Abstract
Cajal bodies (CBs) are nuclear organelles that occur in a variety of organisms, including vertebrates, insects, and plants. They are most often identified with antibodies against the marker protein coilin. Because the amino acid sequence of coilin is not strongly conserved evolutionarily, coilin orthologues have been difficult to recognize by homology search. Here, we report the identification of Drosophila melanogaster coilin and describe its distribution in tissues of the fly. Surprisingly, we found coilin not only in CBs but also in histone locus bodies (HLBs), calling into question the use of coilin as an exclusive marker for CBs. We analyzed two null mutants in the coilin gene and a piggyBac insertion mutant, which leads to specific loss of coilin from the germline. All three mutants are homozygous viable and fertile. Cells that lack coilin also lack distinct foci of other CB markers, including fibrillarin, the survival motor neuron (SMN) protein, U2 small nuclear RNA (snRNA), U5 snRNA, and the small CB-specific (sca) RNA U85. However, HLBs are not obviously affected in coilin-null flies. Thus, coilin is required for normal CB organization in Drosophila but is not essential for viability or production of functional gametes.
INTRODUCTION
The nuclear organelle now known as the Cajal body (CB) was first described in vertebrate neurons in 1903 by the Spanish neurobiologist Ramón y Cajal (Cajal, 1903). Because a CB cannot be identified by morphology alone, it has only recently become clear that CBs exist in organisms as diverse as insects, higher plants, amphibians, mammals, and even budding yeast (reviewed in Gall, 2000; Gall, 2003; Cioce and Lamond, 2005; Matera and Shpargel, 2006). The key to this realization was the discovery of the protein coilin in HeLa cells (Andrade et al., 1991; Raska et al., 1991). Human coilin is a 576-amino acid protein of poorly understood function, which is highly enriched in CBs but also occurs at low concentration throughout the nucleoplasm (Bellini, 2000). Orthologous proteins have been identified by sequence comparison and cloning in Xenopus (Tuma et al., 1993), the mouse (Tucker et al., 2000), and Arabidopsis (Collier et al., 2006), and shown to occur in prominent nuclear bodies. Several attempts to identify Drosophila coilin, either by homology search or by use of heterologous antibodies, met with little success, leading to the suggestion that Drosophila might lack coilin. Nevertheless, it was possible to identify CBs in Drosophila using other molecular markers, particularly splicing small nuclear RNAs (snRNAs), fibrillarin, the survival motor neuron (SMN) protein, and the small CB-specific (sca) RNA U85 (Darzacq et al., 2002; Liu et al., 2006b).
We report here the characterization of Drosophila coilin protein and the coil gene. We also analyzed the phenotype of three mutants in the coil gene. Two are protein nulls that lack coilin in all tissues. Typical CBs are missing from the cells of the null mutants, as judged by the absence of discrete foci of several common CB components. The third mutant has no obvious effect on coilin in somatic tissues, but lacks coilin and CBs in germline cells of the ovary and testis. We conclude that coilin is essential for maintenance of CB composition, but it is not required for viability or production of functional germ cells in Drosophila.
MATERIALS AND METHODS
Fly Stocks
Drosophila melanogaster strains were maintained at 21–23°C on a standard cornmeal-based medium. A y w stock was used for wild type control. Lsm11-EYFP transgenic flies were generated as described previously (Liu et al., 2006b). Transgenic flies that express GFP-labeled polo kinase (CG12306) were derived from a P-element protein trap screen (Buszczak et al., 2007). Several transgenic lines were made that expressed the mouse coilin gene. A Drosophila Kozak sequence was introduced into enhanced green fluorescent protein (EGFP)-mouse coilin by quick change and subcloned into pCaSpeRhs. We made two P-element constructs of full-length D. melanogaster coilin (CG8710), with EYFP (Venus) at either the amino terminus or carboxy terminus of the protein. The P-element was pUASp, in which the cloned protein is under control of the yeast upstream activating sequence (UAS) (Rorth, 1998). pUASp is a modified version of pUASt (Brand and Perrimon, 1993) and was used because it shows enhanced expression in the ovary. P-element transformation was carried out by standard procedures. Twenty different lines were obtained, 10 with EYFP at the amino terminus and 10 at the carboxy terminus. We made similar P-element constructs of the putative short isoform of CG8710, nine with EYFP at the amino terminus and 7 at the carboxy terminus. Two coilin null lines, coil199 and coil203, were generated by site-specific mutation using zinc-finger nucleases (Beumer et al., 2008). A mutant carrying a piggyBac transposon insertion in the coilin gene, PBac{5HPw+}coilinB220, was obtained from the Bloomington Stock Center (Department of Biology, Indiana University, Bloomington, IN; http://fly.bio.indiana.edu/).
Tissue Preparation
Various tissues from D. melanogaster third instars and adult flies were examined as whole mounts. Fresh tissues were isolated in Grace's insect medium (Grace, 1962) and fixed at room temperature for 10 min in 4% paraformaldehyde in phosphate-buffered saline (PBS: 135 mM NaCl, 2.5 mM KCl, 4.3 mM Na2HPO4, and 1.5 mM KH2PO4, pH 7.2). After washing in PBS, samples were used for immunofluorescence or fluorescence in situ hybridization (FISH) immediately. Alternatively, fixed samples were stored at 4°C in 0.5% horse serum and 0.3% Triton X-100 in PBS for immunostaining, or at −20°C in hybridization mix for FISH.
Antibodies
Rabbits and guinea pigs were injected with fragments of D. melanogaster coilin that had been expressed in Escherichia coli. Amino acids 1-146 with a glutathione transferase (GST) tag produced a soluble product. Amino acids 147-634 with a 6-His tag produced an insoluble product. Altogether, four rabbit antibodies were produced, two against the amino-terminal fragment (R1 and R2) and two against the carboxy-terminal fragment (R3 and R4). Similarly, four guinea pig antibodies were made (GP1-4). Crude sera were diluted 1:1000 or 1:2000 for immunostaining or 1:15,000 for Western blots. Other primary antibodies were as follows: rabbit polyclonal serum against D. melanogaster Lsm10 and Lsm11 (Liu et al., 2006b), affinity-purified chicken polyclonal serum against D. melanogaster CID (Blower and Karpen, 2001), rabbit polyclonal serum against Drosophila SMN (Ilangovan et al., 2003); mouse monoclonal antibody (mAb) 72B9 against fibrillarin (Reimer et al., 1987), mouse mAb Y12 against the “Sm” epitope (Lerner et al., 1981), mouse mAb against chicken tubulin (catalog no. T9026; Sigma-Aldrich, St. Louis, MO), mouse mAb against green fluorescent protein (GFP), and rabbit polyclonal serum against GFP (Torrey Pines BioLabs, Houston, TX). Secondary antibodies were goat anti-mouse, goat anti-rabbit, goat anti-chicken, and goat anti-guinea pig labeled with Alexa 488, 568, or 594 (Invitrogen, Carlsbad, CA).
Immunostaining
Whole mounts or cultured cells were stained with a primary antibody overnight, rinsed in PBS, and stained 4 h or overnight with a secondary antibody plus 1 μg/ml the DNA-specific dye 4′,6-diamidino-2-phenylindole (DAPI). To facilitate penetration of reagents into whole tissues, 0.3% Triton X-100 was included in all solutions. Tissues were rinsed in PBS + Triton X-100 and equilibrated for a few minutes in mounting solution (50% glycerol + 1 mg/ml 1,4-diaminobenzene) before mounting under a coverslip on standard 3- × 1-in. glass slides. Coverslips were usually ringed with nail polish. Slides were stored at −20°C for days or weeks without loss of signal.
FISH
Fluorescent RNA probes labeled with Alexa-488-uridine 5′-triphosphate (UTP), Alexa-546-UTP, or Cy5-UTP were prepared by in vitro transcription from DNA clones, polymerase chain reaction (PCR) products, or deoxyoligonucleotides as described previously (Liu et al., 2006a,b). One or more probes were prepared for each specific RNA species (U2, U4, U5, U6, and U7 snRNAs, and U85 scaRNA). Probes were diluted in the following hybridization mix: 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl + 0.15 M Na citrate, pH 7.0), 10 mM citric acid, 50 μg/ml heparin, 500 μg/ml yeast tRNA, and 0.1% Tween 20. Tissues for in situ hybridization were incubated at 42°C for several hours or overnight, depending on the probe size. In many cases, tissues were observed while still in the hybridization mix (with 1 μg/ml DAPI). Otherwise tissues were rinsed in PBS and mounted in 50% glycerol mounting solution.
Fluorescence Microscopy
Images were taken with a 40× (numerical aperture [N.A.] 1.25) or a 63× (N.A. 1.40) plan apochromatic objective on a laser-scanning confocal microscope (SP2 or SP5; Leica, Exton, PA). Images were taken with the laser intensity and photomultiplier gain adjusted so that pixels in the region of interest were not saturated (“glow-over” display). In most cases, contrast and relative intensities of the green (Alexa 488), red (Alexa 546, 568, and 594), far red (Cy5), and blue (DAPI) images were adjusted with Photoshop (Adobe Systems, Mountain View, CA).
RESULTS
Cajal Bodies and Histone Locus Bodies
Before identification of Drosophila coilin, we used other CB markers to study nuclear structures in D. melanogaster. Notably, we found two bodies that contained typical CB components (Liu et al., 2006b). One of these, which we called the CB, contained at least four molecules found in vertebrate CBs—U2 snRNA, U85 scaRNA, SMN, and fibrillarin. The second body contained the U7 small nuclear ribonucleoprotein (snRNP), and because it invariably associated with the histone gene locus, we called it the histone locus body (HLB). CBs and HLBs were frequently close to each other or touching, but they could also be far apart in the nucleus. We were puzzled by the existence of two separate nuclear bodies, each of which contained canonical CB components. Previous studies in mice showed that when coilin is knocked out, the CB is replaced by three separate bodies, each of which contains a subset of CB components (Tucker et al., 2001; Jády et al., 2003). These findings led to the idea that coilin might be the “glue” that holds the CB together and that the presence of two bodies in Drosophila might be due to the lack of coilin (Liu et al., 2006b). To test this hypothesis, we made transgenic flies that expressed EGFP fused to mouse coilin. Despite its expression in a heterologous system, mouse coilin localized to discrete nuclear foci that were often adjacent to HLBs but sometimes were completely separate from them (Figure 1A). Thus, the HLB and the coilin-positive body exhibited the same relationship as in wild-type flies. This observation suggested that mouse coilin interacts with components of Drosophila CBs and/or HLBs. Regardless, CBs and HLBs did not fuse together in the presence of mouse coilin—they retained separate identities. However, one could argue that mouse coilin might not function properly in Drosophila cells (see Supplemental Data). It thus remained imperative to determine whether Drosophila indeed lacked an orthologue of vertebrate coilin.
Figure 1.
Ovarian follicle cells. (A) From a transgenic line expressing EGFP-labeled mouse coilin, stained with anti-GFP (green) to enhance the EGFP signal and with an antibody against Drosophila Lsm11 (red), a protein specific for the U7 snRNP. DNA is stained with DAPI (blue). Only a fraction of nuclei express EGFP-coilin. In these, a single CB (green) and a single HLB (red) occur, often in proximity to each other. (B) From a y w stock, stained with antibodies against Drosophila coilin (green) and Lsm11 (red). The CBs (green) and HLBs (red) are similar in size and distribution to those in the mouse coilin transgenic line.
A Putative Drosophila Orthologue of the Vertebrate Coilin Gene
Human coilin protein and the coilin gene were first identified in 1991 (Andrade et al., 1991; Raska et al., 1991). Since then, orthologous proteins in other vertebrates (Tuma et al., 1993; Tucker et al., 2000) and in Arabidopsis (Collier et al., 2006) have been widely used as specific biochemical markers for the CB. However, attempts to identify coilin in Drosophila, Caenorhabditis, and other invertebrates have been unsuccessful.
Vertebrate coilins show two conserved regions at the ends of the protein with considerable sequence divergence in the middle (Bellini, 2000; Tucker et al., 2000). In earlier BLAST searches of the D. melanogaster genome, neither we nor others could find an orthologue of human, mouse, or Xenopus coilin. However, when we carried out BLAST searches at lower stringency, we found some similarity between the amino- and carboxy termini of vertebrate coilins, especially zebrafish coilin, and the corresponding regions of D. melanogaster gene CG8710 (Figure 2 and Supplemental Data). Encouraged by this finding, we cloned EYFP-tagged versions of CG8710 and made transgenic flies that expressed the protein under control of the GAL4-UAS system. We saw highly specific expression of CG8710 in CBs, leading us to conclude that we had at last found the elusive Drosophila coilin gene.
Figure 2.
Clustal analysis (Thompson et al., 1994) of coilin sequences, displayed in Jalview (Clamp et al., 2004). (A) The first 100 amino acids at the amino terminus are moderately well conserved across species. (B) The most conserved motif occurs near the carboxy terminus, as recognized originally by Bellini (2000). The coil gene has been cloned from Drosophila melanogaster, Arabidopsis thaliana, Xenopus laevis, Homo sapiens, and Mus musculus.
The sequence of CG8710 was used to search for orthologues in other insects. We identified similar sequences in all 12 Drosophila species whose genomes have been sequenced, as well as in three mosquitoes (Culex pipiens, Aedes aegypti, and Anopheles gambiae), and the red flour beetle (Tribolium castaneum). The conserved regions in the amino and carboxy termini of D. melanogaster are shown in Figure 2, along with sequences from several vertebrates, plants and other invertebrates.
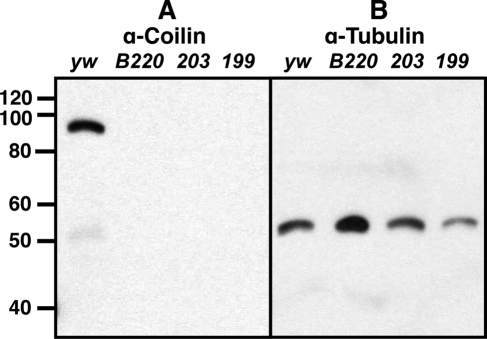
To analyze the distribution of coilin at the cellular level, we produced antibodies against two fragments of CG8710, one from the amino terminus (amino acids [aa] 1-146) and one from the carboxy terminus of the molecule (aa 147-634). On Western blots of ovary extract, each antibody recognizes a band at Mr ∼ 90 kDa, which we presume represents the full-length protein (Figure 3A). CG8710 encodes a 634-amino acid protein with predicted molecular weight (MW) of 70.6 kDa. The discrepancy between the Mr and the predicted MW is not surprising, because a similar discrepancy has been seen with human, mouse, and Xenopus coilin, all of which have Mr ∼ 80 kDa, although their calculated MWs are 59.6 kDa for Xenopus and 62.6 and 62.3 kDa for human and mouse, respectively. The 90-kDa band is missing from Western blots of ovarian tissue from three coilin mutants, which we describe later (Figure 3A). For this reason, we are confident that the 90-kDa band represents full-length Drosophila coilin.
Figure 3.
(A) Western blot of D. melanogaster ovary proteins probed with a guinea pig antibody (GP3) against D. melanogaster coilin. Lane 1, y w; lane 2, coilB220; lane 3, coil203; and lane 4, coil199. The major band at ∼90 kDa presumably represents full-length coilin. No bands are present in the mutant ovaries. (B) The same filter after stripping and reprobing with an anti-tubulin antibody.
Drosophila Coilin in CBs and HLBs
In the absence of a well-defined biochemical function for coilin, further evidence that CG8710 is the Drosophila orthologue of vertebrate coilin comes from analysis of its cellular and tissue distribution. We studied this distribution by antibody staining and by analysis of 20 independent transgenic lines that express EYFP-labeled Drosophila coilin. All of the antibodies showed strong staining of the previously identified CBs in all tissues examined (Figures 4, 5, and 7). Similarly, those transgenic lines in which EYFP was at the amino terminus of coilin were expressed in many tissues in a pattern similar to that seen with the antibodies (Figure 6). For rescue experiments showing that EYFP-coilin can substitute for endogenous coilin, see Supplemental Figure S1. However, those lines with EYFP at the carboxy terminus showed extensive accumulation of labeled protein in multiple aggregates in both the nucleus and cytoplasm. In an earlier study of human coilin, misexpression was found when the GFP label was at the carboxy terminus, but not when it was at the amino terminus (Shpargel et al., 2003), although the effect was not so extreme as in Drosophila. The following description of Drosophila coilin distribution is based on both antibody staining and expression of EYFP-coilin, labeled at the amino terminus. For identification of the HLB, we used FISH for U7 snRNA or immunostaining for Lsm10 and Lsm11 (Liu et al., 2006b), which are specific components of the U7 snRNP.
Figure 4.
Drosophila coilin in CBs of Malpighian tubule cells. (A–C) Coilin (red) colocalizes with a high concentration of snRNPs (green) in a single CB. snRNPs also occur throughout the nucleus. In the overlay, DNA is stained with DAPI (blue). snRNPs are stained with mAb Y12, which recognizes symmetric dimethylarginine (SDMA). (D–F) Coilin (red) colocalizes with fibrillarin (green) in a single CB. The majority of fibrillarin stain is in the nucleolus. In the overlay, DNA is stained with DAPI (blue).
Figure 5.
Germarium from a y w fly stained with antibodies against coilin (red) and Lsm11 (green). DNA is stained with DAPI (blue). (A) Entire germarium with germline stem cells to the left and a stage 1 egg chamber to the right. (B–D) Enlarged view of the two germline stem cells (asterisks), showing a prominent HLB in each nucleus but only diffuse coilin stain without a detectable CB. A CB is present in later stages, often closely associated with the HLB.
Figure 7.
CBs and HLBs in various tissues. (A) Testis from third instar y w larva, stained with antibodies against coilin (green) and Lsm11 (red). DNA is stained with DAPI (blue). The clustered somatic hub cells each display a green CB and a red HLB. The ring of germline stem cells immediately surrounding the hub has a HLB but no evident CB. (B) Anterior end of the larval testis, stained with antibodies against coilin (green) and fibrillarin (red). DNA is stained with DAPI (blue). The large green nuclei on the right are primary spermatocytes in prophase of the first meiotic division. Note the high level of diffuse coilin stain in these nuclei. Fibrillarin stain is prominent in nucleoli through the early spermatocyte stage. (C) Spermatocyte divisions, stained with an antibody against coilin (green) and with DAPI (blue) to show the chromosomes. Note the intense coilin stain in the meiotic spindles. (D) Nuclei from the ejaculatory duct of an adult y w male, stained with antibodies against coilin (green) to show CBs and Lsm 11 (red) to show HLBs. DNA is stained with DAPI (blue). (E) Larval brain stained as in D. In the larval brain, CBs and HLBs are tightly associated with each other. Most of the apparent colocalization of red and green signals is due to orientation of the red and green bodies along the z-axis of the image. (F–H) Nucleus from a cell of the adult Malpighian tubule, stained with antibodies against coilin (red) and Lsm11 (green). DNA is stained with DAPI (blue). Two prominent red CBs are present in this nucleus. Note that the HLB (arrowhead) contains a low level of coilin.
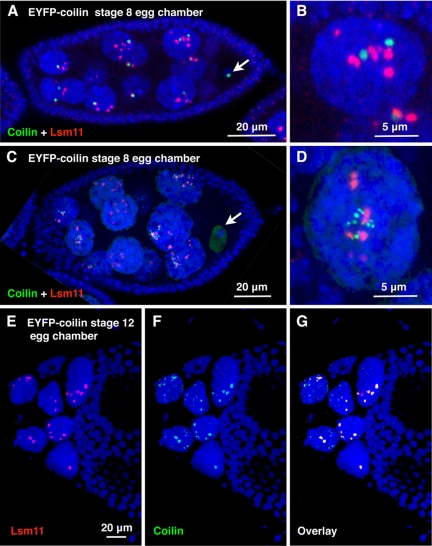
Figure 6.
CBs and HLBs in mid- and late stage egg chambers, from a transgenic line expressing YFP-coilin, stained with anti-GFP (green) to enhance the YFP signal and with anti-Lsm11 (red). DNA is stained with DAPI (blue). (A) This stage 8 egg chamber has a prominent CB in the GV (arrow). There are one or two CBs (green) and multiple HLBs (red) in the nurse cell nuclei. (B) A single nurse cell nucleus from A at higher magnification. (C) This slightly later stage 8 egg chamber no longer has a detectable CB in the GV (arrow). The CBs in the nurse cell nuclei have begun to fragment. (D) A single nurse cell nucleus from C at higher magnification, showing multiple small CBs. The HLBs are similar to those in younger egg chambers. (E–G) Nurse cell nuclei at the anterior end of a stage 12 egg chamber, after nurse cell ”dumping.“ The HLBs (red in E) now display obvious coilin (green in F), but only a few small CBs remain (smaller green dots in F and G that lack red Lsm11).
Ovary.
Coilin is detectable in CBs from follicle cells, nurse cells, and oocytes. Generally, a single CB is observed in each follicle cell nucleus, distinct from the HLB, but often associated with it (Figure 1B). In the germarium, most somatic cells display a single CB and a single HLB. By contrast, the germline stem cells and cystocytes have prominent HLBs, whereas CBs are not evident. Instead, coilin expression is high throughout the nucleoplasm (Figure 5). Separate HLBs and CBs are evident in 16-cell egg chambers. As the egg chamber grows, the oocyte almost invariably displays a single large CB attached to the condensed chromatin (Figure 6A, arrow). This CB eventually gets smaller and usually disappears by about stage 8–9 (Figure 6C, arrow). Nurse cell nuclei in younger egg chambers, up to about stage 7–8, have one or two large CBs, along with multiple HLBs (Figure 6, A and B). Dramatic changes take place in older egg chambers. By stages 8–10, the large CBs in the nurse cells are replaced by multiple small bodies that stain brightly with coilin antibodies in wild-type flies and express EYFP-coilin in transgenic flies (Figure 6, C and D). The number of HLBs remains the same, but they now contain a low level of coilin, again as seen with antibodies in wild-type flies and by EYFP-coilin in the transgenics. At the end of oogenesis, after the nurse cells have dumped their cytoplasm into the oocyte, the degenerating nurse cell nuclei have few or no CBs. However, they retain their full complement of HLBs, which now express a high level of coilin (Figure 6, E–G).
Testis.
In the testis, coilin exhibits striking cell- and stage-specific variation in its intranuclear distribution, particularly well exemplified in testes from third instar larvae. In wild-type larvae, each somatic hub cell at the tip of the testis displays a single focus of coilin stain. In contrast, the germline stem cells immediately around the hub lack such foci (Figure 7A), although coilin is detectable throughout the nucleus. In this respect they resemble the germline stem cells of the ovary. Spermatogonial nuclei also lack discrete foci of coilin stain. Coilin is dramatically up-regulated in early spermatocytes, in which it is widely distributed throughout the nucleus exclusive of the nucleolus (Figures 7B and 9C). A large amount of coilin persists during the spermatocyte divisions, when it decorates the first and second meiotic spindles (Figure 7C). HLBs are evident in the hub cells and early germline cells (Figure 7A), but disappear by the primary spermatocyte stage.
Figure 9.
Absence of coilin from nuclei of coil199 and coil203 flies. (A) Stage 8 egg chamber from a coil203 female, stained with antibodies against coilin (green) and Lsm11 (red). DNA is stained with DAPI (blue). No coilin stain is seen in any nuclei, whereas the Lsm11 stain in the HLBs is normal. (B) Nurse cell nuclei at the anterior end of a stage 12 egg chamber from a coil203 female, after nurse cell dumping. The prominent HLBs (red) fail to stain with the antibody against coilin. Compare with Figure 6, E–G, which shows a comparable stage from a nonmutant ovary. (C and D) Primary spermatocytes stained with antibodies against coilin (green) and fibrillarin (red). In C, from a fly heterozygous for coil199, the entire nucleus stains green for coilin, except for the nucleolus, which stains red for fibrillarin. In D, from a fly homozygous for coil199, only the nucleoli stain. In primary spermatocytes, the DAPI stain is weak, because the DNA is dispersed throughout a very large nucleus. (E and F) Cells of the ejaculatory duct of male flies stained for coilin (green) and Lsm11 (red). CBs (green) and HLBs (red) are evident in E, from a fly heterozygous for coil199, whereas only HLBs (red) are seen in F, from a fly homozygous for coil199. (G and H) Cells of the ejaculatory duct of male flies after in situ hybridization for U7 snRNA (green) and U85 scaRNA (red). HLBs (green) and CBs (red) are evident in G, from a y w fly, whereas only HLBs (green) are seen in H, from a fly homozygous for coil199.
Other Larval and Adult Tissues.
CBs have been seen in all tissues examined, including larval salivary glands, fat bodies, brains (Figure 7E), and imaginal disks, as well as adult accessory glands and ejaculatory duct of the testis (Figure 7D), Malpighian tubules (Figures 4 and 7, F–H), muscles, and gut. In most cases, the HLB is detectable with antibodies against Lsm10 or Lsm11 as a separate structure, often touching the CB or in proximity to it (Figure 7, D and E). As in late stage nurse cell nuclei, HLBs in the large polytene or polyploid nuclei of the salivary glands, fat bodies, and Malpighian tubules often display a low level of coilin stain (Figure 7, F–H).
Coilin in the Cell Cycle
To determine the behavior of coilin during the cell cycle, we performed live cell imaging on early embryos derived from females that expressed EYFP-coilin (Supplemental Movie 1). We began observations at the 10th cell cycle, at which time we saw a clear fluorescent signal in the blastoderm nuclei at the surface of the embryo. During the ensuing nuclear divisions, coilin underwent extensive movements. In interphase, coilin was detectable throughout the nucleoplasm with multiple brighter foci scattered more or less randomly in the nucleus. As the chromosomes condensed during prophase, these foci became more evident, eventually lining up along the metaphase plate. Because of their position and number, these foci seem to be at or near the centromeres of the chromosomes. At the onset of anaphase, the bright foci suddenly disappeared, leaving a faint coilin signal throughout the spindle. For a short period at late telophase, coilin was detectable only in the midbody. Finally, as the nuclei reformed, coilin reappeared in multiple nuclear foci and throughout the nucleoplasm.
We also examined mitosis in cultured S2 cells, in larval brains, and in adult ovarian follicle cells. The pattern of coilin distribution during mitosis was similar to that seen in the blastoderm; namely, coilin occurred as a band of stain or a row of dots lined up along the metaphase plate. To determine more precisely the localization of coilin relative to the chromosomes, we examined mitosis in flies that expressed GFP-labeled polo kinase (Buszczak et al., 2007), a marker for the kinetochores (Figure 8A). We also stained with an antibody against CID, the Drosophila CENP-A protein (Figure 8, B–E). Both polo kinase and CID occurred as double rows of dots external to coilin, which occurred as a single row of dots or band of stain along the center of the metaphase plate. These relationships suggest that coilin lies between the sister chromatids, either at the centromeres themselves or along the entire pericentromeric heterochromatin.
Figure 8.
Coilin on metaphase chromosomes. (A) Metaphase of an ovarian follicle cell, from a fly expressing GFP-labeled polo kinase. Stained with antibodies against coilin (red) and GFP (green). DNA is stained with DAPI (blue). Note that coilin is limited to a single row of dots along the center of the metaphase plate, suggesting that coilin lies between the centromeres of sister chromatids. In contrast, polo kinase occurs in two rows of dots, presumably the sister kinetochores oriented to opposite poles of the spindle. The poles themselves also contain polo kinase (arrows). An arrowhead indicates the plane of the metaphase plate. (B–E) Metaphase from a larval neuroblast stained with antibodies against coilin (red) and CID (green). CID is Drosophila CENP-A and defines the centromeres of the chromosomes. The arrowhead indicates the metaphase plate. Note that coilin is limited to the centromere regions on the metaphase plate and that CID dots lie on either side of the coilin (toward the poles of the spindle), again suggesting that coilin lies between the sister chromatids.
CBs, but not HLBs, Are Disorganized in Coilin Mutants
We have studied three mutants of the Drosophila coil gene. Two of these, coil199 and coil203, were generated by expressing site-specific zinc finger nucleases (ZFNs) in living embryos. In coil199, the third base was deleted from codon #7 (AAG to AA−), whereas in coil203 two bases were removed from codon #7 and C was substituted for G in codon #6 (ATG AAG to ATC A−). A detailed description of the production of these two mutants was recently published (Beumer et al., 2008). The third mutant, coilB220, is an insertion of a piggyBac transposon in the 5′ untranslated region of the gene (http://flybase.bio.indiana.edu/). We used PCR to confirm that the insertion site of the transposon is 22 nt upstream of the ATG translation start site. We are unaware of other coil mutants in public databases. All three coil mutants are homozygous viable and fertile with no obvious morphological phenotype.
Because both coil199 and coil203 lead to frame-shifts in translation at the start of the coding region, we expected them to be protein null mutants. None of the tissues that we have tested from larvae or adults show any staining with antibodies against coilin (Figure 9). Western blots of ovary proteins from these two mutants are completely negative for coilin, whereas proteins from wild-type ovaries show an easily detectable band at ∼90 kDa (Figure 3).
An important question is what effect the absence of coilin has on the organization of CBs and HLBs. In wild-type flies coilin is a prominent component of all CBs, but coilin is also demonstrable in HLBs, usually at a low level (Figure 7, F–H). In late stage nurse cells, however, HLBs stain as brightly as typical CBs with antibodies against coilin (Figure 6, E–G). Because CBs and HLBs are large structures in nurse cell nuclei and both undergo striking changes during development of the wild-type egg chamber, we paid particular attention to the ovary. In the null mutants, all nurse cells and follicle cells of the ovary are negative when stained with antibodies against coilin, and all are negative for U85 scaRNA after FISH. At the same time, all cells show what seem to be normal HLBs when stained with an antibody against Lsm11 or when tested for U7 snRNA by FISH. Examples of nurse cells and oocytes are shown in Figure 9, A and B (coilin and Lsm11) and Figure 10, C and D (U85 and U7).
Figure 10.
In situ hybridization of U7 snRNA and U85 scaRNA. (A) Stage 8 egg chamber from a y w fly. In the giant nurse cell nuclei, HLBs hybridize with the U7 snRNA probe (green) and CBs with the U85 scaRNA probe (red). The green bodies in the nurse cell cytoplasm are U bodies (Liu and Gall, 2007), whereas most of the green in the oocyte at the right end of the egg chamber is due to autofluorescence of yolk. DNA is stained with DAPI (blue). (B) A single nurse cell nucleus from A. (C) Stage 8 egg chamber from a coil203 fly. HLBs hybridize with U7 snRNA (green), but no hybridization is seen with the U85 scaRNA (red) probe. DNA is stained with DAPI (blue). Arrow points to the GV. (D) A single nurse cell nucleus from C.
Somatic cells show essentially the same pattern. Most wild-type cells display one or a few CBs that contain coilin and other CB markers, and one or a few HLBs that contain Lsm11 and U7. By contrast, coilin-null cells lack any foci of CB markers, but display easily detectable HLBs. We have seen these patterns in larval salivary glands, fat bodies, and brains, as well as adult Malpighian tubules (Figure 11 and Supplemental Figure S2), and accessory gland and ejaculatory duct of the testis (Figure 9, E–H).
Figure 11.
CB and HLB components in Malpighian tubule nuclei of wild type (y w, top row) and coilin-null flies (coil199 and coil203, bottom row). CBs are present in wild-type cells but missing from coilin-null cells, whereas HLBs are present in both wild type and coilin-null cells. (A and B) FISH for U7 snRNA (green) and U85 scaRNA (red). In A, the CB and HLB are closely associated. (C and D) FISH for U2 snRNA (green) and U85 scaRNA (red). In the wild-type nucleus in C, U2 is colocalized with U85 in a distinct CB, but it is also diffusely present throughout the nucleus. In the coilin-null nucleus in D, there is no distinct CB, but diffuse U2 snRNA is present. (E and F) FISH for U5 snRNA and U85 scaRNA. In the wild-type nucleus in E, there are two CBs in which U5 and U85 are colocalized. U5 is diffusely distributed in both the wild-type and coilin-null nuclei. No CB is present in the coilin-null nucleus. (G and H) Immunostain for Lsm11 (red) and symmetric dimethylarginine (SDMA, green). A red HLB and a green CB are present in the wild-type nucleus (G) but only a red HLB is seen in the coilin-null nucleus (H). snRNP proteins (SDMA) are diffusely distributed in both wild-type and mutant nuclei. (I and J) Immunostain for Lsm11 and fibrillarin. In the wild-type nucleus (I), most of the fibrillarin is in the large nucleolus (green), with a small amount in a separate CB (green). The single HLB (red) is positive for Lsm11. In the coilin-null nucleus (J), there is a single HLB (red). The nucleolus is stained for fibrillarin, but there is no fibrillarin-positive CB.
The testis is special, because spermatogonia and spermatocytes in wild-type flies do not display typical CBs. Instead, coilin is expressed uniformly throughout the nucleus at a low level in spermatogonia, and at a high level in a patchy pattern in spermatocytes (Figure 9C). In the null mutants, coilin is undetectable by antibody staining in any cells of the testis (Figure 9D).
The coilB220 mutant is quite different from the null mutants. Immunostaining with antibodies against coilin gives normal signals in CBs of all somatic tissues examined from both larvae and adults. By contrast, germline cells in the ovary and testis are completely negative for coilin (Supplemental Figure S3). As in the coilin-null mutants, HLBs are apparently normal in these tissues. Presumably the insertion of the pBac transposon upstream of the coding region affects sequences required for germline specific expression of coilin. In keeping with the staining results, Western blots of ovary proteins from the coilB220 mutant are negative for coilin (Figure 3).
Are There Residual CBs in the Coilin-Null Mutants?
In cells derived from coilin-null mice there are no coilin-positive CBs. Instead, typical CB components are distributed among three “residual” bodies, each of which contains a subset of CB components (Tucker et al., 2001; Jády et al., 2003). To determine whether a similar situation occurs in Drosophila, we examined the distribution of several typical CB components in Malpighian tubule cells in the null flies. Nuclei in Malpighian tubules are large and they contain prominent CBs and HLBs. Often the CB and HLB are well separated from the nucleolus, making analysis of their composition easier than in cells where these bodies closely associate with the nucleolus. As already mentioned, U85 scaRNA no longer occurs in a defined structure in the ovary in null flies, and the same is true for Malpighian tubule cells (Figure 11, A–F). We also examined the following CB components: fibrillarin (associated with small nucleolar RNAs), SMN, U2 snRNA, U5 snRNA, and symmetric dimethylarginine (a marker for snRNPs). In no case were these components detectable in a distinct body within the nuclei of the nulls (Figure 11, C–J and Supplemental Figure S2, A–F). In Malpighian tubule cells of wild-type flies, there is usually a single prominent HLB, which contains U7 snRNA, Lsm11 and a low level of coilin (Figures 7, F–H, and 11, A, G, and I). Despite the absence of coilin in the nulls, there is still a body with Lsm11 and U7 snRNA (Figure 11, B, H, and J).
We find what can be considered a residual CB in one cell type, the oocyte. As already mentioned, the germinal vesicle (GV) displays a prominent coilin-positive CB up to about stage 8–9 (Figure 6A). In the coilin-null mutants, the GV is negative for coilin and U85 at all stages (Figures 9A and 10C). By contrast, strong FISH signals for U4 and U6 snRNAs are seen in these GVs (Supplemental Figure S4). In this respect, the GV differs from all other cell types that we have examined, where the absence of coilin and U85-positive foci correlates with the absence of snRNP-positive foci (Figure 11, D, F, and H, and Supplemental Figure S2D).
In summary, except for the GV, we find no evidence for residual CBs in the nuclei of larval and adult tissues of coilin-null mutants. At the same time, what seem to be typical HLBs occur in the cells we have examined. Thus, the absence of coilin disrupts the normal distribution of CB components, but leaves HLBs apparently intact.
Are There Two Isoforms of Drosophila Coilin?
FlyBase (http://flybase.bio.indiana.edu/) lists two potential isoforms of coilin, based on cDNA sequences. Sequence AY118690 encodes a 634-amino acid protein, with predicted MW 70,558 (“long isoform”). Sequence AY060878 includes an intron that is excluded from AY118690. Translation of AY060878 from the same ATG codon as AY118690 leads to a stop codon near the beginning of the intron. Translation from the second in-frame ATG at position 482 would give rise to a 488-amino acid protein of MW 53,850 (“short isoform”). By reverse transcription-PCR analysis we confirmed the existence of two RNAs in wild-type flies, one RNA with the intron and one RNA without. Among the eight antibodies that we produced, four recognize a region that occurs only in the translation product of the long isoform; the other four recognize a region that would occur in both the long and short isoforms. As mentioned, all our antibodies recognize a protein with Mr ∼ 90 kDa in ovary extracts. If a short isoform exists, it should occur as a band migrating between ∼50 and 70 kDa. It should be detected by the four antibodies directed against the carboxy terminus of coilin, but it should not be detected by the four antibodies targeted to the amino terminus of the full-length protein. So far, we have not identified such a band. Thus, if a short isoform does exist, it must be of low abundance, unstable, or occur only in certain tissues.
To further test for the existence of a short isoform, we made transgenic flies that express EYFP-labeled short isoform. In these flies, fluorescent label was detectable only in the cytoplasm. The cytoplasmic localization could be due to the lack of a nuclear localization signal in the short isoform. Because we have antibodies that recognize only the long isoform, it was possible to probe for the endogenous long isoform in tissues that expressed EYFP-labeled short isoform (Figure 12). We found an inverse relationship between the strength of the cytoplasmic signal (transgenic short isoform) and intensity of antibody stain in the CBs (endogenous long isoform). For example, in follicle cells that strongly expressed the short isoform, no long isoform was seen in the CBs, whereas normal CB stain was evident in nearby cells that did not express the short isoform. Thus, the short isoform, when expressed as an EYFP construct, localizes in the cytoplasm and causes mislocalization of the long isoform. Therefore, it seems unlikely that wild-type cells express any significant amount of the short isoform.
Figure 12.
Follicle cells of a stage 11–12 egg chamber, from a transgenic fly that expressed EYFP-labeled short isoform of coilin. EYFP-labeled protein was enhanced by staining with an antibody against GFP (green). Endogenous coilin was detected with an antibody specific for the long isoform (red). Note that red stain is limited to CBs in the nuclei, whereas green stain, corresponding to the EYFP short isoform, is limited to the cytoplasm. Furthermore, expression of the short isoform is heterogeneous. Endogenous long isoform (red) is detectable only in nuclei of cells that have little or no transgenic short isoform (green) in the cytoplasm.
In summary, the evidence from Western blots and transgenic flies suggests that the predominant isoform of Drosophila coilin is produced from the fully spliced transcript. So far, we have no direct evidence that a protein is produced from the transcript with a retained intron.
DISCUSSION
CG8710 is the Drosophila Orthologue of Vertebrate Coilin
Although vertebrate coilin was identified in 1991 (Andrade et al., 1991; Raska et al., 1991) and has been used as the signature marker for CBs since then, orthologues in invertebrate organisms have been difficult to find for two reasons. First, the amino acid sequence is not strongly conserved (Bellini, 2000); and second, the precise molecular function has been difficult to define. In searching for an orthologue in Drosophila, we relied initially on the relatively weak sequence conservation in the amino and carboxy domains of the protein. Having found what we thought was a likely candidate, CG8710, we made transgenic flies that expressed EYFP-tagged versions of CG8710 protein, and we produced antibodies against two regions of the protein. We found that CG8710 is strongly expressed in CBs, which had been identified previously on the basis of other CB markers (Darzacq et al., 2002; Liu et al., 2006b). For these reasons, we feel confident that we have identified the Drosophila orthologue of mammalian coilin. For the gene itself, we suggest the name coil, corresponding to the name of the mammalian gene (Tucker et al., 2000).
Coilin Mutants
We have analyzed three mutants of the Drosophila coil gene. Two of these, coil199 and coil203, are nulls by two criteria: lack of detectable coilin protein on Western blots and lack of coilin stain in any tissue by immunofluorescence. In wild-type flies, coilin is concentrated in prominent CBs, which are also detectable with a variety of other markers, including fibrillarin, SMN, U2 snRNA, U5 snRNA, SMDA, and U85 scaRNA. In the coilin-null mutants, however, none of these markers are concentrated in defined bodies, suggesting that loss of coilin results in disorganization of CBs. Analysis of a third coil mutant, coilB220, leads to essentially the same conclusion. In this case, coilin stain is absent from germline cells of the ovary and testis, but typical coilin-positive CBs are present in somatic tissues. The absence of coilin from germline cells is correlated with the absence of defined foci of other CB markers.
In contrast to the loss of CBs from nuclei that lack coilin, HLBs seem to be unaffected in the mutants. Even the prominent HLBs of late-stage nurse cells are present in the mutants. These HLBs contain a high level of coilin in wild-type flies, but lack coilin completely in the mutants. Thus, in Drosophila, coilin seems to be necessary for proper assembly of CBs but not for assembly of HLBs.
Coilin mutants have been described in the mouse (Tucker et al., 2001) and in Arabidopsis (Collier et al., 2006). The mouse knockout allele is not complete and could, in principle, express the N-terminal 82 amino acids of coilin, although Western blotting or immunofluorescence did not detect the terminal fragment. Mice homozygous for the knockout allele have reduced viability and fertility, and embryonic fibroblasts derived from these animals lack typical CBs. Instead, there are three residual bodies, each of which contains a subset of typical CB components (Tucker et al., 2001; Jády et al., 2003). In Arabidopsis, the no cajal body 1 (ncb-1) mutant involves a single base substitution in the Arabidopsis coilin gene. It is not known whether the mutants are null for coilin. Plants that are homozygous for ncb-1 show no significant growth defects. However, their cells lack detectable CBs on the basis of three criteria: expression of the CB marker U2B“, staining with an antibody against fibrillarin, and ultrastructure in the electron microscope. In Drosophila, cells of coilin-null mutants lack typical CBs and, except in the GV, residual bodies like those in the mouse are not evident. Although the nature of the coilin mutations is different in the three organisms that have been studied—Drosophila, mouse, and Arabidopsis—in each case coilin is required for normal CB formation, but neither coilin nor a typical CB is essential for viability.
Coilin in the Centromere and Spindle
Although coilin is well known as a constituent of CBs and the nucleoplasm, we were surprised to find easily detectable amounts of coilin in the centromeric regions of the chromosomes (embryonic blastoderm, larval brain, and ovarian follicle cells), and in the spindle (spermatocytes and embryonic blastoderm). To our knowledge, coilin has not been described in these locations in normal mammalian tissues, despite many studies in which such a distribution should have been evident. In contrast, a striking accumulation of coilin, fibrillarin, and SMN at interphase centromeres of HeLa and mouse chromosomes was recently demonstrated in cells that were infected with herpes simplex virus type 1 (Morency et al., 2007). The accumulation of coilin at centromeres was also induced independently of virus infection by knockdown of centromere protein B. It is, therefore, unclear whether coilin plays a role in normal centromere function in mammals or is recruited only when centromere composition is altered. A closer examination of Drosophila coilin during the cell cycle may shed light on this question.
Relationship between CBs and HLBs
Our earlier study of Drosophila nuclear bodies emphasized the fact that typical CB markers were distributed between two bodies, one of which we called the CB and the other the HLB (Liu et al., 2006b). Now that we have identified Drosophila coilin, two important facts emerge. First, coilin is not limited to the CB, but it is found at a low concentration in many if not all HLBs. Second, in late-stage nurse cell nuclei, not only do the HLBs contain a high concentration of coilin but also the CBs that were prominent in earlier stages break down and largely disappear.
These data suggest that a reinterpretation of other CBs may be in order, especially those in the Xenopus GV. Shortly after the discovery of coilin in HeLa cells, an orthologue of human coilin was identified in Xenopus and shown to be a prominent component of the multiple spheres or sphere organelles in the GV (Tuma et al., 1993). Because they contained coilin, the spheres were renamed coiled bodies and later Cajal bodies (Wu et al., 1994; Gall et al., 1999). Well before the discovery of coilin, it had been shown that a few spheres in the GV were attached to the chromosomes at the histone gene loci (Gall et al., 1981; Callan et al., 1991). The U7 snRNP was demonstrated in these bodies and indeed was the major snRNP there (Wu and Gall, 1993; Bellini and Gall, 1998). Thus, the structures called CBs in the Xenopus GV are HLBs by definition, even though they contain coilin. It is possible that they are hybrid bodies combining the features of both CBs and HLBs. Alternatively, the Xenopus GV may have lost its CBs during oocyte growth, ending up with HLBs that contain coilin, like those in the Drosophila nurse cell nuclei. We are currently investigating the early stages of Xenopus oogenesis to clarify this issue.
An interesting possibility is that CBs and HLBs are distinct nuclear bodies in other organisms as well. Earlier studies suggested that the mammalian U7 snRNP is colocalized with coilin in CBs (Frey and Matera, 1995; Shopland et al., 2001). However, it seems that at least in some cases the histone genes themselves are adjacent to CBs, not overlapping them (e.g., Figure 2D in Frey and Matera, 1995). Recent studies also suggest that histone processing factors are found in bodies separate from CBs, as defined by the presence of coilin (Narita et al., 2007; Bongiorno-Borbone et al., 2008). A careful reexamination of the exact relationship between the U7 snRNP, histone genes, and coilin needs to be carried out in a variety of mammalian and nonmammalian cell types.
Despite intensive study over nearly 20 years, the precise molecular function(s) of coilin remains obscure. It is our hope that the identification of Drosophila coilin will permit a variety of genetic, cell biological, and molecular studies that will help clarify not only the molecular functions of coilin but also its role in organizing the CB.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christine Murphy, Jaya Kuchibhotla, and Alison Singer for technical help. This work was supported in part by National Institutes of Health grants GM-33397 (to J.G.G.), GM-53034 and NS-41617 (to A.G.M.), GM-078571 (to D. C.), and in part by the University of Utah Cancer Center support grant. J.G.G. is American Cancer Society Professor of Developmental Genetics.
Glossary
Abbreviations used:
- CB
Cajal body
- HLB
histone locus body
- SMN
survival motor neuron.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0525) on January 21, 2009.
REFERENCES
- Andrade L.E.C., Chan E.K.L., Raska I., Peebles C. L., Roos G., Tan E. M. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M. Coilin, more than a molecular marker of the Cajal (coiled) body. BioEssays. 2000;22:861–867. doi: 10.1002/1521-1878(200009)22:9<861::AID-BIES12>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bellini M., Gall J. G. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol. Biol. Cell. 1998;9:2987–3001. doi: 10.1091/mbc.9.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Trautman J. K., Bozas A., Liu J.-L., Rutter J., Gall J. G., Carroll D. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. USA. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorno-Borbone L., De Cola A., Vernole P., Finos L., Barcaroli D., Knight R. A., Melino G., De Laurenzi V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle. 2008;7:2357–2367. doi: 10.4161/cc.6344. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. Targeted gene expression as a means of altering fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Buszczak M., et al. The Carnegie Protein Trap Library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal S.R.y. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab. Lab. Invest. Biol. 1903;2:129–221. [Google Scholar]
- Callan H. G., Gall J. G., Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Cioce M., Lamond A. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S. M., Barton G. J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Collier S., Pendle A., Boudonck K., van Rij T., Dolan L., Shaw P. A distant coilin homologue is required for the formation of Cajal bodies in Arabidopsis. Mol. Biol. Cell. 2006;17:2942–2951. doi: 10.1091/mbc.E05-12-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Jády B. E., Verheggen C., Kiss A. M., Bertrand E., Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. Eur. Mol. Biol. Organ. J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M. R., Matera A. G. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gall J. G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- Gall J. G., Bellini M., Wu Z., Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Stephenson E. C., Erba H. P., Diaz M. O., Barsacchi-Pilone G. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 1981;84:159–171. doi: 10.1007/BF00399128. [DOI] [PubMed] [Google Scholar]
- Grace T. D. Establishment of four strains of cells from insect tissues grown in vitro. Nature. 1962;195:788–789. doi: 10.1038/195788a0. [DOI] [PubMed] [Google Scholar]
- Ilangovan R., Marshall W. L., Hua Y., Zhou J. Inhibition of apoptosis by Z-VAD-fmk in SMN-depleted S2 cells. J. Biol. Chem. 2003;278:30993–30999. doi: 10.1074/jbc.M303763200. [DOI] [PubMed] [Google Scholar]
- Jády B. E., Darzacq X., Tucker K. E., Matera A. G., Bertrand E., Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. Eur. Mol. Biol. Organ. J. 2003;22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E. A., Lerner M. R., Janeway C. A., Steitz J. A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-L., Gall J. G. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc. Natl. Acad. Sci. USA. 2007;104:11655–11659. doi: 10.1073/pnas.0704977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. L., Buszczak M., Gall J. G. Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res. 2006a;14:465–475. doi: 10.1007/s10577-006-1062-5. [DOI] [PubMed] [Google Scholar]
- Liu J. L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J. G. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006b;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Shpargel K. B. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Morency E., Sabra M., Catez F., Texier P., Lomonte P. A novel cell response triggered by interphase centromere structural instability. J. Cell Biol. 2007;177:757–768. doi: 10.1083/jcb.200612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T., Yung T. M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Raska I., Andrade L.E.C., Ochs R. L., Chan E.K.L., Chang C.-M., Roos G., Tan E. M. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Reimer G., Pollard K. M., Penning C. A., Ochs R. L., Lischwe M. A., Busch H., Tan E. M. Monoclonal autoantibody from a (New Zealand black × New Zealand white) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheumatism. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mechanisms Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Shopland L. S., Byron M., Stein J. L., Lian J. B., Stein G. S., Lawrence J. B. Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol. Biol. Cell. 2001;12:565–576. doi: 10.1091/mbc.12.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel K. B., Ospina J. K., Tucker K. E., Matera A. G., Hebert M. D. Control of Cajal body number is mediated by the coilin C-terminus. J. Cell Sci. 2003;116:303–312. doi: 10.1242/jcs.00211. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K. E., Berciano M. T., Jacobs E. Y., LePage D. F., Shpargel K. B., Rossire J. J., Chan E.K.L., Lafarga M., Conlon R. A., Matera A. G. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K. E., Massello L. K., Gao L., Barber T. J., Hebert M. D., Chan E.K.L., Matera A. G. Structure and characterization of the murine p80 coilin gene, Coil. J. Struct. Biol. 2000;129:269–277. doi: 10.1006/jsbi.2000.4234. [DOI] [PubMed] [Google Scholar]
- Tuma R. S., Stolk J. A., Roth M. B. Identification and characterization of a sphere organelle protein. J. Cell Biol. 1993;122:767–773. doi: 10.1083/jcb.122.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-H.H., Gall J. G. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc. Natl. Acad. Sci. USA. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Murphy C., Gall J. G. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol. Biol. Cell. 1994;5:1119–1127. doi: 10.1091/mbc.5.10.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.