Abstract
Recombination is important for DNA repair, but it can also contribute to genome rearrangements. RecQ helicases, including yeast Sgs1 and human BLM, safeguard genome integrity through their functions in DNA recombination. Sgs1 prevents the accumulation of Rad51-dependent sister chromatid junctions at damaged replication forks, and its functionality seems to be regulated by Ubc9- and Mms21-dependent sumoylation. We show that mutations in Smc5-6 and Esc2 also lead to an accumulation of recombinogenic structures at damaged replication forks. Because Smc5-6 is sumoylated in an Mms21-dependent manner, this finding suggests that Smc5-6 may be a crucial target of Mms21 implicated in this process. Our data reveal that Smc5-6 and Esc2 are required to tolerate DNA damage and that their functionality is critical in genotoxic conditions in the absence of Sgs1. As reported previously for Sgs1 and Smc5-6, we find that Esc2 physically interacts with Ubc9 and SUMO. This interaction is correlated with the ability of Esc2 to promote DNA damage tolerance. Collectively, these data suggest that Esc2 and Smc5-6 act in concert with Sgs1 to prevent the accumulation of recombinogenic structures at damaged replication forks, likely by integrating sumoylation activities to regulate the repair pathways in response to damaged DNA.
INTRODUCTION
DNA damage arising from exogenous or endogenous sources can block the progression of DNA replication and thereby lead to mutations or irresolvable lesions that cause cell death. Appropriate repair of DNA damage is consequently crucial for genome integrity, and posttranslational modifications such as checkpoint dependent phosphorylation, ubiquitylation, and sumoylation have been shown to modulate the recruitment and activities of multiple proteins and pathways that are key to DNA repair and cell survival (Branzei and Foiani, 2008).
Replication forks encountering DNA lesions can restart by repriming downstream of the lesion (Heller and Marians, 2006), generating single-strand gaps behind replication forks (Lehmann and Fuchs, 2006; Lopes et al., 2006). Two pathways of gap filling have been proposed. One pathway uses a combination of replicative and translesion synthesis polymerases to replicate across the lesion, and in such situations the bypass can occur either in error-free or error-prone manners (Lehmann and Fuchs, 2006; Branzei and Foiani, 2007). The other gap-filling mechanism, referred to as the template switch (TS) pathway, is essentially error-free and uses the undamaged information of the sister DNA duplex by using a mechanism that shares similarities with homologous recombination (HR) (Higgins et al., 1976; Goldfless et al., 2006; Branzei and Foiani, 2007).
The TS process gives rise to transient, hemicatenane-like or pseudodouble Holliday Junction intermediates that were shown to require the activity of the RecQ helicase Sgs1/BLM and Top3 for their resolution (Wu and Hickson, 2003; Liberi et al., 2005; Suski and Marians, 2008). Accordingly, cruciform, X-shaped intermediates with biochemical properties of pseudodouble Holliday junctions have been shown to accumulate at damaged replication forks in mutants affecting the functionality of the Sgs1-Top3 complex (Liberi et al., 2005; Mankouri et al., 2007). Recently, it was proposed that the ability of Sgs1 to promote dissolution of the hemicatenane-like structures formed during replication of damaged templates is regulated by Ubc9- and Mms21-dependent sumoylation events (Branzei et al., 2006). Notably, budding yeast Sgs1 and its functional counterpart in human cells, BLM, are sumoylated (Eladad et al., 2005; Branzei et al., 2006), but Sgs1 sumoylation is independent of the SUMO ligase activity of Mms21 (Branzei et al., 2006), suggesting that there must be other SUMO-targets involved in this process.
To gain further insight into the factors that may act in coordination with Sgs1 to prevent the accumulation of hemicatenane-like structures at damaged replication forks (Liberi et al., 2005; Branzei et al., 2006), we have examined in this study several yeast mutants affecting DNA repair or chromosome metabolism processes. In this way, we established that mutations in SMC6 and ESC2 result in impaired ability to resolve these repair intermediates. Structural maintenance of chromosome (Smc) proteins play fundamental roles in chromosome organization and dynamics as well as in DNA repair (Losada and Hirano, 2005), and Smc orthologues have been identified in all eukaryotic organisms studied. There are six Smc proteins that act in pairs to form the core of three SMC protein complexes: Cohesin, Condensin, and the Smc5-6 complex (Losada and Hirano, 2005).
The Smc5-6 complex is central for repair of DNA damage, functioning in the same pathway as Rad51 and Rad52 (Lehmann et al., 1995; Verkade et al., 1999; Morikawa et al., 2004; Onoda et al., 2004) and plays a role in the rescue of stalled and collapsed replication forks (Fousteri and Lehmann, 2000; Morikawa et al., 2004; Ampatzidou et al., 2006; Lindroos et al., 2006). In both budding and fission yeast, the Smc5-6 complex consists of Smc5, Smc6, and six non-Smc proteins: Nse1, Nse2 (Mms21), Nse3 (Ydr288W), Nse4 (Qri2), Nse5 (YML023c), and Nse6 (Kre29) (Hazbun et al., 2003; Sergeant et al., 2005; Pebernard et al., 2006). Mms21 (Nse2) has SUMO ligase activity and promotes sumoylation of several proteins, including Smc5-6 (Andrews et al., 2005; Potts and Yu, 2005; Zhao and Blobel, 2005). In budding yeast, YML023c (Nse5) interacts by two-hybrid with Ubc9, Smt3 (yeast SUMO), and Slx5 (Hazbun et al., 2003), which forms a heterodimer with Slx8 (Yang et al., 2006). Slx5-Slx8 complex is functionally associated with the SUMO pathway (Hannich et al., 2005; Wang et al., 2006; Burgess et al., 2007) and displays ubiquitin ligase activity (Ii et al., 2007b; Uzunova et al., 2007; Xie et al., 2007). SUMO attachment to a substrate stimulates Slx5-Slx8 dependent ubiquitination, primarily through direct noncovalent interactions between SUMO and Slx5 (Ii et al., 2007a; Ii et al., 2007b; Uzunova et al., 2007; Xie et al., 2007).
The Schizosaccharomyces pombe DNA repair factor Rad60 and the Saccharomyces cerevisiae Esc2 protein contain SUMO-like domains (Novatchkova et al., 2005), and they may represent candidate targets for SUMO-mediated Slx5-Slx8-dependent proteolysis (Prudden et al., 2007; Sun et al., 2007). Intriguingly, Rad60 physically associates with the Smc5-6 complex (Boddy et al., 2003), and based on genetic consideration, it has been proposed to act in coordination with Smc5-6 to promote the repair of structures that may arise during replication (Morishita et al., 2002; Boddy et al., 2003; Miyabe et al., 2006). rad60 mutants are synthetic lethal with mutations in SMC6 (Morishita et al., 2002; Miyabe et al., 2006), and similar to smc6 mutants, they are synthetic lethal with the S. pombe RecQ (Sgs1) orthologue, rqh1 (Miyabe et al., 2006). In S. cerevisiae, mutation in esc2 has also been reported to be slow growing in combination with sgs1 (Tong et al., 2001) and to affect gene silencing (Dhillon and Kamakaka, 2000; Cuperus and Shore, 2002; Andrulis et al., 2004) and sister chromatid cohesion and life span (Ohya et al., 2008).
In our effort to uncover other factors implicated together with Sgs1 and sumoylation in preventing the accumulation of recombinogenic structures during replication, we identified Smc6 and Esc2. Furthermore, we have characterized their relationships to Sgs1 and the SUMO pathway. Our results suggest that Esc2 and Smc6 act to promote damage tolerance during replication.
MATERIALS AND METHODS
Yeast Strains and Plasmids
The yeast strains used in this study are derivatives of DF5 or W303 and the relevant genotypes are shown in Supplemental Table 1. The plasmids BD-TOP3, AD-SMT3, AD-UBC9, and AD-SGS1 have been described previously (Branzei et al., 2006). The BD-ESC2 fusion plasmids were gifts from D. Shore (University of Geneva, Switzerland) and R. Sternglanz (Stony Brook University, Stony Brook, NY) and are described in Cuperus and Shore (2002) and Andrulis et al. (2004).
Growing Conditions, Cell Cycle Arrest, and Drug Treatments
Synchronization with α-factor or nocodazole and release from the cell cycle arrests were performed as described previously (Liberi et al., 2005; Branzei et al., 2006). Unless otherwise indicated, cells were grown at 25°C and released at 30°C, and methyl-methane sulfonate (MMS) and hydroxyurea (HU) concentrations were used at a final concentration of 0.033% (vol/vol) and 0.2 M, respectively.
DNA Extraction, Two-dimensional (2D) Gel Technique, and Fluorescence-activated Cell Sorting (FACS) Analysis
Purification of DNA intermediates, FACS analysis, and 2D gel procedure were carried out as described previously (Branzei et al., 2006). The DNA samples were digested with HindIII and EcoRV and analyzed by 2D gel with probes against ARS305.
Spot Assays of Drug Sensitivity
Logarithmic phase cells were counted and 10-fold series dilutions were spotted on plates containing the indicated concentrations of drugs and incubated at 25°C or at the indicated temperatures for 3 d.
Two-Hybrid Assays
The strain Y190 (FY0101) or its derivatives, Y190 his3Δ (HY0803) and Y190 esc2Δ (ΗY0705), were transformed with combinations of two plasmids, containing genes fused to the binding domain (BD) or the activation domain (AD) of the GAL4 protein. The β-galactosidase (β-gal) assay was performed as described previously (Branzei et al., 2006).
Protein Techniques (Protein Extracts, Antibodies, Fast-Performance Liquid Chromatography Gel Filtration, and Coimmunoprecipitation)
Western blot analysis, trichloroacetic acid extraction of yeast proteins, gel filtration chromatography, and coimmunoprecipitation experiments were performed as described previously (Chiolo et al., 2005). 9Myc-Sgs1, 13Myc-Esc2, and 3FLAG-Smc6 were analyzed using the monoclonal antibodies 9E11 (Neo-Markers, Fremont, CA) for α-Myc and M2 (Sigma) for α-FLAG. Rad53 was detected with the mouse monoclonal EL7 antibody (a gift from A. Pellicioli, University of Milan, Italy), proteins fused to Gal4BD with the rabbit polyclonal sc577 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and sumoylation with a α-SUMO rabbit antibody (a gift from X. Zhao, Sloan-Kettering Cancer Center, New York, NY).
RESULTS
Smc6 Prevents Accumulation of Sister Chromatid Junctions at Damaged Replication Forks
We previously found that the Ubc9-Mms21 sumoylation pathway prevents the accumulation of X-shaped structures (pseudodouble Holliday junctions) at damaged replication forks likely by affecting Sgs1 functionality (Liberi et al., 2005; Branzei et al., 2006). Sgs1 is sumoylated in vivo, but in a manner independent of Mms21 (Branzei et al., 2006), and it is still unclear whether Sgs1 sumoylation per se is required for Sgs1 ability to resolve such structures (Branzei et al., 2006). Together, the previous findings suggest that other SUMO targets are involved in this process.
As an approach to identify such factors, we examined various S. cerevisiae mutant strains impaired in chromosome metabolism processes. Thus, by 2D gel electrophoresis, we analyzed the profile of replication intermediates formed during replication in the presence of MMS at ARS305, an early efficient replication origin on chromosome III (Figure 1A). The mutants tested were selected on the basis of them being hypersensitive to MMS or having well-documented functions in DNA repair (rad18, rad5, exo1, rad55, rad59, cdc2, pol32, esc4, rrm3, chl1, smc5, and smc6), being defective in chromosome cohesion, segregation, or condensation processes (eco1, scc1, smc1, smc3, smc2, top2, and pds5), or mutated in a way that could affect SUMO metabolic processes (slx5, slx8, and esc2). As described below, of the mutants tested, only smc6 and esc2 recapitulated the X-molecule accumulation phenotype of ubc9-1, mms21, and sgs1 cells.
Figure 1.
smc6 mutants accumulate X-molecules during replication of damaged templates. (A) Representation of the genomic region containing the ARS305 origin on chromosome III. E and H stand for EcoRV and HindIII, respectively. (B) Left, profile of replication intermediates of wild type (SY2234) and smc6-9 (SY2232) replicating in MMS. Right, diagram of the replication intermediates visualized by 2D gel. (C) Schematic representation of the mutations identified in smc6-9 (SY2232) and smc6-56 (FY1103) alleles. (D) Replication intermediates of wild type (SY2234), smc6-9 (SY2232), and smc6-56 (FY1103) in MMS.
First, we found that a mutation in SMC6, smc6-9, which causes hypersensitivity to DNA damaging agents (Torres-Rosell et al., 2005), accumulates X-shaped molecules at replication forks during replication in the presence of sublethal doses of MMS (Figure 1B). Previously, at restrictive temperatures, smc6 mutants were shown to accumulate branched recombination structures at ribosomal DNA (rDNA) regions (Torres-Rosell et al., 2005) that could also account for the material remaining in wells by pulse-field gel electrophoresis (Lindroos et al., 2006). However, we found that in high-temperature conditions that inactivate Smc6 function, replication forks arising at normal origins of replication, such as ARS305, do not display an aberrant pattern of replication intermediates and do not show an accumulation of branched molecules (Supplemental Figure 1). Thus, although Smc6 is required to protect the integrity of rDNA (Torres-Rosell et al., 2005), which is a particular zone, prone to recombination events, its role at maintaining the integrity of replication forks generated at normal origins of replication is only apparent in the presence of genotoxic replication stress (Figure 1B).
The smc6-9 mutant is hypersensitive to both DNA-damaging agents and replication inhibitors (Torres-Rosell et al., 2005). As reported for sgs1 (Liberi et al., 2005), ubc9-1 and mms21 mutants (Branzei et al., 2006), a change in the profile of replication intermediates was only detected in smc6-9 cells at damaged replication forks, but not at forks stalled by HU treatment, or, as mentioned above, when cells were grown in the absence of genotoxic agents at high temperatures (Supplemental Figure 1). Conversely, S. pombe smc6 mutants were shown to accumulate X-shaped intermediates in response to HU-induced replication stress, although only in conditions when the replication checkpoint kinase Cds1 was inactivated (Ampatzidou et al., 2006).
Sequencing the smc6-9 allele revealed it to contain two point mutations in the Smc6 C-terminal coiled-coil region (Gln903Gly and Ser908Pro; Figure 1C). Another SMC6 mutant, smc6-56, was reported previously to display DNA damage hypersensitivity as a consequence of mutations in the Smc6 N-terminal coiled-coil region (Onoda et al., 2004) (Figure 1C). As mutations in the Smc6 complex in S. pombe were shown to be allele-specific for certain phenotypes (Sheedy et al., 2005), we tested the effect of the smc6-56 mutation on the X-molecule accumulation phenotype at damaged replication forks. We found it to be similar to that of smc6-9 (Figure 1D). Importantly, as is the case for sgs1, ubc9-1, and mms21 mutants (Liberi et al., 2005; Branzei et al., 2006), we found that the X-molecule accumulation in the smc6 mutants required Rad51, revealing it to depend on homologous recombination events (Supplemental Figure 2).
We further examined whether the X-molecule accumulation of smc6 mutants can be complemented by the wild-type SMC6. We constructed a diploid smc6-9/SMC6 and compared its phenotype in MMS with the one of the WT (SMC6/SMC6) diploid. We found that the X molecule accumulation phenotype of smc6-9 was not manifest when the other allele of SMC6 was present as a wild-type copy (Supplemental Figure 3). We thus conclude that the accumulation of X-shaped molecules present in smc6 mutants is due to a loss of function of SMC6. Likewise, the temperature and MMS sensitivity of smc6-9 seemed to be recessive traits (data not shown).
Esc2 Is a Novel Regulator of Recombinogenic Events at Replication Forks
One of the mutants included in our screen was esc2. Esc2 belongs to the recently identified family of proteins RENi, named based on its most prominent members: Rad60 (S. pombe), Esc2 (S. cerevisiae), and mouse/human NIP45 (Novatchkova et al., 2005). A key feature of this class of proteins is the presence of one or two SUMO-like domains in their C-terminal regions (Novatchkova et al., 2005).
In addition to this characteristic, Esc2 has several SUMO-binding motifs (SBMs) that could mediate noncovalent interaction with SUMO (Song et al., 2004; Hannich et al., 2005; Hecker et al., 2006; Kerscher et al., 2006; Raffa et al., 2006), and several SUMO consensus sites that may be targets for SUMO conjugation (Figure 2A). Similar to smc6, mms21, and sgs1 mutants, esc2 accumulated X-shaped structures at damaged replication forks (Figure 2B) in a Rad51-dependent manner (data not shown; see the accompanying article Mankouri et al., 2009). We also obtained evidence that esc2 mutant cells might accumulate spontaneous DNA damage. Consistent with previous reports (Alvaro et al., 2007), we found that esc2 mutants exhibited an increased number of spontaneously-arising Rad52 foci (Supplemental Figure 4A). Furthermore, esc2 cells showed constitutive phosphorylation of the replication checkpoint kinase Rad53, although to a lesser extent than the Rad53 phosphorylation triggered by RAD52 deletion (Supplemental Figure 4B). Furthermore, similar to sgs1 mutant cells, esc2 mutants displayed a sixfold elevation in the frequency of mitotic recombination measured between two direct tandem repeats (Aguilera and Klein, 1989) (Supplemental Figure 5). The double mutant esc2 sgs1 showed a synergistic increase in the recombination frequency compared with the single mutants (Supplemental Figure 5). Our results on Esc2 suggest that Esc2 and Sgs1 may be involved, at least partly, in different pathways that prevent spontaneous accumulation of DNA damage and the subsequent repair of these lesions by recombination (Figure 2 and Supplemental Figures 4 and 5). Our findings are also consistent with the results reported in the accompanying article by Mankouri et al. (2009).
Figure 2.
esc2 mutants accumulate X-molecules during replication of damaged templates. (A) Schematic representation of Esc2 protein with the two SUMO-like domains, the predicted coiled-coil domain, the SUMO-binding motifs and the predicted SUMO-consensus sites. The SUMO-binding motifs of consensus 1 that are followed by an acidic patch are pictured as big stars, whereas the motifs without an acidic patch are represented as small stars. (B) Wild-type (SY2234) and esc2Δ (FY1081) cells were synchronized with α-factor and released in medium containing MMS, and the replication intermediates were analyzed by 2D gel.
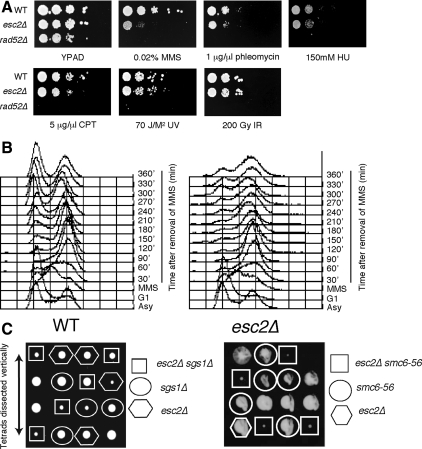
When we further examined the sensitivity of ESC2-deleted cells toward several types of DNA-damaging agents and replication inhibitors, we found that they were hypersensitive to MMS, mildly hypersensitive to phleomycin, but not hypersensitive to HU, camptothecin (CPT), UV light, or ionizing radiation (IR; Figure 3A), supporting the idea that Esc2 may be specifically required to repair lesions arising during S phase. In addition, we found that esc2 cells are defective in recovery from transient MMS treatment (Figure 3B). Thus, when G1-synchronized wild-type and esc2 cells were released into medium containing sublethal doses of MMS (0.033%) for 1 h, both slowed S-phase progression, as expected for cells proficient in the intra-S phase DNA damage checkpoint (Paulovich et al., 1997). After MMS removal, wild-type cells seemed to accumulate in G2 after 1 h, began to reenter the cell cycle after 2 h, and regained an asynchronous FACS profile after 4–5 h, indicating normal progression through the cell cycle. By contrast, esc2 cells only seemed to reenter the cell cycle after 3 h, and moreover, after 6 h they still displayed a large proportion of cells residing in the S-G2 phase (Figure 3B). This delay in recovery seemed to be specific to DNA damage, because esc2 mutants did not display defects in resuming replication after HU treatment (Supplemental Figure 6). Together, these results indicate that Esc2 promotes replication through damaged templates.
Figure 3.
esc2 mutants are sensitive to MMS and defective in recovery from transient MMS-induced damage. (A) Wild-type (SY2234) and esc2Δ (FY1081) strains were analyzed for sensitivity against different damaging agents. (B) Logarithmic cultures of wild-type (SY2234) and esc2Δ (FY1081) cells were synchronized in G1 and released from arrest into fresh YPD medium for 10 min at 30°C before addition of MMS (0.033%). After 60 min in MMS, cells were washed in YPD containing sodium thiosulphate (2.5%, wt/vol) and released in fresh medium without MMS. Cells were collected at the indicated times following removal of MMS for FACS analysis. (C) Tetrad analysis of esc2 sgs1 and esc2 smc6-56 mutants.
Esc2 Does Not Seem to Physically Interact with Sgs1-Top3 or Smc6
The phenotype of esc2 mutants at damaged replication forks (Figure 2B) was similar to those of ubc9-1, mms21, and sgs1 mutants (Liberi et al., 2005; Branzei et al., 2006), suggesting that Esc2 might also influence the ability of Sgs1-Top3 to resolve the pseudodouble Holliday junctions arising during template-switch replication. This possibility is also highlighted by the synthetic slow growth interactions reported in S. pombe and to some extent in S. cerevisiae between Esc2/Rad60, Sgs1/Rqh1, and Smc5-6 (Tong et al., 2001; Boddy et al., 2003; Torres-Rosell et al., 2005; Miyabe et al., 2006) that we also confirmed in our study (Figure 3C).
We therefore considered the scenario that Sgs1-Top3 might physically interact with Esc2 and/or Smc6. We first addressed this by using gel filtration chromatography, an approach that resolves proteins and protein complexes based on their relative sizes. Thus, we prepared crude extracts from wild-type cells that had been treated with MMS (0.033% for 3 h), resolved them on a Superose 6 column, and analyzed the resulting fractions by Western immunoblotting with antibodies against epitope-tagged Sgs1, Smc6, and Esc2. Consistent with previous reports (Chiolo et al., 2005), we found that under these conditions, Sgs1 eluted mainly in fractions 12–14 (Figure 4A). By contrast, the broad elution profile of Smc6 peaked in fraction 8, near the void volume of the column, whereas Esc2 eluted mainly in fractions 17–19 (Figure 4A), suggesting that the three proteins are not part of the same complex. Consistent with this interpretation, we were unable to detect interactions between Sgs1 and Smc6 by coimmunoprecipitation experiments performed either with whole cell extracts or with fractions obtained by gel filtration, in which either of these proteins was enriched (Supplemental Figure 7, A and B; also see Figure 4A). Furthermore, yeast two-hybrid experiments did not identify a physical interaction between Esc2 and Sgs1 (Supplemental Figure 7C). Also in accord with the above-mentioned data, we found that ESC2 deletion did not abolish the interaction between Sgs1 and Top3 as detected by two-hybrid analysis, although the intensity of the interaction seemed somewhat decreased (Figure 4B). Collectively, these results therefore suggest that Esc2 does not robustly interact with Sgs1-Top3 or with Smc5-Smc6, although we cannot rule out the possibility that such interactions do exist but are too weak or transient to be detected. We note that S. pombe Rad60 was shown to weakly interact with Smc5-6 (Boddy et al., 2003).
Figure 4.
Esc2, Smc6, and Sgs1 are not part of the same complex. (A) Crude protein extracts were prepared from the wild-type strains CY7676 (SGS1-9Myc and SMC6-3FLAG) and FY1028 (ESC2-13Myc) in the presence of MMS and fractionated by gel filtration. The collected fractions were analyzed by Western blotting using specific antibodies against 9Myc-tagged Sgs1, 3FLAG-tagged Smc6, and 13Myc-tagged Esc2. The elution peaks of standards are indicated by arrows. (B) BD-TOP3 in combination with AD-SGS1 and pGAD424 (empty vector) was transformed in wild-type (FY0101) or esc2Δ (HY0705) strains, and the interaction was examined by the β-gal assay.
Interaction between Esc2, SUMO, and Ubc9 and Physiological Significance of the Esc2-SUMO Interaction
The presence of SUMO-like domains in Esc2 (Figure 2A) suggests that this protein might interact with enzymes of the SUMO pathway and/or affect sumoylation events. Indeed, we found that Esc2 interacts with Ubc9 and the yeast SUMO, Smt3 (Figure 5A). Nevertheless, immunoprecipitation experiments did not reveal a clear sumoylated isoform of Esc2, although this could be due to the fact that only a very small percentage of Esc2 is sumoylated or that the interaction between Esc2 and SUMO is mainly mediated by hydrophobic and noncovalent bonding (data not shown). We further addressed whether Esc2 may affect sumoylation of other targets such as Sgs1. However, Sgs1 interactions with Ubc9 and SUMO were not abolished by ESC2 deletion (Figure 5B). Mutations affecting the functionality of SUMO enzymes often lead to a synthetic phenotype when combined. The esc2 mutation was reported to be synthetic lethal with mutations in SLX5/SLX8 (Tong et al., 2001), encoding SUMO and ubiquitin ligases (Burgess et al., 2007; Xie et al., 2007) (Uzunova et al., 2007), and we confirmed this result (Figure 5C). We further found that esc2 cells were slightly slow growing in combination with mms21 mutations (Figure 5D) and that esc2 siz1, esc2 siz2, and esc2 mms21 (esc2 mms21-CH, esc2 mms21-11) had higher levels of MMS sensitivity than the corresponding single mutants (Figure 5D).
Figure 5.
Genetical and physical interactions between Esc2 and the SUMO-pathway. (A) BD-ESC2 in combination with AD-SMT3, AD-yUBC9, or pGAD424 (empty vector) was transformed in the wild-type strain (HY0803), and the interactions examined by the β-gal assay. (B) Two-hybrid analysis of Sgs1 interaction with Ubc9 and SUMO. BD-SGS1 in combination with AD-UBC9, AD-SMT3, and pGAD424 was transformed in wild-type (FY0101) or esc2Δ (HY0705) strains, and the interaction was examined by the β-gal assay (C) Tetrad analysis of esc2 slx8 strains. (D) Spot assay of the MMS sensitivity of wild-type (SY2234), esc2Δ (FY1081), mms21-CH (FY1003), mms21-11 (FY1012), siz1Δ (HY0628), siz2Δ (HY0634), esc2Δ mms21-CH (RDY354), esc2Δ mms21-11 (RDY355), esc2Δ siz1Δ (RDY137), and esc2Δ siz2Δ (RDY358) strains.
Next, we examined the domains in Esc2 that mediate its interaction with SUMO and Ubc9. For this purpose, we used a set of Esc2 truncations fused to the Gal4 DNA binding domain (Gal4BD; Cuperus and Shore, 2002; Andrulis et al., 2004) in two-hybrid analyses (Figure 6A). We first tested the stability of these truncated Esc2 derivatives by Western immunoblotting with an anti-Gal4BD antibody (Figure 6B), and we found that most of the fusion proteins were stable and expressed at similar levels, with the exceptions of the derivative containing Esc2 residues 1-260 that was not expressed at detectable levels, and a derivative containing the C-terminal part of Esc2 from amino acids 389-456 that was expressed at low levels (Figure 6B). Notably, whereas the full-length (1-456) construct displayed interactions with both SUMO and Ubc9 (Figure 6C; also see Figure 5A), deletion of the N terminus abolished interaction with SUMO but not with Ubc9. Furthermore, the Esc2 C-terminal constructs containing residues 254-456 and 389-456 interacted with Ubc9, whereas the 254-389 construct did not (Figure 6C), suggesting that the C-terminal domain of Esc2 (389-456) containing the SUMO-like domains is required for the interaction with Ubc9 (Figure 2A). Conversely, we found that the Esc2 C terminus is not required for the interaction of Esc2 with SUMO, because an Esc2 derivative containing residues 1-389 was proficient in SUMO interaction (Figure 6C). Significantly, the 254-456 construct, which robustly interacted with Ubc9, was not able to interact with SUMO, and even deletion of the first 26 amino acid residues of Esc2 diminished somewhat this interaction (Figure 6C). As in this N-terminal region lies the core of a very well conserved SBM of consensus 1 that is followed by an acidic patch (Figure 2A), this suggests that it may be potent at promoting a hydrophobic interaction with SUMO (Kerscher et al., 2006). From these data, we thus conclude that the SBM located between Esc2 residues 21-48 is required to sustain Esc2 interaction with SUMO. The SBMs are known to mediate hydrophobic, noncovalent interactions with SUMO, which often serve to promote sumoylation of the protein containing the SBM or some of its interacting proteins (Kerscher et al., 2006).
Figure 6.
Mapping of the interaction domains of Esc2 with SUMO and Ubc9. (A) Schematic representation of the Esc2 protein fragments used for the two-hybrid assay. The SUMO-binding motif of consensus 1 located between the 21–48 residues and the C-terminal SUMO like domains are represented. (B) The stability of the Esc2 protein fragments was analyzed by Western blotting using a specific antibody against the GAL4-binding (BD) domain. (C) The interaction between the Esc2 constructs and SUMO or Ubc9 was assayed by β-gal assay. (D) WT (SY2234) or esc2Δ (FY1081) strains were transformed with the Esc2 constructs shown in (A) and examined for MMS sensitivity. (E) Schematics summarizing the efficiency of interaction between Esc2 constructs and SUMO, Ubc9, and ability to complement the MMS sensitivity of the different fragments of Esc2. (F) Summary panel illustrating the physical interactions reported or found in this study among Sgs1/Top3, Esc2, Smc5/6, Slx5/Slx8, and Ubc9/Smt3 in S. cerevisiae or in S. pombe. N/A, not applicable; S.p., S. pombe; S.c., S. cerevisiae.
Based on the above-mentioned findings, we addressed whether there is any correlation between the ability of Esc2 to promote tolerance to MMS and its interaction with SUMO or Ubc9. To do this, esc2 cells were transformed with various Esc2 fusion constructs (Figure 6A), and the cells were plated on medium containing MMS (Figure 6D). Notably, only the full-length and the N-terminal (residues 1-389) fusions were able to rescue the MMS sensitivity of esc2 cells, suggesting that the lack of interaction with SUMO, but not the one with Ubc9, may be responsible for the MMS sensitivity of esc2 mutant cells (Figure 6, D and E).
Genetic Interactions in the Presence of DNA Damage between ESC2, SGS1, and SMC6
In view of our other findings and the reported physical interactions (Figure 6F), we examined whether SGS1, SMC6, and ESC2 work in the same or different genetic pathways to promote resistance to MMS. To this end, we combined esc2 and sgs1 deletion mutations with each other, as well as with smc6-9 and smc6-56 alleles. ESC2 deletion produced a slow-growth phenotype in combination with either sgs1 or smc6 mutations, although to a different degree: the esc2 and smc6-56 combination was almost synthetic lethal (Figure 3C), whereas the esc2 smc6-9 double mutant was slightly slow growing (Figure 7C); the double mutant sgs1 esc2 was slow growing, but in our background was not synthetic lethal as has been reported previously by synthetic genetic array (SGA) analysis (Tong et al., 2001) (Figure 3C). When we analyzed the MMS sensitivity of these various mutants, we found that all combinations of double mutants were somewhat more sensitive to MMS than the single mutants (Figure 7), suggesting that at least partly, Sgs1, Smc5-6 and Esc2 have independent functions in response to genotoxic treatment.
Figure 7.
Epistasis tests between esc2, smc6, sgs1, and rad51. (A) Wild-type (SY2234), esc2Δ (FY1081), sgs1Δ (FY1060), and sgs1Δ esc2Δ (HY1124) were grown to logarithmic phase and analyzed for MMS sensitivity by spot assay. (B) Wild-type (SY2234), smc6-9 (SY2232), sgs1Δ (FY1060), and sgs1Δ smc6-9 (CY8997) were grown to logarithmic phase and analyzed for MMS sensitivity by spot assay. (C) Wild-type (SY2234), esc2Δ (FY1081), smc6-9 (SY2232), esc2Δ smc6-9 (CY8696), esc2Δ smc6-9 rad51Δ (HY111), rad51Δ (SY2083), and esc2Δ rad51Δ (FY1027) strains were grown to logarithmic phase and analyzed for MMS sensitivity by spot assay.
We also noted that esc2 sgs1 and smc6-9 sgs1 are genetically unstable and have a tendency to accumulate suppressors during normal growth. Sgs1 and Smc6 were reported to function in the Rad52 homologous recombination pathway (Onoda et al., 2001, 2004) and consistent with previous reports in S. pombe (Morishita et al., 2002), we find that esc2 is epistatic to rad51 (Figure 7C). As also reported for S. pombe (Miyabe et al., 2006), we found that the slow growth phenotype of esc2 smc6-9 is rescued by RAD51 deletion (Figure 7C). It is thus possible that some of the suppressors spontaneously arising in esc2 sgs1 and sgs1 smc6-9 may affect the functionality of RAD51. Together, these results suggest that Smc6 and Esc2 have roles in homologous recombination mediated DNA repair, and their functionality becomes crucial in the absence of Sgs1.
DISCUSSION
A common feature of the Smc5-6 complex and the eukaryotic RecQ helicases Sgs1, Rqh1, and BLM is their involvement in the restart of stalled replication forks and DNA repair through HR. Studies in S. pombe were the first to draw the attention to this similarity: smc6 and rqh1 mutants were originally described as factors that operate during S phase to promote bypass of the UV-induced damaged sites, in a manner distinct from nucleotide excision repair, but involving the products of the HR machinery (Lehmann et al., 1995; Murray et al., 1997). Accordingly, in S. cerevisiae, both Smc6 and Sgs1 have been reported to work in concert with recombination proteins to promote repair of MMS-induced damage (Onoda et al., 2001, 2004; Torres-Rosell et al., 2005) and nse1 mutants of the Smc5-6 complex were recently shown to be defective in promoting the gap-filling repair of UV-damaged DNA (Santa Maria et al., 2007). The accumulation of cruciform structures during replication of damaged templates in sgs1 mutant cells is evidence for Sgs1 contribution to gap-filling repair (Liberi et al., 2005).
The sister chromatid junctions visualized at damaged replication forks have the biochemical properties of pseudodouble Holliday junctions, require HR function for their formation (Liberi et al., 2005), and it is envisaged that, if they fail to be resolved, they may generate strand breaks and trigger recombinogenic events (Branzei and Foiani, 2007). This physical evidence is congruous with previous genetic models that proposed a dual role for Sgs1 in recombination: in maturating certain recombination structures that become substrates for Top3 (Gangloff et al., 1999; Shor et al., 2002) and also in preventing certain recombinogenic events (Hickson, 2003; Ira et al., 2003; Robert et al., 2006), such as those represented by the cruciform molecules accumulating in sgs1 mutants at damaged replication forks (Liberi et al., 2005).
Recently, it was shown that Ubc9- and Mms21-dependent sumoylation act in concert with Sgs1 to prevent the detrimental accumulation of recombinogenic X-shaped structures at replication forks (Branzei et al., 2006). However, Sgs1 sumoylation is independent of Mms21, suggesting that other SUMO targets are implicated in this process. Besides its involvement with Sgs1 during replication of damaged templates, SUMO modification has also been connected to regulating other recombination processes as well (Branzei and Foiani, 2008). For example, the recombination factor Rad52 is sumoylated in yeast and mammals (Ho et al., 2001; Sacher et al., 2006), and proliferating cell nuclear antigen sumoylation acts to prevent recombination-mediated repair (Papouli et al., 2005; Pfander et al., 2005). Here, we found that the X-molecule accumulation phenotype at damaged forks found in sgs1, ubc9-1, and mms21 cells (Liberi et al., 2005; Branzei et al., 2006) is mimicked by mutations in SMC6 and ESC2. Our data suggest that these two newly identified factors are connected to each other and to the sumoylation pathway and that they act in parallel or together with Sgs1 in promoting repair events during replication.
Smc5 and Smc6 are sumoylated by Mms21 in response to DNA damage (Andrews et al., 2005; Potts and Yu, 2005; Zhao and Blobel, 2005) and Esc2 belongs to a family of proteins containing SUMO-like domains in their C terminus (Novatchkova et al., 2005) (Figure 2A). Here, we also show that Esc2 interacts physically with Ubc9 and SUMO, primarily through a C-terminal SUMO-like domain and an N-terminal SBM, respectively (Figure 6). The S. pombe protein Rad60, containing SUMO-like domains in its C terminus, also interacts with SUMO by means of an SBM, and with the ubiquitin ligase complex Rfp-Slx8 (Slx5-Slx8 in S. cerevisiae) (Prudden et al., 2007) (Figure 6F), implicated in regulating SUMO homeostasis in both budding and fission yeasts (Burgess et al., 2007; Prudden et al., 2007; Sun et al., 2007; Uzunova et al., 2007; Xie et al., 2007). Our data suggest that the Esc2 function in promoting DNA-damage tolerance is correlated to its ability to physically interact with SUMO (Figure 6E). A similar conclusion was reached in S. pombe: Rad60 with mutations in the N-terminal SBM are defective in tolerating HU-induced replication stress (Raffa et al., 2006), and the DNA repair defects of rad60 resemble the ones of slx8 (Prudden et al., 2007). Intriguingly, rad60 also shares most of the smc6 phenotypes and functional interactions, and genetic evidence suggested that Rad60 and Smc6 act in concert to promote DNA repair of replication-induced lesions (Morishita et al., 2002; Miyabe et al., 2006). Thus, Rad60 is functionally connected to both Slx5 and Smc6. To close this triangle, in budding yeast, Smc5-6 physically associates with Slx5 (Hazbun et al., 2003) (Figure 6F). It is important to note that all of these factors, Smc5-6, Esc2, and Slx5, display functional interactions with Sgs1 and with each other, revealed as synthetic sickness/lethality interactions or synergism toward MMS (Mullen et al., 2001; Tong et al., 2001, 2004) (Figures 3C and 7). Unlike sgs1, esc2, and smc6 mutants, we found that slx5 and slx8 mutants do not accumulate X-shaped molecules at damaged replication forks (our unpublished data), suggesting that the role of Slx5 and Slx8 in intra-S repair is different from the roles of Sgs1, Esc2, and Smc6 or that they might mainly mediate repair of different types or lesions, such as double-strand breaks (DSBs) (Nagai et al., 2008). Based on the evidence obtained in S. pombe (Prudden et al., 2007; Sun et al., 2007) and the phenotypes we have uncovered for Esc2 in this study, we propose that the association between Smc5-6 and Slx5-mediated repair processes is likely to be regulated by Esc2.
Interestingly, one of the Smc5-6 subunits, Nse1, contains a structural domain indicating that it works as an E3 ligase for ubiquitylation (McDonald et al., 2003). Ubiquitin ligase activity for Nse1 has not yet been demonstrated, but a point mutation in the ligase sequence motif, nse1 C274A, renders cells sensitive to DNA-damaging agents, suggesting that it may affect DNA repair (Santa Maria et al., 2007). Another possibility is that Nse1 and Smc5-6 define a differently regulated ubiquitylation pathway from Esc2 and Slx5-8 (Uzunova et al., 2007; Xie et al., 2007), and they may act in parallel to promote sumoylation-induced degradation of substrates to facilitate DNA repair. This could also explain the additive effect in MMS sensitivity of esc2 smc6-9 mutants (Figure 7C), which according to the classical interpretation of epistasis data, suggests that Smc6 and Esc2 have independent functions in promoting repair of MMS-induced lesions. Because esc2, sgs1, and smc6 mutants are epistatic to rad51/rad52 for MMS sensitivity (Figure 7C; Onoda et al., 2001, 2004), we conclude that they are all implicated in homologous recombination repair.
The wealth of genetic data clearly points at a role for Smc5-6 in coordinating DNA repair activities. In S. pombe, the SMC6 allele smc6-74 is suppressed by multicopy Brc1 (Sheedy et al., 2005; Lee et al., 2007), a protein consisting of six consecutive BRCA1 C-terminal domains (Verkade et al., 1999). This suppression is allele specific, induced by DNA damage, and requires several DNA repair nucleases, such as Slx1-Slx4 and Mus81 (Sheedy et al., 2005; Lee et al., 2007), which in both budding and fission yeast have been identified to act redundantly with Sgs1 and Rqh1, respectively (Mullen et al., 2001; Doe et al., 2002; Coulon et al., 2004). Also in budding yeast, the homologue of Brc1, Esc4, plays a role in MMS-induced repair and associates to several DNA repair proteins, including HR factors and the Slx4 nuclease (Rouse, 2004; Chin et al., 2006; Roberts et al., 2006). There is no evidence of a physical association between Brc1 and Smc6 in S. pombe, but using mass spectrometry we found that Esc4 associates with Smc6 (Sollier, Foiani, and Branzei, unpublished data). It is thus possible that nucleases and repair factors that may promote inappropriate repair events are controlled by means of SUMO-mediated ubiquitin ligase degradation processes. Indeed, the Slx5-Slx8 complex was shown to negatively regulate Rad51-independent recombination by affecting the sumoylation of enzymes implicated in the early steps of this process (Burgess et al., 2007) and to promote repair of DSBs, which might result also from endonuclease-mediated processing of the X-shaped cruciform structures forming during damage bypass replication, by targeting them to nuclear pores for repair (Nagai et al., 2008).
In conclusion, we have shown that different enzymes associated with the SUMO pathway act in a manner similar with Sgs1 to promote DNA repair, and to prevent the accumulation of recombinogenic structures at damaged replication forks. Considering that these structures resemble the catenanes formed at replication termination, when two replicons fuse together, it will be of interest in the future to study the contribution of these enzymes (Esc2 and Smc6) to unperturbed replication and to understand whether their role in facilitating chromosome segregation is related to their function in promoting the resolution of such catenane-like structures that arise during replication-related processes. A cross-talk between ubiquitin and SUMO pathways in promoting protein degradation has been recently suggested or reported (Burgess et al., 2007; Prudden et al., 2007; Sun et al., 2007; Uzunova et al., 2007; Xie et al., 2007). The finding that enzymes affecting activities implicated in ubiquitin- and SUMO-mediated processes (Slx5-Slx8, Esc2, and Smc5-6) are brought together with repair factors (Rad51 and Sgs1), implicated in the initiation or maturation/resolution of DNA repair intermediates, suggests that the SUMO or ubiquitin ligase activities of these complexes may act to regulate the repair functions of various factors in response to damaged DNA. Future challenges are to identify the factors targeted by these complexes and to understand whether this multifaceted regulation is triggered by protein–protein interactions or rather by the formation of certain repair intermediates or DNA structures in particular cellular contexts.
Supplementary Material
ACKNOWLEDGMENTS
We thank I. Hickson for discussion and for communicating unpublished data on Esc2. We thank X. Zhao, L. Aragon (MRC Clinical Sciences Center, London, United Kingdom), A. Aguilera (CABIMER, Universidad de Sevilla, Spain), T. Enomoto, M. Seki (Tohoku University, Sendai, Japan), D. Shore, R. Sternglanz, R. Rothstein (Columbia University Medical Center, New York, NY), M. Lisby (University of Copenhagen, Denmark), A. Pellicioli, and N. Lowndes (National University of Ireland, Galway, Ireland) for yeast strains, plasmids, or antibodies; and all members of our laboratories for discussions. We thank M. Fumasoni in D. Branzei's laboratory for technical support. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (to D. B.), Association for International Cancer Research and GENICA (to M. F. and D. B.), and the European Community DNA Repair grant (to M. F.). J. S. was supported by the European Molecular Biology Organization grant ALTF 283-2006. R. D. is supported by a UK Biotechnology and Biological Sciences Research Council Cooperative Awards in Science and Engineering studentship with KuDOS Pharmaceuticals and by a Cancer Research UK program grant (to S. J.). Work in S. J.'s laboratory was also supported by grants from DNA Repair and GENICA.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0875) on January 21, 2009.
REFERENCES
- Aguilera A., Klein H. L. Genetic and molecular analysis of recombination events in Saccharomyces cerevisiae occurring in the presence of the hyper-recombination mutation hpr1. Genetics. 1989;122:503–517. doi: 10.1093/genetics/122.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro D., Lisby M., Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampatzidou E., Irmisch A., O'Connell M. J., Murray J. M. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell. Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews E. A., Palecek J., Sergeant J., Taylor E., Lehmann A. R., Watts F. Z. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E. D., Zappulla D. C., Alexieva-Botcheva K., Evangelista C., Sternglanz R. One-hybrid screens at the Saccharomyces cerevisiae HMR locus identify novel transcriptional silencing factors. Genetics. 2004;166:631–635. doi: 10.1534/genetics.166.1.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Shanahan P., McDonald W. H., Lopez-Girona A., Noguchi E., Yates I. J., Russell P. Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol. Cell. Biol. 2003;23:5939–5946. doi: 10.1128/MCB.23.16.5939-5946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Rep. 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Burgess R. C., Rahman S., Lisby M., Rothstein R., Zhao X. The slx5-slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 2007;27:6153–6162. doi: 10.1128/MCB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. K., Bashkirov V. I., Heyer W. D., Romesberg F. E. Esc4/Rtt107 and the control of recombination during replication. DNA Rep. 2006;5:618–628. doi: 10.1016/j.dnarep.2006.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I., Carotenuto W., Maffioletti G., Petrini J. H., Foiani M., Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S., Gaillard P. H., Chahwan C., McDonald W. H., Yates J. R., 3rd, Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol. Biol. Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus G., Shore D. Restoration of silencing in Saccharomyces cerevisiae by tethering of a novel Sir2-interacting protein, Esc8. Genetics. 2002;162:633–645. doi: 10.1093/genetics/162.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N., Kamakaka R. T. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell. 2000;6:769–780. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- Doe C. L., Ahn J. S., Dixon J., Whitby M. C. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- Eladad S., Ye T. Z., Hu P., Leversha M., Beresten S., Matunis M. J., Ellis N. A. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- Fousteri M. I., Lehmann A. R. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 2000;19:1691–1702. doi: 10.1093/emboj/19.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., de Massy B., Arthur L., Rothstein R., Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfless S. J., Morag A. S., Belisle K. A., Sutera V. A., Jr, Lovett S. T. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol. Cell. 2006;21:595–604. doi: 10.1016/j.molcel.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- Hazbun T. R., et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- Heller R. C., Marians K. J. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- Hickson I. D. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Kato K., Strauss B. A model for replication repair in mammalian cells. J. Mol. Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- Ho J. C., Warr N. J., Shimizu H., Watts F. Z. SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52. Nucleic Acids Res. 2001;29:4179–4186. doi: 10.1093/nar/29.20.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii T., Fung J., Mullen J. R., Brill S. J. The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity. Cell Cycle. 2007a;6:2800–2809. doi: 10.4161/cc.6.22.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii T., Mullen J. R., Slagle C. E., Brill S. J. Stimulation of in vitro sumoylation by Slx5-Slx 8, evidence for a functional interaction with the SUMO pathway. DNA Rep. 2007b;6:1679–1691. doi: 10.1016/j.dnarep.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J. E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Lee K. M., Nizza S., Hayes T., Bass K. L., Irmisch A., Murray J. M., O'Connell M. J. Brc1-mediated rescue of Smc5/6 deficiency: requirement for multiple nucleases and a novel Rad18 function. Genetics. 2007;175:1585–1595. doi: 10.1534/genetics.106.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R., Fuchs R. P. Gaps and forks in DNA replication: rediscovering old models. DNA Rep. 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Lehmann A. R., Walicka M., Griffiths D. J., Murray J. M., Watts F. Z., McCready S., Carr A. M. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 1995;15:7067–7080. doi: 10.1128/mcb.15.12.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J. E., Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos H. B., Strom L., Itoh T., Katou Y., Shirahige K., Sjogren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Lopes M., Foiani M., Sogo J. M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- Mankouri H. W., Ngo H. P., Hickson I. D. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell. 2007;18:4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri H. W., Ngo H. P., Hickson I. D. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009;20:1683–1694. doi: 10.1091/mbc.E08-08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald W. H., Pavlova Y., Yates J. R., 3rd, Boddy M. N. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 2003;278:45460–45467. doi: 10.1074/jbc.M308828200. [DOI] [PubMed] [Google Scholar]
- Miyabe I., Morishita T., Hishida T., Yonei S., Shinagawa H. Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol. Cell. Biol. 2006;26:343–353. doi: 10.1128/MCB.26.1.343-353.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa H., Morishita T., Kawane S., Iwasaki H., Carr A. M., Shinagawa H. Rad62 protein functionally and physically associates with the Smc5/Smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell. Biol. 2004;24:9401–9413. doi: 10.1128/MCB.24.21.9401-9413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T., Tsutsui Y., Iwasaki H., Shinagawa H. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 2002;22:3537–3548. doi: 10.1128/MCB.22.10.3537-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Kaliraman V., Ibrahim S. S., Brill S. J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. M., Lindsay H. D., Munday C. A., Carr A. M. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M. B., Roberts T. M., Brown G. W., Varela E., Hediger F., Gasser S. M., Krogan N. J. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M., Bachmair A., Eisenhaber B., Eisenhaber F. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinformatics. 2005;6:22. doi: 10.1186/1471-2105-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya T., Arai H., Kubota Y., Shinagawa H., Hishida T. A SUMO-like domain protein, Esc2, is required for genome integrity and sister chromatid cohesion in Saccharomyces cerevisiae. Genetics. 2008;180:41–50. doi: 10.1534/genetics.107.086249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoda F., Seki M., Miyajima A., Enomoto T. Involvement of SGS1 in DNA damage-induced heteroallelic recombination that requires RAD52 in Saccharomyces cerevisiae. Mol. Gen. Genet. 2001;264:702–708. doi: 10.1007/s004380000358. [DOI] [PubMed] [Google Scholar]
- Onoda F., Takeda M., Seki M., Maeda D., Tajima J., Ui A., Yagi H., Enomoto T. SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in Saccharomyces cerevisiae. DNA Rep. 2004;3:429–439. doi: 10.1016/j.dnarep.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Papouli E., Chen S., Davies A. A., Huttner D., Krejci L., Sung P., Ulrich H. D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Paulovich A. G., Margulies R. U., Garvik B. M., Hartwell L. H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S., Wohlschlegel J., McDonald W. H., Yates J. R., 3rd, Boddy M. N. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Potts P. R., Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., Boddy M. N. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa G. D., Wohlschlegel J., Yates J. R., 3rd, Boddy M. N. SUMO-binding motifs mediate the Rad60-dependent response to replicative stress and self-association. J. Biol. Chem. 2006;281:27973–27981. doi: 10.1074/jbc.M601943200. [DOI] [PubMed] [Google Scholar]
- Robert T., Dervins D., Fabre F., Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. M., Kobor M. S., Bastin-Shanower S. A., Ii M., Horte S. A., Gin J. W., Emili A., Rine J., Brill S. J., Brown G. W. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol. Biol. Cell. 2006;17:539–548. doi: 10.1091/mbc.E05-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J. Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J. 2004;23:1188–1197. doi: 10.1038/sj.emboj.7600129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Pfander B., Hoege C., Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 2006;8:1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- Santa Maria S. R., Gangavarapu V., Johnson R. E., Prakash L., Prakash S. Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:8409–8418. doi: 10.1128/MCB.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant J., Taylor E., Palecek J., Fousteri M., Andrews E. A., Sweeney S., Shinagawa H., Watts F. Z., Lehmann A. R. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol. Cell. Biol. 2005;25:172–184. doi: 10.1128/MCB.25.1.172-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy D. M., Dimitrova D., Rankin J. K., Bass K. L., Lee K. M., Tapia-Alveal C., Harvey S. H., Murray J. M., O'Connell M. J. Brc1-mediated DNA repair and damage tolerance. Genetics. 2005;171:457–468. doi: 10.1534/genetics.105.044966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Gangloff S., Wagner M., Weinstein J., Price G., Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Leverson J. D., Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suski C., Marians K. J. Resolution of converging replication forks by RecQ and topoisomerase III. Mol. Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J., Machin F., Farmer S., Jarmuz A., Eydmann T., Dalgaard J. Z., Aragon L. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 2005;7:412–419. doi: 10.1038/ncb1239. [DOI] [PubMed] [Google Scholar]
- Uzunova K., et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- Verkade H. M., Bugg S. J., Lindsay H. D., Carr A. M., O'Connell M. J. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell. 1999;10:2905–2918. doi: 10.1091/mbc.10.9.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Jones G. M., Prelich G. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics. 2006;172:1499–1509. doi: 10.1534/genetics.105.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Hickson I. D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Xie Y., Kerscher O., Kroetz M. B., McConchie H. F., Sung P., Hochstrasser M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- Yang L., Mullen J. R., Brill S. J. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 2006;34:5541–5551. doi: 10.1093/nar/gkl685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.