFP, colocalization was clear (Figure 5A). Distance intensity measurements (Figure 5B) indicate that Ysc84 arrives several seconds after Las17 at the endocytic site; however, it seems to move inward further than Las17 and continues to remain at the patch after Las17 has disassembled. In addition, the peak of Ysc84 intensity is reached only at the point at which invagination commences. The lifetime of Ysc84 at the patches is 24.25 ± 1.1 s compared with 30.9 ± 0.93 s for Las17 (errors are SEM). Ysc84 arrives earlier than Sac6, supporting the idea that Ysc84 may function early in the actin polymerization process. Both proteins show clear inward movement including fast movement away from the membrane.
Previous work demonstrated that although Ysc84 colocalizes with cortical actin patches, deletion of YSC84 has no effect on fluid phase endocytosis as determined in a Lucifer yellow fluorescence uptake assay (Dewar et al., 2002). However, since this earlier study, more sensitive assays have been developed using live cell imaging and the analysis of the kinetic behavior of proteins involved in endocytosis (Kaksonen et al., 2003). Many proteins previously observed to have no endocytic phenotype are now considered core proteins in the process; these proteins include capping protein and the bundling proteins Abp1 and Scp1 (Kaksonen et al., 2005; Gheorghe et al., 2008). Three proteins were analyzed in the presence and absence of ysc84. These were Sla1 (marker of early–mid endocytosis), Abp1 (marker of mid–late endocytosis) and Las17 (actin nucleation promoting factor). Analysis of the lifetimes of complexes containing these proteins revealed that in all three cases the patch lifetime was reduced (Figure 6A).
Kymographs comparing wild-type and Δysc84 cells (Figure 6B) reveal that although many patches do show inward movement of Sla1GFP, this movement is markedly reduced. Quantitation of patch movements from the plasma membrane indicates a reduction of inward movement from 275 ± 20.5 to 202 ± 15.4 nm (p value 0.0055). We also considered the proportion of patches that fail to show any internalization at all in the mutant. A failure to internalize at all would also strongly indicate a role in early stage of actin polymerization as actin is the major driving force for invagination. In wild-type cells, 5% of Sla1GFP spots that form on the plasma membrane showed no movement away from the membrane (n = 158). In the absence of ysc84, the number of failed endocytic events increased to 15% (n = 123). Thus, lack of ysc84 leads to a threefold increase in the number of spots that form but that are subsequently unable to move inward into the cell.
We also considered the potential role of the Ysc84 homologue Lsb3. Again this protein has been shown to localize to actin patches in vivo and interacts with both Las17 and Sla1 in two-hybrid studies (Madania et al., 1999; Dewar et al., 2002). Thus, Lsb3 might have a function that overlaps with that of Ysc84. We generated strains in which both ysc84 and lsb3 were deleted to determine the effect on Sla1GFP endocytic patch lifetimes. We examined the time from when the patch began to move, to the point of disassembly in detail as our analysis above indicated that this was the primary difference between wild-type and Δysc84 patch movement. As shown in Figure 6C, the effect of the additional deletion of LSB3 seems additive indicating that there is a level of overlap between the functions of these proteins within the endocytic patch.
DISCUSSION
Recent years have seen great advances in our understanding of the process of endocytosis. In particular, studies in yeast have yielded a wealth of information about key stages in the assembly and disassembly of several complexes important in driving the invagination of plasma membrane and the formation of endocytic vesicles. However, the endocytic process is highly complex, and the function of many proteins that localize to the sites of endocytosis is almost completely unknown.
In this study, we present data revealing that Ysc84 is a conserved protein that is able to regulate actin dynamics. The actin-binding region of Ysc84 lies at its N terminus and is 48% identical to this domain in its human counterpart SH3yl-1. Given that the level of homology between yeast and human profilin is 34%, cofilin is 38%, and capping protein is 30%, this 180-amino acid domain is one of the most highly conserved domains that interacts directly with actin. Interestingly, the domain contains no recognizable homologies, indicating a completely new class of actin-binding proteins. We have called the domain the YAB domain for Ysc84 Actin Binding.
In vitro approaches were used to investigate the actin-binding properties of the N-terminal domain of Ysc84. We demonstrate that the Ysc84 N-terminal region (1-218), including the YAB domain (1-184), can bind and bundle actin. This activity was not typical of many bundling proteins, however, because binding was only apparent if the domain was present and potentially loaded onto filaments during polymerization. Surprisingly, full-length Ysc84 did not bind actin at all, suggesting that another protein binds Ysc84 to activate its actin-binding function. We identified a region in the yeast WASP homologue Las17 as this activator of Ysc84 actin binding. Binding occurs between Ysc84 SH3 domain and the polyproline region of Las17. Additional experiments indicated that this region of Las17 is also the preferred binding region for SH3 domain of the Ysc84 related protein Lsb3 (Supplemental Figure 3). Intriguingly, the construct carrying Las17(292-422) interacts with Ysc84, whereas Las17(292-384) does not. The former contains a single PXXPXR site (387PPPPPR392), which fits a consensus binding site identified for Ysc84 (Landgraf et al., 2004; Beltrao and Serrano, 2005). The latter contains four sites that still fit the overall consensus PXXPXR indicating that the SH3 domain of Ysc84 is able to show discrimination beyond this broad consensus, or that elements outside of the consensus can promote specific binding. This is the first SH3 binding site mapped to a specific binding motif within the polyproline region of Las17. It highlights the possibility that despite the plethora of SH3 and WW domain consensus sites with apparently overlapping specificities there is the possibility for much greater specificity than previously reported and these interactions could be both competitive or cooperative in nature. Interestingly, there are only two other proteins in the yeast proteome containing PPPPPR—Abp1 and Scd5—both proteins also found at endocytic sites. Our previous work suggested a possible role for Abp1 in localizing Ysc84 (Dewar et al., 2002), but a physical interaction between Ysc84 and either Abp1 or Scd5 has not been reported.
The Las17 interaction with Ysc84 does not simply allow Ysc84 to bind actin, this complex also promotes actin polymerization. Neither the Las17 fragment alone, nor Ysc84Nt are able to increase levels of actin polymerization, demonstrating that this is a function only possible when the proteins are present together with actin. Importantly, this action of Las17 does not require Arp2/3 nor does it involve regions previously identified to interact with actin. The polymerization driven by Ysc84-Las17 is markedly different from that driven by Arp2/3 alone or in conjunction with Las17. First, the levels of Ysc84 are relatively high (micromolar) compared with Arp2/3 (nanomolar). This might indicate that in association with Las17, Ysc84 is potentially organizing actin in a conformation that favors polymerization but that this conformation is less productive for driving polymerization than that generated by Arp2/3. Second, many of the resulting filaments polymerized in the presence of Las17 and Ysc84 are loosely bundled, and they do not show branching as observed with Arp2/3. These data highlight a new role for Las17 in an Arp2/3-independent actin polymerization pathway.
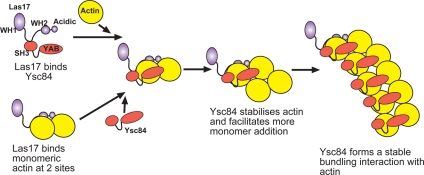
The question then arises, how is this fragment of Las17 functioning to promote actin polymerization when it lacks known actin binding sites and when Ysc84 alone does not bind actin monomer? Three pieces of data indicate that the Las17 polyproline fragment is capable of interacting with monomeric actin. First, Las17(300-422) seems to shift a portion of actin (yeast or rabbit muscle) from pellet to the supernatant in a high-speed pelleting assay (Figure 4A and Supplemental Figure 4B). The same fragment of Las17 also inhibits polymerization in a pyrene actin assay (Figure 4D). Finally, a two-hybrid assay demonstrates an interaction between Las17(292-536) and actin. These data provide crucial data for the function of Las17 as they suggest that Las17 can potentially bind two actin monomers—one at the site delineated by 300-422 and one at its C-terminal WH2 domain (Li, 1997; Winter et al., 1999; Rodal et al., 2005). This result could then explain previous data in which the effect of the Las17 WA fragment (WH2 + acidic domains, amino acids 547-633) on Arp2/3 polymerization of actin was considerably less than when full-length Las17 was incubated with Arp2/3 (Rodal et al., 2003). Figure 7 outlines a model to describe how Las17 and Ysc84 could function together to promote actin polymerization in the absence of Arp2/3. Briefly, Las17 could present two actin monomers to Ysc84, which, when bound to Las17 can bind these monomers, so generating a stable dimer. This dimer can then presumably be extended to form filaments with Ysc84 remaining associated with the extending filament and acting as a bundling protein. The generation of a stable dimer rather than trimer may explain differences in the efficiency of Ysc84 versus Arp2/3 in actin polymerization assays. In vivo, the two monomeric binding sites on Las17 could potentially promote both Ysc84-based actin polymerization and Arp2/3 polymerization. It is also possible in vivo that Arp2/3 and Ysc84 polymerization mechanisms are not mutually exclusive. This might be supported by a recent report of an interaction between Ysc84 and Arc40 (Tarassov et al., 2008).
Figure 7.
Model for actin polymerization by Las17 and Ysc84. Our data indicate that in cells, Las17 is able to bind two actin monomers—one monomer at its previously recognized WH2, actin-binding domain and one monomer at the region defined in this study encompassed by amino acids 300-422. When Ysc84-Las17 and actin associate in a ternary complex, Ysc84 is able to bind to and stabilize the actin dimer presented by Las17 and thereby promote polymerization. Ysc84 can then remain associated with actin and in this way actin becomes bundled. In the absence of Las17, Ysc84 is not able to interact with actin most likely because its own SH3 domain sterically blocks binding.
In vivo we reveal that Ysc84 arrives at the endocytic site several seconds after Las17 but before Sac6 and also before Arp2/3 (Kaksonen et al., 2003). This suggests the possibility that a Las17-Ysc84–dependent actin polymerization event could precede an Arp2/3–Las17 interaction. In addition, and contrary to previous observations (Dewar et al., 2002), there is an effect of ysc84 deletion on endocytosis. Three proteins involved in endocytosis were all shown to have reduced lifetimes in the absence of YSC84. This phenotype was exacerbated in the absence of the Ysc84 homologue Lsb3, indicating that these proteins have at least partially overlapping functions (Figure 6). The endocytic phase that corresponds to the defect is the invagination step, when actin is being actively polymerized and organized to provide a framework to support inward movement. In the absence of ysc84, the inward movement is reduced, indicating that invagination is less effective. Furthermore, the number of failed endocytic events that show no invagination at all is increased threefold. Given that actin is providing the inward force for invagination, this phenotype is entirely consistent with the idea that Ysc84 functions to contribute to initial stages of actin polymerization required to drive inward membrane movement.
In summary, our data presents important new findings that will have significant implications for the study of actin function in yeast, and also in the studies of homologues of Ysc84 and Las17 in other organisms. We identify a new class of actin-binding proteins with a distinct actin-binding domain termed YAB (Ysc84 actin binding), and we demonstrate a role for Ysc84 during polymerization stages of filament formation. Furthermore, new insight into the role of the WASP homologue Las17 is revealed. First, Las17 regulates Ysc84 actin association and second we identify a second actin monomer binding site. Together, these activities allow Las17 to function in driving actin polymerization in the absence of Arp2/3.
Supplementary Material
ACKNOWLEDGMENTS
We thank Phil Mitchell for critical reading of the manuscript, John Rafferty and Chatrudee Suwannachart (University of Sheffield) for untagged Ysc84, Klaus Natter (University of Graz) and Peter Piper (University of Sheffield) for yeast strains, and Gianni Cesareni (University Roma) for GST-Ysc84-SH3 domain plasmid. The work was supported by Medical Research Council Senior nonclinical fellowships G117/394 and G0601600 (to K.R.A.) and Biotechnology and Biological Sciences Research Council studentships (to A. R., A. S., and F. G.). The light microscopy-imaging center at the University of Sheffield was funded by grant GR077544AIA from the Wellcome Trust.
Abbreviations used:
- SH3
Src homology 3
- YAB
Ysc84 actin binding.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0982) on January 21, 2009.
REFERENCES
- Amberg D. C., Basart E., Botstein D. Defining protein interactions with yeast actin in-vivo. Nat. Struct. Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Aoki N., Ito K., Ito M. A novel mouse gene, Sh3yl1, is expressed in the anagen hair follicle. J. Invest. Dermatol. 2000;114:1050–1056. doi: 10.1046/j.1523-1747.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D. G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P., Serrano L. Comparative genomics and disorder prediction identify biologically relevant SH3 protein interactions. PLoS Comput. Biol. 2005;1:e26. doi: 10.1371/journal.pcbi.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R., Ayscough K. R. Interactions between Sla1p, Lsb5p and Arf3p in yeast endocytosis. Biochem. Soc. Trans. 2005;33:1273–1275. doi: 10.1042/BST0331273. [DOI] [PubMed] [Google Scholar]
- Dewar H., Warren D. T., Gardiner F. C., Gourlay C. G., Satish N., Richardson M. R., Andrews P. D., Ayscough K. R. Novel proteins linking the actin cytoskeleton to the endocytic machinery in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:3646–3661. doi: 10.1091/mbc.E02-05-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta B. J., Chuang D. Y., Cooper J. A. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 2008;6:72–85. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A. C., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gheorghe D. M., Aghamohammadzadeh S., Smaczynska de-Rooij I. I., Allwood E. G., Winder S. J., Ayscough K. R. Interactions between the yeast SM22 homologue Scp1 and actin demonstrate the importance of actin bundling in endocytosis. J. Biol. Chem. 2008;283:15037–15046. doi: 10.1074/jbc.M710332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C. W., Dewar H., Warren D. T., Costa R., Satish N., Ayscough K. R. An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J. Cell Sci. 2003;116:2551–2564. doi: 10.1242/jcs.00454. [DOI] [PubMed] [Google Scholar]
- Hagan I. M., Ayscough K. R. Fluorescence microscopy in yeast. In: Allan V. J., editor. Protein Localization by Fluorescence Microscopy: A Practical Approach. Oxford, NY: Oxford University Press; 2000. pp. 179–205. [Google Scholar]
- Huckaba T. M., Gay A. C., Pantalena L. F., Yang H. C., Pon L. A. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 2004;167:519–530. doi: 10.1083/jcb.200404173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Kals M., Natter K., Thallinger G. G., Trajanoski Z., Kohlwein S. D. YPL.db(2): the Yeast Protein Localization database, version 2.0. Yeast. 2005;22:213–218. doi: 10.1002/yea.1204. [DOI] [PubMed] [Google Scholar]
- Kim K., Galletta B. J., Schmidt K. O., Chang F. S., Blumer K. J., Cooper J. A. Actin-based motility during endocytosis in budding yeast. Mol. Biol. Cell. 2006;17:1354–1363. doi: 10.1091/mbc.E05-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübler E., Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf C., Panni S., Montecchi-Palazzi L., Castagnoli L., Schneider-Mergener J., Volkmer-Engert R., Cesareni G. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2004;2:94–103. doi: 10.1371/journal.pbio.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J. Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Madania A., Dumoulin P., Grava S., Kitamoto H., Scharer-Brodbeck C., Soulard A., Moreau V., Winsor B. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell. 1999;10:3521–3538. doi: 10.1091/mbc.10.10.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S., Rohrer J., Crausaz F., Riezman H. end3 and end 4, two mutants defective in receptor-mediated endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal A. A., Manning A. L., Goode B. L., Drubin D. G. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr. Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- Rodal A. A., Sokolova O., Robins D. B., Daugherty K. M., Hippenmeyer S., Riezman H., Grigorieff N., Goode B. L. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- Smythe E., Ayscough K. R. Actin regulation in endocytosis. J. Cell Sci. 2006;119:4589–4598. doi: 10.1242/jcs.03247. [DOI] [PubMed] [Google Scholar]
- Tarassov K., Messier V., Landry C. R., Radinovic S., Molina M.M.S., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S. W. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Tong A.H.Y., et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- Warren D. T., Andrews P. D., Gourlay C. G., Ayscough K. R. Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J. Cell Sci. 2002:115. doi: 10.1242/jcs.115.8.1703. [DOI] [PubMed] [Google Scholar]
- Winder S. J., Hemmings L., Maciver S. K., Bolton S. J., Tinsley J. M., Davies K. E., Critchley D. R., Kendrick Jones J. Utrophin actin-binding domain—analysis of actin-binding and cellular targeting. J. Cell Sci. 1995;108:63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- Winter D., Lechler T., Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.