Abstract
Coordinated regulation of PI3-kinase (PI3K) and the tumor suppressor phosphatase and tensin homologue deleted on chromosome 10 (PTEN) plays a pivotal role in various cell functions. PTEN is deficient in many cancer cells, including Jurkat human leukemia. Here, we demonstrate that the status of PTEN determines cellular susceptibility to oxidative stress through antioxidant-responsive element (ARE)-mediated transcription of detoxification genes. We found that ferritin H transcription was robustly induced in tert-butylhydroquinone (t-BHQ)-treated Jurkat cells via an ARE, and it was due to PTEN deficiency. Chromatin immunoprecipitation assays revealed that p300/CREB-binding protein (CBP) histone acetyltransferases and Nrf2 recruitment to the ARE and Bach1 release were blocked by the PI3K inhibitor LY294002, along with the partial inhibition of Nrf2 nuclear accumulation. Furthermore, acetylations of histone H3 Lys9 and Lys18, and deacetylation of Lys14 were associated with the PI3K-dependent ARE activation. Consistently, PTEN restoration in Jurkat cells inhibited t-BHQ–mediated expression of ferritin H and another ARE-regulated gene NAD(P)H:quinone oxidoreductase 1. Conversely, PTEN knockdown in K562 cells enhanced the response to t-BHQ. The PTEN status under t-BHQ treatment affected hydrogen peroxide-mediated caspase-3 cleavage. The PI3K-dependent ferritin H induction was observed by treatment with other ARE-activating agents ethoxyquin and hemin. Collectively, the status of PTEN determines chromatin modifications leading to ARE activation.
INTRODUCTION
Oxidative stress is caused by a metabolic imbalance between the formation and detoxification of oxidants during various housekeeping and pathological processes of cellular reactions, such as mitochondrial respiratory function, inflammatory response, and xenobiotic metabolism. Xenobiotic exposure leading to the formation of pro-oxidants/reactive oxygen species (ROS), and subsequent oxidative cell damage are important mechanisms critically associated with various human disorders, including neurodegenerative disease and cancer (Andersen, 2004).
Xenobiotic metabolism is coordinately carried out by two broadly classified groups of genes, termed xenobiotic-metabolizing phase I and phase II genes. Proteins encoded by phase I genes, such as cytochrome P450 monooxygenases, transiently activate a variety of xenobiotics through oxidation reactions, which is the necessary step for subsequent detoxification and excretion of xenobiotics in phase II reactions (Nebert et al., 2000). Phase II genes, such as glutathione transferases and NAD(P)H quinone oxidoreductase-1 (NQO1), encode proteins which increase the water-solubility of phase I reactive metabolites via conjugation reactions or destruction of active centers (Li et al., 2004). Thus, tight regulation of phase II detoxification genes is crucial to protect cells from the toxic effects of reactive metabolites and ROS. A battery of phase II detoxification genes are transcriptionally regulated during xenobiotic metabolism via an antioxidant-responsive element (ARE), an enhancer containing a conserved TGAC/TnnnGCA motif (Nguyen et al., 2003).
Ferritin is a substantially conserved iron storage multimeric protein composed of 24 subunits of H and L types (Arosio and Levi, 2002). The ferritin H has ferroxidase activity, converting Fe2+ to Fe3+, whereas the ferritin L subunit stabilizes the ferritin shell and facilitates iron uptake (Theil, 2003). Knockout of the ferritin H gene in mice causes early embryonic lethality between 3.5 and 9.5 d of development (Ferreira et al., 2001). Knockdown of ferritin H with siRNA affected intracellular iron availability and induced susceptibility to H2O2- or rotenone-mediated cytotoxicity (Cozzi et al., 2004; MacKenzie et al., 2008), whereas ferritin L knockdown did not have a significant impact on intracellular iron availability (Cozzi et al., 2004). Reciprocally, we and others demonstrated that ferritin H is an antioxidant detoxification gene by showing that cells overexpressing ferritin H are more resistant to pro-oxidant-mediated cytotoxicity (Picard et al., 1996; Epsztejn et al., 1999; Cozzi et al., 2000; Orino et al., 2001).
The iron-mediated translational regulatory mechanism of ferritin H and L was proven by substantial evidence of protein–mRNA interactions between iron regulatory proteins and iron-responsive element in the 5′-nontranslated region of both H and L mRNAs (Hentze et al., 2004; Papanikolaou and Pantopoulos, 2005; MacKenzie et al., 2008). In addition, an increase in the stability of the ferritin H mRNA in cells treated with phorbol 12-myristate 13-acetate or the calcium ionophore, ionomycin, was observed (Pang et al., 1996; MacKenzie and Tsuji, 2008). Ferritin is also regulated at the transcriptional level under oxidative conditions (Hintze and Theil, 2006; MacKenzie et al., 2008), and both ferritin H and L genes belong to the ARE-regulated gene family. We and others demonstrated that H2O2, hemin, and tert-butylhydroquinone (t-BHQ), a phenolic antioxidant that also has the potential to produce ROS, transcriptionally activates mouse and human ferritin H and L genes via a far-upstream enhancer element containing the conserved ARE motif (Wasserman and Fahl, 1997; Tsuji et al., 2000; Hintze and Theil, 2005; Tsuji, 2005; Iwasaki et al., 2006). ARE-activating agents seem to trigger various signaling pathways that lead to transcriptional activation of ferritin and other phase II genes via the recruitment of Nrf2/Maf and several b-zip family transcription factors to the ARE (Itoh et al., 1999; Li et al., 2004; Motohashi and Yamamoto, 2004); however, a particular signaling pathway leading to epigenetic ARE activation through coactivator recruitment and histone modifications has not been investigated.
To elicit the transcriptional activation or repression of a specific gene in response to external stimuli, regulatory enhancer elements need to recruit specific chromatin remodelling factors to their immediate vicinity, which ultimately determines the accessibility of nascent transcription factors and RNA polymerases to the specific enhancer and promoter regions (Marmorstein, 2001). N-terminal tails of core histones have lysine-rich sequences and are subject to covalent posttranslational modifications such as acetylation and methylation, leading to chromatin relaxation or condensation (Strahl and Allis, 2000). Histone lysine acetylation generally induces transcriptional activation by way of neutralization of the positive charges on lysines and subsequent chromatin relaxation. It remains unknown as to how epigenetic mechanisms of chromatin environment and signaling pathways are associated with the regulation of an ARE enhancer.
Phosphatidylinositol 3-kinase (PI3K) and the tumor suppressor phosphatase and tensin homologue deleted on chromosome 10 (PTEN) coordinately regulate the PI3K pathway that relays signals via AKT phosphorylation on Thr308 and Ser473 to downstream target proteins involved in cell proliferation, survival, motility, and transformation (Bader et al., 2005; Cully et al., 2006). The tumor suppressor PTEN serves as a negative regulator of the PI3K pathway by preferentially dephosphorylating phosphatidylinositol trisphosphate (PIP3) to generate phosphatidylinositol bisphosphate, resulting in inactivation of the downstream AKT pathway (Cully et al., 2006). Genetic inactivation of PTEN has been detected in many human cancers, including brain, breast, and prostate cancers (Li et al., 1997). PTEN deficiency due to mutations in the PTEN gene cause constitutively activated PI3K-AKT signaling pathways (Wu et al., 1998), resulting in cell proliferation and survival in the absence of external stimuli. Jurkat human leukemic T cells were shown to have constitutive localization of Pleckstrin homology domain-containing kinase Itk to the plasma membrane (Shan et al., 2000). This is the result of a PTEN deficiency caused by frame shift mutations in exon 7, which leads to increased levels of PIP3 in the plasma membrane; this in turn results in a constitutively active PI3K-AKT signaling pathway (Shan et al., 2000). Besides the major function of PTEN as a phosphatase that negatively regulates the PI3K pathway in plasma membrane, emerging evidence suggests that nuclear PTEN plays a critical role in the regulation of cell proliferation and transformation (Baker, 2007; Tamguney and Stokoe, 2007). Nuclear PTEN functions as a tumor suppressor function because loss of nuclear PTEN is associated with enhanced cell proliferation and transformation (Tamguney and Stokoe, 2007). Mono- and polyubiquitination of PTEN by NEDD4-1 regulate nuclear import and protein stability of PTEN, respectively (Trotman et al., 2007; Wang et al., 2007), although it is still in debate (Fouladkou et al., 2008). The function of nuclear PTEN seems to be mediated through its physical interaction with nuclear proteins. For example, PTEN in the nucleus associates with E2F-1 and enhances transcription of a double-strand DNA repair gene Rad51, resulting in the maintenance of chromosomal integrity (Shen et al., 2007).
Although some ARE-activating electrophilic chemicals were shown to activate diverse signaling pathways including the PI3K pathway (Nakaso et al., 2003; Li et al., 2004, 2006), it has not been elucidated how the PI3K pathway and PTEN status determine the activation of an ARE through chromatin environment and posttranslational core histone modifications. This study aimed to elucidate the roles of PTEN in histone modifications associated with ARE-mediated gene transcription.
MATERIALS AND METHODS
Cell Culture and Reagents
Jurkat human T cell leukemia cells (clone E6-1) were purchased from American Type Culture Collection (Manassas, VA). They were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS; Mediatech, Manassas, VA), 10 mM HEPES, 2 mM l-glutamine, 1 mM sodium pyruvate, 4.5 g/l glucose, 100 U/ml penicillin, and 100 μg/ml streptomycin. The tet-inducible PTEN Jurkat PIJ17 and noninducible Con18 cells were kindly provided by Drs. Wange and Zack Howard (Seminario et al., 2003; Gao et al., 2005). They were maintained in Jurkat RPMI 1640 growth medium containing 400 μg/ml each of G418 and hygromycin B (AG Scientific, San Diego, CA). For induction of PTEN expression, PIJ17 and Con18 were treated with 1 μg/ml doxycycline (Clontech, Mountain View, CA) for 24 h before treatment with t-BHQ (Sigma-Aldrich, St. Louis, MO). K562 human erythroleukemia cells, purchased from American Type Culture Collection, were cultured in RPMI 1640 medium supplemented with 25 mM HEPES, 0.3g/l l-glutamine, and 10% FBS. Ethoxyquin and hemin were purchased from MP Biochemicals (Irvine, CA) and Fluka Biochemika (Buchs, Switzerland), respectively.
Luciferase Reporter Assay and Transfection
pBluescript SK(−)-4.5kb ARE (+)- and -4.4kb ARE (−)-h-ferritin H-luciferase reporter plasmids were described previously (Tsuji, 2005). Transient DNA transfection into Jurkat cells was carried out by electroporation (X-Cell; Bio-Rad, Hercules, CA) with an optimized preset condition by Bio-Rad for Jurkat cells (exponential decay, 1000 μF; 140 V; 100-μl cell suspension in a cuvette with a 0.2-cm gap). After electroporation of luciferase reporters into 10 × 106 Jurkat cells, they were plated at a density of ∼2.5 × 106 cells/35-mm plate containing 2 ml of culture medium. As a transfection internal control, 10 ng of pRL-null (Promega, Madison, WI) was simultaneously cotransfected. After incubation for 20–24 h, the cells were treated with various concentrations of t-BHQ for 24 h. pcDNA3.1-T7AKT plasmid DNA was kindly provided by Dr. Sellers (Ramaswamy et al., 1999) via Addgene. Luciferase assays were performed using Dual-Luciferase assay reagents (Promega). Firefly luciferase expression driven by the ferritin H gene was normalized by Renilla luciferase activity.
Western Blotting
All antibodies used in this study are listed in Supplemental Table 1. Proteins separated on 10% or 12.5% SDS-polyacrylamide gel electrophoresis (PAGE) were transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore, Billerica, MA) and incubated at 4°C overnight with the antibodies listed in Supplemental Table 1. For Nrf2 nuclear localization experiments, cell fractionation was carried out using a nuclear extract kit (Active Motif, Carlsbad, CA), and the purity of each fraction was verified by Western blotting with anti-lamin B or anti-lactate dehydrogenase (LDH) antibody. In the experiments for caspase-3 cleavage after hydrogen peroxide treatment, PIJ17 tet-inducible Jurkat cells were treated with 1 μg/ml doxycycline for 24 h, followed by treatment with 10 or 30 μM t-BHQ for 48 h and then treatment with 100 μM hydrogen peroxide for 6 h. Total cell lysates were subjected to Western blotting by using anti-caspase-3 antibody. After incubation with secondary antibodies conjugated with horseradish peroxidase, proteins were visualized using an ECL detection kit (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) or HyGLO (Denville Scientific, Metuchen, NJ).
Northern Blotting
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Two to 10 μg of total RNA was separated on a 1% agarose gel containing 5% formaldehyde in 3-(N-morpholino)propanesulfonic acid buffer, followed by capillary transfer of separated RNA to an Immobilon-NC nitrocellulose membrane (Millipore). 32P-Labeled cDNA probe for human ferritin H, ferritin L, or NQO1 was used for hybridization at 42°C overnight. Membranes were then washed in 0.5× SSC (0.07 M sodium chloride, 0.007 M sodium citrate) at 52°C for 0.5–1 h and subjected to autoradiography.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were carried out as described previously (Iwasaki et al., 2007) by using the ChIP assay kit (Millipore), with some minor modifications. Briefly, in total 1–3 × 106 Jurkat or K562 cells/100-mm plates were treated with 10 or 50 μM t-BHQ for indicated incubation periods, followed by chromatin cross-linking and preparation of cell lysates. In PI3K inhibitor experiments, cells were pretreated with 50 μM LY294002 for 1 h before treatment with t-BHQ and incubated for 3–6 h in the continuous presence of LY294002. Approximately 1/10 aliquots of cell lysate containing sheared DNA by sonication (Iwasaki et al., 2007) were immunoprecipitated with 1–2 μg each of antibodies listed in Supplemental Table 1. PCR was performed in 50-μl reactions containing [32P]dCTP, Advantage 2 PCR polymerase mix (Clontech), and a pair of primers (sequences in the Supplemental Table 1) to amplify human ferritin H ARE-containing 0.15 kb region or a 2-kb downstream non-ARE 0.2-kb region. The PCR samples were loaded and separated on an 8% acrylamide gel and subjected to autoradiography. Quantitative analysis of ChIP DNA bands was done using ImageJ software (National Institutes of Health, Bethesda, MD).
Small Interfering RNA (siRNA) Transfection
In PTEN siRNA experiments, 1 × 107 K562 cells were electroporated with 100 pmol of siPTEN (D-003023-05; Dharmacon RNA Technologies, Lafayette, CO) by using Gene Pulser X-Cell (Bio-Rad), preoptimized setting for K562 cells, in FBS- and antibiotic-free media. After incubation of electroporated K562 cells in the cuvette for 10 min at room temperature, the cells were suspended in 10 ml of FBS-, antibiotic-free Opti-MEM (Invitrogen), and incubated for 24 h in a 100-mm dish. Then, 2 ml of fresh FBS was added and incubated for another 24 h. For ChIP assay experiments, 22 ml of new RPMI-1640 medium containing FBS was added and divided into 3, 100-mm dishes, each containing 11 ml of cell suspension. After 24-h incubation, cells were treated with 10 or 50 μM t-BHQ for 6 h, and 1 ml of 11 ml was taken for whole cell lysate preparation for Western blotting. Ten milliliters of electroporated cells was subjected to ChIP assays. For Northern blotting experiments, cells were divided into six-well plates (2 ml/well), treated with t-BHQ for 24 h, and 4 μg of total RNA was hybridized with 32P-labeled human ferritin H and NQO1 cDNA probes.
RESULTS
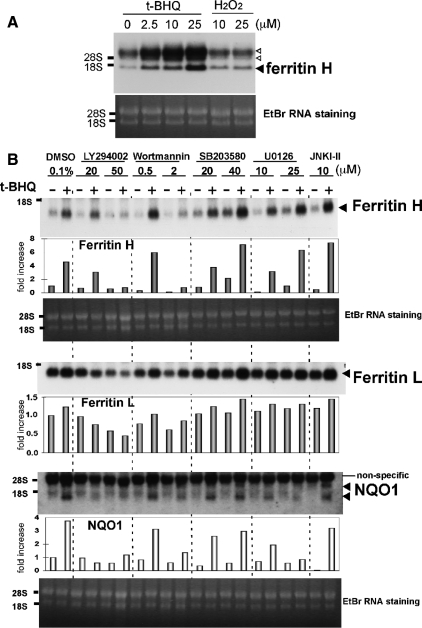
Inhibition of t-BHQ-mediated Ferritin and NQO1 mRNA Induction by PI3K Inhibitors
We reported that the phenolic antioxidant t-BHQ induced ferritin H mRNA in several different cell types, with approximately two- to threefold induction at 50–250 μM t-BHQ (Tsuji et al., 2000; Tsuji, 2005; Iwasaki et al., 2006). However, treatment of human T cell leukemia Jurkat cells with much lower concentrations of t-BHQ (2.5–25 μM) reproducibly induced ferritin H mRNA by 7- to 10-fold (7.8-fold at 25 μM t-BHQ, Figure 1A). This enhanced induction of ferritin H mRNA was not observed by treatment with H2O2 (Figure 1A), another activator of ferritin H transcription in several different cell types (Tsuji et al., 2000; Tsuji, 2005). We investigated why the induction of ferritin H mRNA by t-BHQ is so enhanced in Jurkat cells. Wange and his colleagues demonstrated that Jurkat cells have a defect in the tumor suppressor PTEN due to a frame-shift insertion of nucleotides in exon 7, resulting in constitutively activated PI3K-AKT pathways (Shan et al., 2000). To investigate whether the elevated PI3K pathway in Jurkat cells is involved in the enhanced induction of ferritin H, we pretreated Jurkat cells with LY294002 or wortmannin, both PI3K inhibitors, or with various mitogen-activated protein kinase (MAPK) inhibitors for 1 h, and then we treated the cells with 10 μM t-BHQ for 24 h. Indeed, the PI3K inhibitor LY294002 and wortmannin completely blocked t-BHQ–mediated ferritin H mRNA induction in Jurkat cells, whereas p38, extracellular signal-regulated kinase, and c-Jun NH2-terminal kinase (JNK) MAPK inhibitors (SB203580, U0126, and JNK inhibitor-II, respectively) failed (Figure 1B). Induction of NQO1, a phase II detoxification gene, by t-BHQ treatment was similarly inhibited by pretreatment with the PI3K inhibitors (Figure 1B). Ferritin L mRNA was induced to a lesser extent than ferritin H by t-BHQ; this induction was also blocked by the PI3K inhibitors (Figure 1B). These results suggest that t-BHQ may activate the PI3K pathway to induce transcription of ferritin and NQO1 genes.
Figure 1.
Induction of ferritin mRNA by t-BHQ treatment is PI3K-dependent in Jurkat cells. (A) Growing Jurkat cells were treated with t-BHQ (2.5, 10, and 25 μM) or H2O2 (10 and 25 μM) for 24 h and ferritin H Northern blot was carried out (top). The band below 18S RNA is the ferritin H mRNA (a filled arrowhead). The hybridized bands above 28S RNA (open arrowheads) were unknown but proportionally increased by t-BHQ treatment. RNA staining with ethidium bromide is shown on the bottom for verification of the quality and comparative loading of RNA. Representative results are shown from four independent experiments. (B) Growing Jurkat cells were pretreated with 0.1% dimethyl sulfoxide (DMSO), 20 and 50 μM LY294002, 0.5 and 2 μM wortmannin, 20 and 40 μM SB203580, 10 and 25 μM U0126, or 10 μM JNK inhibitor-II for 1 h, followed by 10 μM t-BHQ treatment for 24 h, and ferritin H, ferritin L, or NQO1 Northern blot was carried out. RNA staining with ethidium bromide is shown below. Representative results and the -fold increase in mRNA expression by densitometry are shown from three independent experiments.
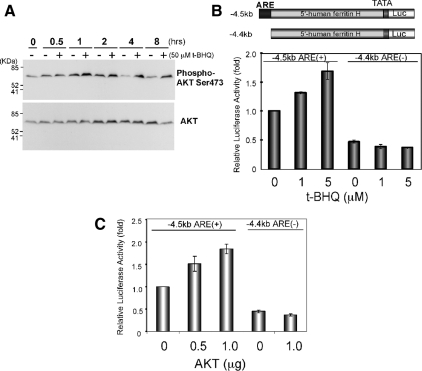
Role of AKT in Transcriptional Activation of the Ferritin H Gene via the ARE
PI3K activation results in activation of the downstream serine/threonine kinase AKT through phosphorylation of Thr308 and Ser473 by PDK (Bader et al., 2005; Cully et al., 2006). To examine whether t-BHQ treatment activates AKT, Jurkat cells treated with t-BHQ were analyzed for AKT phosphorylation by Western blotting with anti-phospho Ser473 AKT antibody. Untreated Jurkat cells showed phosphorylated Ser473-AKT (Figure 2A); t-BHQ treatment further increased this phosphorylation at 1 h and sustained the activation (Figure 2A). Pretreatment with the PI3K inhibitor LY294002 completely blocked basal and t-BHQ–mediated AKT phosphorylation (data not shown, but see Supplemental Figure S2). t-BHQ activates transcription of the ferritin H gene through the ARE (Tsuji et al., 2000; Tsuji, 2005). We observed in Jurkat cells that t-BHQ treatment activated luciferase expression driven by −4.5kb ferritin H ARE (+), but not by −4.4kb ferritin H ARE (−) (Figure 2B). This t-BHQ effect was mimicked by cotransfection of an AKT expression plasmid (Figure 2C). Collectively, these results suggest that the ferritin mRNA induction by t-BHQ is regulated by the PI3K-AKT signaling pathway via the ARE, and it seems to be amplified in Jurkat cells because of the PTEN deficiency.
Figure 2.
Ferritin H ARE is activated by t-BHQ treatment or AKT in Jurkat cells. (A) Growing Jurkat cells were either untreated (−) or treated with 50 μM t-BHQ (+), and Western blot was carried out for phospho-Ser473 AKT (top) and AKT (bottom). Representative results from four independent experiments are shown. (B) Jurkat cells (1–2 × 107) were transfected with 1 μg of −4.5kbARE(+)- or −4.4kbARE(−)-ferritin H luciferase and then treated with 1 or 5 μM t-BHQ for 24 h, or (C) along with 0.5 or 1 μg of pcDNA3.1T7-AKT1 transfection. Luciferase expression from −4.5kbARE(+)-luciferase without t-BHQ treatment in B, or with 1 μg of pcDNA3.1T7 empty vector in C was set to 1.0. Means ± SEs from three independent experiments are shown.
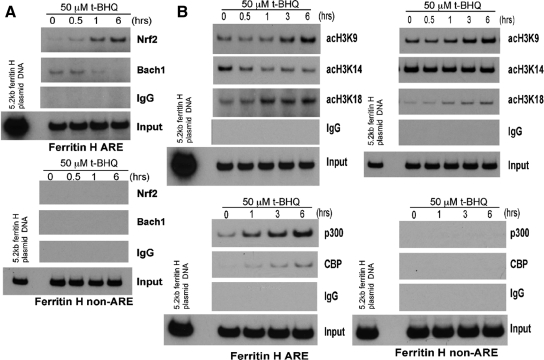
The PI3K Pathway Is Involved in the Formation of ARE Binding Complexes and Posttranslational Histone Modifications
Alterations in the activity of both transcription factors and the chromatin environment juxtaposed to an enhancer element are crucial for the overall enhancer activity. To elucidate these mechanisms, we first performed ChIP assays and tested whether the binding of transcription factors to the ARE is altered after t-BHQ treatment in Jurkat cells. The binding of Nrf2, a key ARE transcriptional activator (Motohashi and Yamamoto, 2004), to the ferritin H ARE but not to a non-ARE region was induced by 50 μM t-BHQ treatment within 1 h and was further increased until 6 h (Figure 3A). By contrast, the specific binding of Bach1 to the ARE, a transcriptional repressor of the heme oxygenase-1 gene (Sun et al., 2002), was diminished in a time-dependent manner after t-BHQ treatment (Figure 3A).
Figure 3.
Alterations in histone H3 acetylation along with recruitment of Nrf2 and p300/CBP to the ferritin H ARE in t-BHQ treated Jurkat cells. (A) Jurkat cells (1–2 × 106) were treated with 50 μM t-BHQ and subjected to chromatin immunoprecipitation with rabbit immunoglobulin G (IgG), anti-Nrf2 or anti-Bach1 antibody as described in Materials and Methods. The immunoprecipitates were subjected to semiquantitative PCR by using the ferritin H ARE and non-ARE primer sets. 0.5 pg of −5.2kb ferritin H-luciferase plasmid DNA was used as a template of the PCR-positive control and size marker of the PCR-amplified 0.15-kb (ARE) or 0.2-kb (non-ARE) DNA. Nonimmunoprecipitated DNA was also PCR amplified to assess the amount of input DNA for the ChIP assay. A representative of three independent experiments is shown. (B) Jurkat cells (1–2 × 106) treated with 50 μM t-BHQ were similarly subjected to ChIP assays with rabbit IgG or antibodies against acetyl histone H3-K9, -K14, -K18, p300, and CBP. A representative of three independent experiments is shown.
Acetylation of core histones, in particular the N terminus of histone H3, has been shown to be associated with activation of gene transcription. t-BHQ treatment increased overall histone H3 acetylation in Jurkat cells (Supplemental Figure 1). We then asked whether t-BHQ treatment induces acetylation of histone H3 juxtaposed to the ferritin H ARE by ChIP assays. As shown in Figure 3B, acetylation of histone H3 Lys9 (H3K9) and Lys18 (H3K18) on the ARE was induced at 1–3 h after t-BHQ treatment, showing a time course similar to Nrf2 binding to the ferritin H ARE. In addition, we observed recruitment of p300 and CBP histone acetyltransferases to the ferritin H ARE (Figure 3B). Conversely, histone H3K14 acetylation around the ferritin H ARE was inhibited by t-BHQ treatment (Figure 3B). Because histone modifications including acetylation are globally distributed in the genome, we next investigated these t-BHQ-induced chromatin modifications in a non-ARE region located 2 kb downstream from the ferritin H ARE. As shown in Figure 3B (right), Nrf2 and p300/CBP binding was specific to the ARE, whereas increased acetylation of H3K9 and K18 by t-BHQ treatment was not, being detected in the non-ARE region as well. This is consistent with the results that p300 recruitment to various enhancer elements serves as an epicenter that dictates a global distribution of increased histone acetylation in the human genome (Heintzman et al., 2007). The decrease in H3K14 acetylation observed in the ARE region was not significant in the non-ARE region (Figure 3B). These results suggest that t-BHQ treatment induces dynamic alterations in the ARE and non-ARE chromatin environments along with the DNA sequence-specific recruitment of Nrf2 and p300/CBP to the ARE in Jurkat cells.
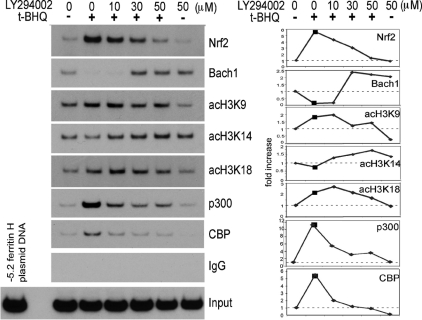
Next, we tested whether t-BHQ–mediated alterations in Nrf2 and Bach1 ARE binding and histone H3 acetylation were PI3K dependent. Our ChIP assays showed that pretreatment of Jurkat cells with the PI3K inhibitor LY294002 completely blocked t-BHQ–induced Nrf2 and p300/CBP binding to the ferritin H ARE (Figure 4). Decreased Bach1 binding to the ferritin H ARE after t-BHQ treatment was overridden by the pretreatment of cells with LY294002 (Figure 4). Furthermore, LY294002 overrode both the increased acetylation of histone H3K9 and K18 and the decreased H3K14 acetylation on the ARE (Figure 4), as well as the 2-kb downstream non-ARE region (data not shown) after t-BHQ treatment, suggesting alterations in histone acetylation in a relatively global region of the ferritin H gene. We observed that LY294002 pretreatment showed only partial inhibition of Nrf2 nuclear accumulation induced by t-BHQ treatment (Supplemental Figure 2A), suggesting that the PI3K pathway activated by t-BHQ treatment may be more important in the nuclear formation of the ARE binding complex and posttranslational modifications of core histones.
Figure 4.
PI3K inhibitor abrogates t-BHQ-mediated transcriptional events on the ferritin H ARE in Jurkat cells. Jurkat cells (1–2 × 106) were pretreated with 0–50 μM LY294002 for 1 h, followed by treatment with or without 50 μM t-BHQ for 3 h. ChIP assays using antibodies against Nrf2, Bach1, acetyl histone H3-K9, -K14, -K18, p300, or CBP were performed along with rabbit IgG as a negative control. A representative autoradiograph and quantified ChIP DNA bands (-fold increases in y-axis) are shown from three independent experiments.
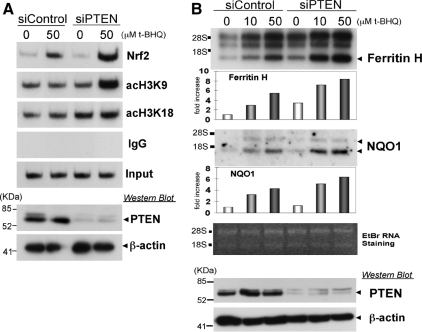
PTEN Knockdown or Forced Expression of PTEN Affects ARE Activation after t-BHQ Treatment
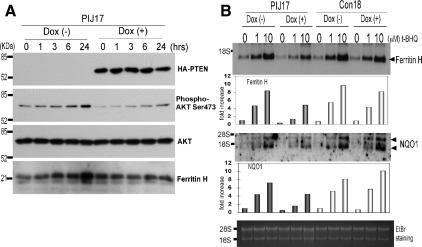
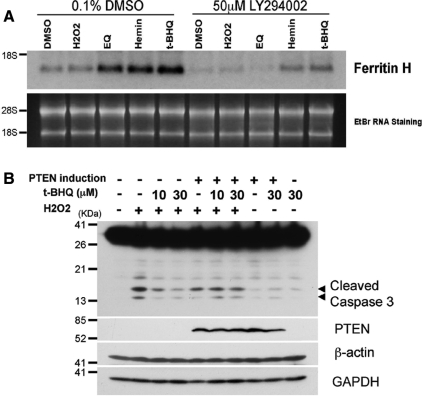
If the deficiency of the PTEN tumor suppressor enhances t-BHQ–mediated ARE activation and ARE-dependent gene expression, PTEN knockdown in wild-type PTEN expressing cells should enhance the cellular response to t-BHQ. To test this possibility, K562 human erythroleukemia cells were transiently transfected with nontargeting siRNA (siControl) or PTEN-targeting siRNA (siPTEN) and subsequently treated with t-BHQ; ChIP assays were then carried out to examine the histone acetylation status and Nrf2 binding to the ferritin H ARE. As shown in Figure 5A, the t-BHQ-mediated increase in Nrf2 binding to the ARE was enhanced in PTEN-knockdown K562 cells. Furthermore, enhanced histone H3K9 and K18 acetylation by t-BHQ treatment were observed in PTEN-knockdown K562 cells (Figure 5A). Consistently, PTEN knockdown significantly enhanced t-BHQ-mediated induction of ferritin H and NQO1 mRNA expression (Figure 5B). Conversely, forced expression of the PTEN tumor suppressor in tet-inducible PTEN Jurkat PIJ17 cells inhibited t-BHQ-induced ferritin H and NQO1 mRNA expression (Figure 6B), along with decreased Ser473 phosphorylated AKT (Figure 6A) and a partial inhibition of Nrf2 nuclear accumulation (Supplemental Figure 2B). The PI3K-dependent mechanism of ferritin H mRNA was shared by several ARE-activating agents, such as hemin (ferriprotoporphyrin IX) and ethoxyquin (6-ethoxy-1,2-dihydro-2,2,4-trimethylquinoline) (Figure 8A). Collectively, these results suggest that the status of the PTEN tumor suppressor determines the magnitude of ARE-regulated gene transcription.
Figure 5.
Effect of PTEN knockdown in K562 cells on t-BHQ–mediated histone acetylation, Nrf2 binding to the ARE, and ferritin H and NQO1 mRNA expression. (A) K562 cells (1 × 107) were electroporated with 100 pmol of nontargeting (siControl) and PTEN-targeting siRNA (siPTEN). After 60–70 h of transfection, the cells were treated with 50 μM t-BHQ for 6 h followed by ChIP assays using indicated antibodies. (B) K562 cells (1 × 107) transfected with siControl or siPTEN for 48 h were treated with 10 and 50 μM t-BHQ for 24 h, and 4 μg of total RNA was subjected to ferritin H and NQO1 Northern blot analyses. RNA staining with ethidium bromide is shown for comparative loading of RNA. In both A and B, 20 and 2 μg of whole cell lysates isolated in the same experiment were analyzed by Western blotting using anti-PTEN and anti-β-actin antibodies, respectively. Representative results and the -fold increase in mRNA expression by densitometry are shown from two independent experiments.
Figure 6.
Restoration of PTEN expression and t-BHQ–mediated induction of ferritin and NQO1 mRNAs in Jurkat cells. (A) tet-inducible PTEN Jurkat cell line (PIJ17) (5 × 106) were pretreated with or without 1 μg/ml doxycycline for 24 h, followed by treatment with 10 μM t-BHQ for 0, 1, 3, 6, or 24 h and preparation of whole cell lysates. We subjected 50 μg of whole cell lysates to Western blotting with anti-HA, anti-phospho AKT (Ser473), anti-AKT, or anti-ferritin H antibody. (B) PIJ17 and control cell line (Con18) (5 × 106) were pretreated with or without 1 μg/ml doxycycline for 24 h, followed by treatment with 0, 1, or 10 μM t-BHQ for 24 h. After the treatment, total RNA was isolated, and 10 μg of RNA was subjected to Northern blotting for ferritin H or NQO1 mRNA. Representative results and the -fold increase in mRNA expression by densitometry are shown from four independent experiments.
Figure 8.
Effect of PTEN expression on t-BHQ–mediated cytoprotection from hydrogen peroxide-induced apoptosis. (A) Jurkat cells (2 × 106) were pretreated with 0.1% DMSO or 50 μM LY294002 for 1 h, followed by treatment with 0.25% DMSO, 25 μM H2O2, 50 μM ethoxyquin (EQ), 20 μM hemin, or 10 μM t-BHQ for 16 h. Total RNA was isolated and 5 μg each of RNA was subjected to Northern blotting analysis with human ferritin H cDNA probe. RNA staining with ethidium bromide is shown for comparative loading of RNA. Representative results from three independent experiments are shown. (B) PIJ17 cells (5 × 106) were treated with 1 μg/ml doxycycline for 24 h, followed by pretreatment with 10 or 30 μM t-BHQ for 48 h before 100 μM hydrogen peroxide challenge for 6 h. We analyzed 50 μg of whole cell lysates by Western blotting using anti-caspase-3, anti-PTEN, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), or anti-β-actin antibody.
Nuclear PTEN Does Not Seem to Play a Role in the Regulation of the Ferritin H ARE
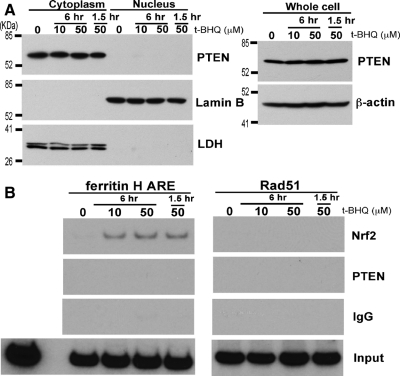
In addition to PTEN′s crucial function as a negative regulator of the PI3K pathway in the plasma membrane, PTEN was also found in the nucleus (Gimm et al., 2000; Perren et al., 2000), where it plays a role as a tumor suppressor including the maintenance of chromosomal stability through association with centromeres and transcriptional regulation of a double strand DNA-repair gene, Rad51 (Shen et al., 2007). Our results in this study show that PTEN is a negative regulator of the ferritin H ARE (Figures 5 and 6). To test whether endogenous nuclear PTEN is detectable and shuttles between nucleus and cytoplasm after t-BHQ treatment, K562 cells were treated with 10 and 50 μM t-BHQ for 1.5 and 6 h and subjected to cytoplasmic and nuclear fractionation for Western blotting. As shown in Figure 7A, nuclear PTEN was undetectable under this experimental condition before or after t-BHQ treatment with no effect of total PTEN expression levels. Because PTEN was shown to associate with a Rad51 promoter region and regulate Rad51 transcription (Shen et al., 2007), ChIP assays were performed to examine whether endogenous PTEN interacts with the ferritin H ARE in K562 cells. Under this experimental condition in K562 cells, t-BHQ treatment induced Nrf2 binding to the ferritin H ARE but not to a region containing a consensus E2F-1 binding site in the Rad51 promoter (Figure 7B) where PTEN indirectly associates via interaction with E2F-1 (Shen et al., 2007). Our ChIP assays in K562 cells by using the same anti-PTEN antibody used in the Rad51 study failed to detect endogenous PTEN binding to the Rad51 and the ferritin H ARE (Figure 7B). Collectively, we concluded that nuclear PTEN does not play a role in the ferritin H ARE in K562 cells and that the major negative regulatory role of PTEN in t-BHQ-induced ARE activation is the inhibition of the PI3K pathway.
Figure 7.
Assessment of Nuclear PTEN and its association with the ferritin H ARE in K562 cells. (A) K562 cells (3 × 106) were treated with 0, 10, or 50 μM t-BHQ for 1.5 or 6 h and subjected to isolation of whole cell lysate, cytoplasmic, and nuclear fractions. Samples (50 μg) were analyzed for protein expression by Western blotting with anti-PTEN, anti-Lamin B, anti-LDH antibodies. (B) K562 cell (1 × 106) were treated with 0, 10, or 50 μM t-BHQ for 1.5 or 6 h, and ChIP assays were performed using control IgG, anti-Nrf2, or anti-PTEN antibody. The immunoprecipitates were subjected to semiquantitative PCR by using primer sets for the ferritin H ARE or the PTEN-regulated Rad51 promoter (Shen et al., 2007).
The PTEN Status Affects Cellular Susceptibility to Hydrogen Peroxide-induced Apoptosis
Activation of the PI3K pathway is involved in cell survival through interaction with multiple signaling networks. Given the results in this study of the enhanced ARE activation in PTEN-deficient Jurkat cells by t-BHQ, we then asked whether t-BHQ treatment may promote cell survival against apoptosis and whether the PTEN status affects the efficacy. To address these questions, PTEN tet-inducible PIJ17 Jurkat cells were pretreated with t-BHQ and challenged with hydrogen peroxide to induce apoptosis. As shown in Figure 8B, hydrogen peroxide induced the cleavage of caspase 3 in PIJ17 cells, and this cleavage was blocked by t-BHQ treatment. On doxycycline treatment of PIJ17 for PTEN induction, the inhibition of caspase-3 cleavage by t-BHQ was diminished. These results suggest that t-BHQ pretreatment promotes cell survival and that PTEN status affects cellular susceptibility to apoptosis, at least in part, through ARE-regulated antioxidant detoxification genes, including ferritin H and NQO1.
DISCUSSION
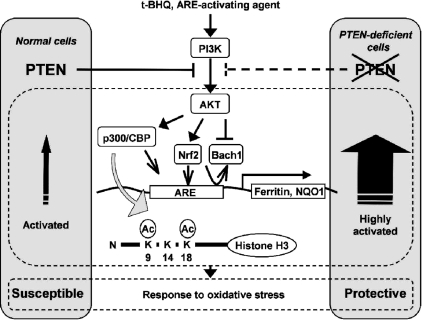
PI3K has been shown to facilitate Nrf2 nuclear accumulation and subsequent expression of ARE-regulated antioxidant genes (Lee et al., 2001; Nakaso et al., 2003; Harrison et al., 2006; Li et al., 2006); however, nuclear events downstream of the PI3K-PTEN pathway, including coordinated regulation of transcription factors, coactivators, and histone modifications leading to ARE enhancer activation, have not been elucidated. In this study, we have attempted to advance our understanding of the sequential events leading to activation of the ARE. The results obtained in this study are schematically summarized in Figure 9. We found that the PI3K-PTEN pathway regulates, in response to t-BHQ, the subsequent recruitment of transcriptional coactivator p300 and CBP, the binding of transcription factors (Nrf2 and Bach1), and histone H3 acetylation juxtaposed to the human ferritin H ARE. The binding of Nrf2, Bach1, and p300/CBP in response to t-BHQ was an event specific to the ARE, whereas alterations in the status of histone H3 acetylation were induced in both the ARE and a non-ARE region of the human ferritin H gene in a PI3K-dependent manner (Figures 3B and 4). Even a 6.5-kb downstream region from the ARE within the ferritin H coding region showed similar histone H3 acetylation after t-BHQ treatment in Jurkat cells (data not shown). This is consistent with the report that distal p300 binding sites of many enhancer elements govern global but unique chromatin signatures, including elevated histone H3 and H4 acetylation and enriched H3K4 monomethylation (Heintzman et al., 2007). Similarly, dynamic posttranslational modifications of histones seem to be induced after t-BHQ treatment in a global region of the ferritin H gene and probably in other specific regions in the genome.
Figure 9.
Model for the role of the tumor suppressor PTEN in antioxidant responsive element-mediated transcription of ferritin and NQO1 genes, associated histone modifications, and cellular antioxidant response (see text for details).
Our study shows that acetylation of histone H3 K9 and K18 were induced within 1–3 h after t-BHQ treatment in accordance with increased p300/CBP and Nrf2 binding to the ARE (Figure 3B). H3K9 methylation in heterochromatin was shown to recruit HP1 leading to chromatin compaction and gene silencing (Bannister et al., 2001), whereas H3K9 acetylation was shown to be critical for the recruitment of TFIID and subsequent gene activation (Agalioti et al., 2002). Thus, the increase in H3K9 acetylation in the vicinity of the ARE after t-BHQ treatment may allow the association between the far upstream ferritin H ARE region and the TATA box through TFIID. p300 and CBP are structurally and functionally similar proteins that have a wider range of highly overlapped histone acetylation activity (Schiltz et al., 1999; Roth et al., 2001). We observed the recruitment of both p300 and CBP to the ferritin H ARE at 1 h after t-BHQ treatment, which was followed by H3K9 and K18 acetylation (Figure 3B). Interestingly, the PI3K inhibitor LY294002 abrogated p300 and CBP recruitment and the acetylation of H3K9 and H3K18 in a dose-dependent manner (Figure 4), implying the possible involvement of phosphorylated p300 and CBP via the PI3K-AKT pathway in the acetylation of H3K9 and K18. AKT has recently been shown to phosphorylate p300 at Ser1834 and induce histone acetylation and transcriptional activation (Huang and Chen, 2005; Liu et al., 2006). The AKT phosphorylation motif in p300 (RRRMAS-Ser1834) is conserved in CBP (RRRMAT-Thr1871). Currently, antibodies to detect these specific phosphorylated p300 and CBP motifs are not available; however, in our future studies it will be pursued as to whether Ser1834 phosphorylated p300 as well as Thr1871 phosphorylated CBP are recruited to the ferritin H ARE after t-BHQ treatment. In addition to p300 and CBP, ARE binding complexes seem to be composed of more histone acetyltransferasess and chromatin remodeling proteins, including monocytic leukemia zinc-finger protein (Ohta et al., 2007), and Brahma-related gene 1, a catalytic subunit of SWI2/SNF2-like chromatin remodeling complexes (Zhang et al., 2007). Collectively, these results suggest that relatively global chromatin modifications induced by the recruitment of functionally overlapped histone acetyltransferases and chromatin remodeling complexes to the ARE in conjunction with specific binding of Nrf2 and other binding proteins to the ARE may allow transcriptional activation of ferritin H and some other ARE-regulated genes in response to t-BHQ and related electrophilic compounds.
In contrast to t-BHQ–induced acetylation of H3K9 and H3K18, acetylation of H3K14 was diminished in a PI3K-dependent manner (Figures 3B and 4). This is an unexpected result because H3K14 acetylation along with H3K9 acetylation has been associated with gene activation in yeast and mammalian cells (Agalioti et al., 2002; Pokholok et al., 2005). H3K9 and K14 acetylation seems to stimulate trimethylation of H3K4 (Milne et al., 2002), which has been observed to correlate with gene activation (Ruthenburg et al., 2007). Further investigation will be necessary to identify downstream events including characterization of proteins preferentially associated with these acetylated and methylated lysines on the histone H3 tail.
PI3K was shown to regulate nuclear localization of Nrf2 after hemin-, t-BHQ-, or insulin-mediated transcriptional activation of heme oxygenase-1 or NQO1 gene (Lee et al., 2001; Nakaso et al., 2003; Harrison et al., 2006), although the precise mechanism behind Nrf2 nuclear localization facilitated by the PI3K pathway remains unknown. In this study, the PI3K inhibitor LY294002 completely blocked Nrf2 and p300/CBP recruitment to the ARE of the ferritin gene, Bach1 dissociation from the ARE, and acetylation of histone H3 K9 and K18 upon t-BHQ treatment in Jurkat cells (Figure 4); however, it caused only partial inhibition of Nrf2 nuclear accumulation (Supplemental Figure 2A), suggesting that the major role of the PI3K pathway is the formation of the ARE binding complex and posttranslational modifications of core histones. Competitive binding of Bach1 and Nrf2 on a Maf-recognition element (MARE) or an ARE has been demonstrated previously (Sun et al., 2004; Dhakshinamoorthy et al., 2005), in which Bach1 plays a major role in determining Nrf2 binding to an ARE (Reichard et al., 2007). Bach1, a transcriptional repressor via dimerization with Maf family b-zip transcription factors on MARE and ARE sequences, was shown to be directly inactivated by heme by direct binding to heme-binding motifs in the C-terminal region (Ogawa et al., 2001) and subsequent Crm1-dependent nuclear export (Suzuki et al., 2004). Recent in vitro studies demonstrated that Bach1 directly binds to the ferritin L ARE and other ARE sequences in the absence of small Maf proteins and that hemin reversed their interactions (Hintze et al., 2007). Furthermore, recent identification of intracellular heme chaperones that control heme homeostasis (Rajagopal et al., 2008) will shed light on the roles of these heme responsive gene products in the regulation of Bach1. Bach1 is also subject to redox regulation, demonstrated by its inactivation by oxidants (Ishikawa et al., 2005). In addition to these direct inactivation mechanisms of Bach1 by heme and some oxidants, our results suggest that Bach1 is subject to a PI3K-dependent inactivation after t-BHQ treatment (Figure 4), including the possibility of phosphorylation of Bach1 and/or its binding proteins.
Growing evidence has indicated that PTEN plays a new tumor suppressor role in the nucleus (Baker, 2007; Tamguney and Stokoe, 2007). PTEN has recently been shown to associate with a Rad51 promoter region in mouse embryonic fibroblasts and PTEN-transfected PC-3 human prostate cancer cells, in which PTEN interacts with E2F-1 and enhances E2F-1–mediated transcriptional activation of the Rad51 gene (Shen et al., 2007). Our results in this study show that PTEN is a negative regulator of ARE-regulated ferritin H and NQO1 genes (Figures 4–6). To test the possibility of nuclear PTEN-mediated negative regulation of the ferritin H ARE, we performed PTEN Western and ChIP experiments in K562 cells (Figure 7). Nuclear PTEN was detected in various cell types and undetectable or lower nuclear PTEN seems to be associated with malignant transformation (Gimm et al., 2000; Perren et al., 2000; Trotman et al., 2007). The mechanisms of PTEN nuclear import and PTEN stability via mono- and polyubiquitination have been reported previously (Trotman et al., 2007; Wang et al., 2007). In our experimental condition, endogenous PTEN was exclusively detected in the cytoplasmic fraction of K562 erythroleukemia cells (Figure 7A). In our ChIP assay, t-BHQ treatment induced Nrf2 binding to the ferritin H ARE in K562 cells, but the association of PTEN with the ferritin H ARE or the Rad51 promoter that contains the E2F-1 binding site was undetectable regardless of the t-BHQ treatment (Figure 7B). These results suggest that K562 erythroleukemic cells that express the Bcr-Abl tyrosine kinase (Lozzio and Lozzio, 1975) may have an impaired PTEN nuclear import system. These results support our conclusion that the major negative regulatory effect of PTEN on t-BHQ-induced ARE activation is the inhibition of the PI3K pathway.
Our results suggest that the genetic status of the PTEN gene can determine ferritin H transcription through the ARE. PTEN mutations have been detected in many human cancers including breast, brain, and prostate cancers (Li et al., 1997). Several studies suggest a relationship between ferritin synthesis and cancer. Serum and/or tissue ferritin levels are frequently elevated in patients with cancer (Guner et al., 1992; Elliott et al., 1994) and cancer cells (Vaughn et al., 1987; Modjtahedi et al., 1992), and a correlation was observed between high ferritin levels and advanced stages of cancer (Guner et al., 1992). Autocrine growth factors purified from culture media of lung cancer and erythroleukemia cells contained ferritin H, suggesting that ferritin H protein may have a growth-promoting function (Kikyo et al., 1994). Although the molecular mechanism of increased ferritin expression in cancer remains poorly understood, it is likely that PTEN deficiency in some of these cancer cells may contribute to the increased expression of tissue/serum ferritin and regulation of cell growth.
In PTEN-deficient Jurkat cells, t-BHQ and other ARE-activating agents such as hemin and ethoxyquin showed PI3K-dependent induction of the ferritin H gene (Figure 8A). Hemin was shown to activate PI3K leading to Nrf2 activation in human dopaminergic neuroblastoma SHSY-5Y cells (Nakaso et al., 2003). Ethoxyquin, like t-BHQ, is an antioxidant used as a food preservative that has been shown to activate Nrf2 and induce ARE-regulated detoxification genes (McMahon et al., 2001). These antioxidants have the potential to produce ROS. To examine whether the activation of the ARE is dependent on the ROS production and oxidative stress induced by t-BHQ, we pretreated Jurkat cells with 2 mM N-acetylcysteine (NAC) and examined ferritin H mRNA expression after t-BHQ treatment. Our results showed that NAC treatment failed to block t-BHQ-mediated induction of ferritin H and NQO1 mRNA expression (Supplemental Figure 3), suggesting that the ROS production may not play a key role in the activation of the PI3K pathway and the ARE in Jurkat cells.
Activation of the PI3K pathways promotes cell survival through interaction with multiple signaling networks such as p53, the Bcl2 family and NF-κB (Bader et al., 2005; Cully et al., 2006). Our results show that t-BHQ enhanced cell survival in the face of hydrogen peroxide-induced apoptosis. This suggests an alternative cell survival mechanism for the PI3K pathway; activation of this pathway by ARE-activating agents or oxidative stress conditions results in transcriptional activation of ARE-regulated antioxidant detoxification genes, including ferritin H and NQO1. This conforms to the known cytoprotective roles of ferritin and other ARE-regulated detoxification genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. R. L. Wange and O. M. Zack Howard (National Institutes of Health) for kindly providing us with the PTEN-inducible Jurkat cell lines PIJ17 and Con18. We are grateful to Paul Ray for reading this manuscript thoroughly. This work was supported in part by National Institutes of Health research grant DK-60007 (to Y. T.).
Abbreviations used:
- ARE
antioxidant-responsive element
- CBP
CREB-binding protein
- ChIP
chromatin immunoprecipitation
- LDH
lactate dehydrogenase
- NAC
N-acetyl cysteine
- NQO1
NAD(P)H:quinone oxidoreductase 1
- PI3K
phosphatidylinositol 3-kinase
- PTEN
phosphatase and tensin homologue deleted on chromosome 10
- ROS
reactive oxygen species
- t-BHQ
tert-butylhydroquinone.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0762) on January 21, 2009.
REFERENCES
- Agalioti T., Chen G., Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Andersen J. K. Oxidative stress in neurodegeneration: cause or consequence? Nat. Med. 2004;10(suppl):S18–S25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- Arosio P., Levi S. Ferritin, iron homeostasis, and oxidative damage. Free Radic. Biol. Med. 2002;33:457–463. doi: 10.1016/s0891-5849(02)00842-0. [DOI] [PubMed] [Google Scholar]
- Bader A. G., Kang S., Zhao L., Vogt P. K. Oncogenic PI3K deregulates transcription and translation. Nat. Rev. Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- Baker S. J. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Cozzi A., Corsi B., Levi S., Santambrogio P., Albertini A., Arosio P. Overexpression of wild type and mutated human ferritin H-chain in HeLa cells: in vitro role of ferritin ferroxidase activity. J. Biol. Chem. 2000;275:25122–25129. doi: 10.1074/jbc.M003797200. [DOI] [PubMed] [Google Scholar]
- Cozzi A., Corsi B., Levi S., Santambrogio P., Biasiotto G., Arosio P. Analysis of the biologic functions of H- and L-ferritins in HeLa cells by transfection with siRNAs and cDNAs: evidence for a proliferative role of L-ferritin. Blood. 2004;103:2377–2383. doi: 10.1182/blood-2003-06-1842. [DOI] [PubMed] [Google Scholar]
- Cully M., You H., Levine A. J., Mak T. W. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- Dhakshinamoorthy S., Jain A. K., Bloom D. A., Jaiswal A. K. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- Elliott R. L., Head J. F., McCoy J. L. Relationship of serum and tumor levels of iron and iron-binding proteins to lymphocyte immunity against tumor antigen in breast cancer patients. Breast Cancer Res. Treat. 1994;30:305–309. doi: 10.1007/BF00665972. [DOI] [PubMed] [Google Scholar]
- Epsztejn S., Glickstein H., Picard V., Slotki I. N., Breuer W., Beaumont C., Cabantchik Z. I. H-ferritin subunit overexpression in erythroid cells reduces the oxidative stress response and induces multidrug resistance properties. Blood. 1999;94:3593–3603. [PubMed] [Google Scholar]
- Ferreira C., Santambrogio P., Martin M. E., Andrieu V., Feldmann G., Henin D., Beaumont C. H ferritin knockout mice: a model of hyperferritinemia in the absence of iron overload. Blood. 2001;98:525–532. doi: 10.1182/blood.v98.3.525. [DOI] [PubMed] [Google Scholar]
- Fouladkou F., Landry T., Kawabe H., Neeb A., Lu C., Brose N., Stambolic V., Rotin D. The ubiquitin ligase Nedd4–1 is dispensable for the regulation of PTEN stability and localization. Proc. Natl. Acad. Sci. USA. 2008;105:8585–8590. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Wange R. L., Zhang N., Oppenheim J. J., Howard O. M. Negative regulation of CXCR4-mediated chemotaxis by the lipid phosphatase activity of tumor suppressor PTEN. Blood. 2005;106:2619–2626. doi: 10.1182/blood-2004-08-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimm O., et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am. J. Pathol. 2000;156:1693–1700. doi: 10.1016/s0002-9440(10)65040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guner G., Kirkali G., Yenisey C., Tore I. R. Cytosol and serum ferritin in breast carcinoma. Cancer Lett. 1992;67:103–112. doi: 10.1016/0304-3835(92)90132-f. [DOI] [PubMed] [Google Scholar]
- Harrison E. M., McNally S. J., Devey L., Garden O. J., Ross J. A., Wigmore S. J. Insulin induces heme oxygenase-1 through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in renal cells. FEBS J. 2006;273:2345–2356. doi: 10.1111/j.1742-4658.2006.05224.x. [DOI] [PubMed] [Google Scholar]
- Heintzman N. D., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hentze M. W., Muckenthaler M. U., Andrews N. C. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- Hintze K. J., Katoh Y., Igarashi K., Theil E. C. Bach1 repression of ferritin and thioredoxin reductase1 is heme-sensitive in cells and in vitro and coordinates expression with heme oxygenase1, beta-globin, and NADP(H) quinone (oxido) reductase1. J. Biol. Chem. 2007;282:34365–34371. doi: 10.1074/jbc.M700254200. [DOI] [PubMed] [Google Scholar]
- Hintze K. J., Theil E. C. DNA and mRNA elements with complementary responses to hemin, antioxidant inducers, and iron control ferritin-L expression. Proc. Natl. Acad. Sci., USA. 2005;102:15048–15052. doi: 10.1073/pnas.0505148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintze K. J., Theil E. C. Cellular regulation and molecular interactions of the ferritins. Cell Mol. Life Sci. 2006;63:591–600. doi: 10.1007/s00018-005-5285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. C., Chen C. C. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol. Cell Biol. 2005;25:6592–6602. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Numazawa S., Yoshida T. Redox regulation of the transcriptional repressor Bach1. Free Radic. Biol. Med. 2005;38:1344–1352. doi: 10.1016/j.freeradbiomed.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., Hailemariam K., Tsuji Y. PIAS3 interacts with ATF1 and regulates the human ferritin H gene through an antioxidant-responsive element. J. Biol. Chem. 2007;282:22335–22343. doi: 10.1074/jbc.M701477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., Mackenzie E. L., Hailemariam K., Sakamoto K., Tsuji Y. Hemin-mediated regulation of an antioxidant-responsive element of the human ferritin H gene and role of Ref-1 during erythroid differentiation of K562 cells. Mol. Cell Biol. 2006;26:2845–2856. doi: 10.1128/MCB.26.7.2845-2856.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikyo N., Suda M., Kikyo N., Hagiwara K., Yasukawa K., Fujisawa M., Yazaki Y., Okabe T. Purification and characterization of a cell growth factor from a human leukemia cell line: immunological identity with ferritin. Cancer Res. 1994;54:268–271. [PubMed] [Google Scholar]
- Lee J. M., Hanson J. M., Chu W. A., Johnson J. A. Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J. Biol. Chem. 2001;276:20011–20016. doi: 10.1074/jbc.M100734200. [DOI] [PubMed] [Google Scholar]
- Li J., Lee J. M., Johnson D. A., Johnson J. A. Antioxidant responsive element activation by quinones: antioxidant responsive element target genes, role of PI3 kinase in activation. Methods Enzymol. 2004;378:238–258. doi: 10.1016/S0076-6879(04)78019-2. [DOI] [PubMed] [Google Scholar]
- Li J., et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Li M. H., Cha Y. N., Surh Y. J. Peroxynitrite induces HO-1 expression via PI3K/Akt-dependent activation of NF-E2-related factor 2 in PC12 cells. Free Radic. Biol. Med. 2006;41:1079–1091. doi: 10.1016/j.freeradbiomed.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Liu Y., Denlinger C. E., Rundall B. K., Smith P. W., Jones D. R. Suberoylanilide hydroxamic acid induces Akt-mediated phosphorylation of p300, which promotes acetylation and transcriptional activation of RelA/p65. J. Biol. Chem. 2006;281:31359–31368. doi: 10.1074/jbc.M604478200. [DOI] [PubMed] [Google Scholar]
- Lozzio C. B., Lozzio B. B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- MacKenzie E. L., Iwasaki K., Tsuji Y. Intracellular iron transport and storage: from molecular mechanisms to health implications. Antioxid. Redox. Signal. 2008;10:997–1030. doi: 10.1089/ars.2007.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie E. L., Ray P. D., Tsuji Y. Role and regulation of ferritin H in rotenone-mediated mitochondrial oxidative stress. Free Radic. Biol. Med. 2008;44:1762–1771. doi: 10.1016/j.freeradbiomed.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie E. L., Tsuji Y. Elevated intracellular calcium increases ferritin H expression through an NFAT-independent post-transcriptional mechanism involving mRNA stabilization. Biochem. J. 2008;411:107–113. doi: 10.1042/BJ20071544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R. Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell Biol. 2001;2:422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- McMahon M., Itoh K., Yamamoto M., Chanas S. A., Henderson C. J., McLellan L. I., Wolf C. R., Cavin C., Hayes J. D. The Cap‘n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- Milne T. A., Briggs S. D., Brock H. W., Martin M. E., Gibbs D., Allis C. D., Hess J. L. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Modjtahedi N., Frebourg T., Fossar N., Lavialle C., Cremisi C., Brison O. Increased expression of cytokeratin and ferritin-H genes in tumorigenic clones of the SW 613-S human colon carcinoma cell line. Exp. Cell Res. 1992;201:74–82. doi: 10.1016/0014-4827(92)90349-d. [DOI] [PubMed] [Google Scholar]
- Motohashi H., Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Nakaso K., Yano H., Fukuhara Y., Takeshima T., Wada-Isoe K., Nakashima K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003;546:181–184. doi: 10.1016/s0014-5793(03)00517-9. [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Roe A. L., Dieter M. Z., Solis W. A., Yang Y., Dalton T. P. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- Nguyen T., Sherratt P. J., Pickett C. B. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- Ogawa K., et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Ohigashi M., Naganawa A., Ikeda H., Sakai M., Nishikawa J., Imagawa M., Osada S., Nishihara T. Histone acetyltransferase MOZ acts as a co-activator of Nrf2-MafK and induces tumour marker gene expression during hepatocarcinogenesis. Biochem. J. 2007;402:559–566. doi: 10.1042/BJ20061194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orino K., Lehman L., Tsuji Y., Ayaki H., Torti S. V., Torti F. M. Ferritin and the response to oxidative stress. Biochem. J. 2001;357:241–247. doi: 10.1042/0264-6021:3570241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J. H., Wu C. J., Chau L. Y. Post-transcriptional regulation of H-ferritin gene expression in human monocytic THP-1 cells by protein kinase C. Biochem. J. 1996;319:185–189. doi: 10.1042/bj3190185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolaou G., Pantopoulos K. Iron metabolism and toxicity. Toxicol. Appl. Pharmacol. 2005;202:199–211. doi: 10.1016/j.taap.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Perren A., Komminoth P., Saremaslani P., Matter C., Feurer S., Lees J. A., Heitz P. U., Eng C. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am. J. Pathol. 2000;157:1097–1103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard V., Renaudie F., Porcher C., Hentze M. W., Grandchamp B., Beaumont C. Overexpression of the ferritin H subunit in cultured erythroid cells changes the intracellular iron distribution. Blood. 1996;87:2057–2064. [PubMed] [Google Scholar]
- Pokholok D. K., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Rajagopal A., et al. Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature. 2008;453:1127–1131. doi: 10.1038/nature06934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Nakamura N., Vazquez F., Batt D. B., Perera S., Roberts T. M., Sellers W. R. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard J. F., Motz G. T., Puga A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Denu J. M., Allis C. D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Ruthenburg A. J., Allis C. D., Wysocka J. Methylation of lysine 4 on histone H 3, intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Schiltz R. L., Mizzen C. A., Vassilev A., Cook R. G., Allis C. D., Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- Seminario M. C., Precht P., Wersto R. P., Gorospe M., Wange R. L. PTEN expression in PTEN-null leukaemic T cell lines leads to reduced proliferation via slowed cell cycle progression. Oncogene. 2003;22:8195–8204. doi: 10.1038/sj.onc.1206872. [DOI] [PubMed] [Google Scholar]
- Shan X., Czar M. J., Bunnell S. C., Liu P., Liu Y., Schwartzberg P. L., Wange R. L. Deficiency of PTEN in Jurkat T cells causes constitutive localization of Itk to the plasma membrane and hyperresponsiveness to CD3 stimulation. Mol. Cell Biol. 2000;20:6945–6957. doi: 10.1128/mcb.20.18.6945-6957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. H., Balajee A. S., Wang J., Wu H., Eng C., Pandolfi P. P., Yin Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun J., Brand M., Zenke Y., Tashiro S., Groudine M., Igarashi K. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl. Acad. Sci. USA. 2004;101:1461–1466. doi: 10.1073/pnas.0308083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Tashiro S., Hira S., Sun J., Yamazaki C., Zenke Y., Ikeda-Saito M., Yoshida M., Igarashi K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004;23:2544–2553. doi: 10.1038/sj.emboj.7600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamguney T., Stokoe D. New insights into PTEN. J. Cell Sci. 2007;120:4071–4079. doi: 10.1242/jcs.015230. [DOI] [PubMed] [Google Scholar]
- Theil E. C. Ferritin: at the crossroads of iron and oxygen metabolism. J. Nutr. 2003;133:1549S–1553S. doi: 10.1093/jn/133.5.1549S. [DOI] [PubMed] [Google Scholar]
- Trotman L. C., et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24:7567–7578. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y., Ayaki H., Whitman S. P., Morrow C. S., Torti S. V., Torti F. M. Coordinate transcriptional and translational regulation of ferritin in response to oxidative stress. Mol. Cell Biol. 2000;20:5818–5827. doi: 10.1128/mcb.20.16.5818-5827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn C. B., Weinstein R., Bond B., Rice R., Vaughn R. W., McKendrick A., Ayad G., Rockwell M. A., Rocchio R. Ferritin content in human cancerous and noncancerous colonic tissue. Cancer Invest. 1987;5:7–10. doi: 10.3109/07357908709020300. [DOI] [PubMed] [Google Scholar]
- Wang X., et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W. W., Fahl W. E. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Senechal K., Neshat M. S., Whang Y. E., Sawyers C. L. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. USA. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem. J. 2007;404:459–466. doi: 10.1042/BJ20061611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.