Abstract
The dioxin receptor (AhR) modulates cell plasticity and migration, although the signaling involved remains unknown. Here, we report a mechanism that integrates AhR into these cytoskeleton-related functions. Immortalized and mouse embryonic fibroblasts lacking AhR (AhR−/−) had increased cell area due to spread cytoplasms that reverted to wild-type morphology upon AhR re-expression. The AhR-null phenotype included increased F-actin stress fibers, depolarized focal adhesions, and enhanced spreading and adhesion. The cytoskeleton alterations of AhR−/− cells were due to down-regulation of constitutive Vav3 expression, a guanosine diphosphate/guanosine triphosphate exchange factor for Rho/Rac GTPases and a novel transcriptional target of AhR. AhR was recruited to the vav3 promoter and maintained constitutive mRNA expression in a ligand-independent manner. Consistently, AhR−/− fibroblasts had reduced Rac1 activity and increased activation of the RhoA/Rho kinase (Rock) pathway. Pharmacological inhibition of Rac1 shifted AhR+/+ fibroblasts to the null phenotype, whereas Rock inhibition changed AhR-null cells to the AhR+/+ morphology. Knockdown of vav3 transcripts by small interfering RNA induced cytoskeleton defects and changes in adhesion and spreading mimicking those of AhR-null cells. Moreover, vav3−/− MEFs, as AhR−/− mouse embryonic fibroblasts, had increased cell area and enhanced stress fibers. By modulating Vav3-dependent signaling, AhR could regulate cell shape, adhesion, and migration under physiological conditions and, perhaps, in certain pathological states.
INTRODUCTION
A rapidly increasing number of studies consistently recognize that the aryl hydrocarbon (dioxin) receptor (AhR) serves a physiological function in the cell. AhR has been best characterized by its ability to mediate the toxic and carcinogenic effects of environmental toxicants such as 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) (Shimizu et al., 2000; Safe, 2001; Marlowe and Puga, 2005). The mechanism of regulation of AhR during this xenobiotic-dependent process is relatively well known. Under basal conditions, AhR remains sequestered in the cytosol in a heteromolecular complex containing Hsp90, XAP2, p23, and, possibly, additional proteins. On binding to TCDD or other ligands, AhR moves to the nucleus where it interacts with the aryl hydrocarbon receptor nuclear translocator and with a partially characterized set of coactivators and/or corepressors. These protein complexes regulate the expression of genes having xenobiotic responsive elements (XREs) in their promoters (Whitlock, 1999; Barouki et al., 2007; Furness et al., 2007). As expected, many of the AhR target genes code for xenobiotic-metabolizing enzymes (Hankinson, 1995; Nebert et al., 2004). However, it is now known that AhR regulates other gene subsets with roles not directly linked to xenobiotic responses (Kolluri et al., 2001; Matikainen et al., 2001; Ogi et al., 2001; Santiago-Josefat et al., 2004; Chang et al., 2007; Frericks et al., 2007). The expanding array of AhR target genes, together with increasing evidence of AhR-regulated physiological functions in different animal models (Fernandez-Salguero et al., 1995; Schmidt et al., 1996; Mimura et al., 1997; Barouki et al., 2007; Furness et al., 2007; McMillan and Bradfield, 2007), has fostered research aimed at defining new signaling pathways requiring AhR activity.
Several reports have implicated AhR in the regulation of cell morphology and migration in mammals and in Caenorhabditis elegans (Qin and Powell-Coffman, 2004; Barouki et al., 2007), although the mechanisms involved remain rather obscure. Recent data have shown that the effects of AhR on the shape and motility of MCF-7 breast cancer cells are mediated via the c-Jun N-terminal kinase (Diry et al., 2006), a downstream element of the Rac1 pathway (Bustelo et al., 2007). We have also recently shown that immortalized mouse fibroblasts lacking AhR expression (T-FGM AhR−/−) display a flattened morphology, increased stress fibers, and impaired migration (Mulero-Navarro et al., 2005). Moreover, T-FGM AhR−/− cells had lower focal adhesion kinase phosphorylation and decreased Rac1, phosphatidylinositol 3-kinase, and extracellular signal-regulated kinase activities (Mulero-Navarro et al., 2005).
Cell morphology is the result of complex signaling pathways that act in concert to regulate F-actin dynamics and the turnover of cell–cell and cell–substratum interactions (Mitra et al., 2005). The strength and extent of cell–substratum interactions significantly determines cell migration and can be highly relevant in processes requiring major changes in plasticity such as the epithelial-to-mesenchymal transition (EMT) (Li et al., 2005). Many proteins coordinately interact next to the plasma membrane to regulate cell adhesion and migration. Rho/Rac GTPases such as RhoA, Rac1, and Cdc42 are relevant players in this process (Clark et al., 1998; Nobes and Hall, 1999). Thus, it has been shown that RhoA regulates the formation of F-actin stress fibers and focal adhesions (Ridley and Hall, 1992; Chen et al., 2002), that Rac1 promotes the formation of lamellipodia and membrane ruffles (Ridley et al., 1992; Nobes and Hall, 1999), and that Cdc42 induced de formation of filopodia (Fukata et al., 2003; Disanza et al., 2006). To ensure coordinated cytoskeleton changes, there are multiple cross-talks between RhoA, Rac1, and other GTPases. For example, it is known that Rac1 stimulation leads to the down-modulation of RhoA activity and, as a consequence, to the reduction in stress fibers and focal adhesions in certain cell types (Rottner et al., 1999; Sander et al., 1999). As most Ras family members, Rho/Rac proteins have to cycle between an inactive (guanosine diphosphate [GDP]-bound) and an active (guanosine triphosphate [GTP]-bound) states. This cycling is modulated by different regulators. GDP/GTP exchange factors (GEFs) catalyze the exchange of GDP by GTP on the GTPases, thus favoring the rapid transition of these GTPases to their activated state during cell signaling. Instead, GTPase-activating proteins (GAPs) enhance the intrinsic GTPase activity of Rho/Rac proteins, a process that is essential for the hydrolysis of the bound GTP and therefore for the transition from the active to the inactive state at the end of the stimulation cycle. Rho/Rac proteins are also regulated by other processes, such as cytosolic sequestration (via GDP dissociation inhibitor proteins), ubiquitin-mediated degradation, gene expression, and phosphorylation (Bustelo et al., 2007). When in the active state, these proteins bind to effector molecules that modulate many different biological processes such as cytoskeleton changes, gene transcription, vesicle trafficking, and cell proliferation (Bustelo et al., 2007).
To shed further light in the mechanism by which AhR mediates these cytoskeleton-related events, we have studied the effect of AhR gene deficiency in cell shape, adhesion, and F-actin structures of both immortalized and primary fibroblasts. Furthermore, the use of microarray- and signaling-based strategies has allowed us to pinpoint the connection of AhR with the vav3 proto-oncogene product, a phosphorylation-dependent exchange factor for Rho/Rac GTPases.
MATERIALS AND METHODS
Cell Culture
The AhR−/− cells used in this study were obtained from AhR-null mice that were generated by gene knockout as reported previously (Fernandez-Salguero et al., 1995). Immortalized T-FGM AhR+/+ and T-FGM AhR−/− mouse fibroblasts were produced and characterized as described previously (Mulero-Navarro et al., 2005). T-FGM fibroblasts were grown in DMEM/F-12 medium containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 μg/ml gentamicin, and 11 mM d-glucose. Primary AhR+/+ and AhR−/− MEF cells were isolated from 14.5 dpc mouse embryos as indicated (Santiago-Josefat et al., 2001). Vav3-null and wild-type mice were produced as reported previously (Sauzeau et al., 2006) and used to isolate primary mouse embryonic fibroblast (MEF) cells. MEFs were cultured in DMEM supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Culturing conditions were 37°C and 5% CO2 atmosphere. To induce actin stress fibers and to analyze changes in morphology, cells were maintained for 12 h in the absence of fetal bovine serum before experimentation.
Antibodies and Reagents
Antibody to paxillin was from BD Biosciences Transduction Laboratories (Lexington, KY). Anti-vinculin and anti-β-actin were from Sigma-Aldrich (St. Louis, MO). Anti-RhoA and anti-Rac1 were purchased from Cell Signaling Technology (Beverly, MA). Anti-Net1 was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rhodamine-labeled phalloidin was from Invitrogen (Carlsbad, CA). Taq DNA polymerase was from Ecogen (Barcelona, Spain). SuperScript II reverse transcriptase was purchased from Invitrogen. SYBR Green I was obtained from Invitrogen and QTaq DNA Polymerase mix from BD Biosciences (San Jose, CA). NSC23766 and Y27632 were obtained from Calbiochem (San Diego, CA). The expression construct for the constitutively active AhR (CA-AhR) was a generous gift from Dr. Fujii-Kuriyama (University of Tsukuba, Japan), and it was produced from the wild-type AhR by deleting the minimal PAS-B motif and by ligating the resulting construct to the green fluorescent protein (EGFP) (Ito et al., 2004).
SDS-Polyacrylamide Gel Electrophoresis (PAGE) and Western Immunoblotting
SDS-PAGE and Western blotting were performed using total cell extracts, as described previously (Santiago-Josefat et al., 2004; Mulero-Navarro et al., 2005).
Measurement of Cell Area, Immunocytochemistry, and Rhodamine-Phalloidin Staining
Cell area and the minor/major axis ratio were measured in AhR+/+ and AhR−/− T-FGM cells and MEFs by blinded analysis in different random fields for each cell culture. Cellular contour and axis were determined using the ImageJ software (National Institutes of Health, Bethesda, MD). Significance of the data was determined as indicated below (see Statistical Analyses). Fluorescence-activated cell cytometry (FACS) was used to determine that T-FGM AhR+/+ and T-FGM AhR−/− had similar cell volume. For immunocytochemistry, cultures were fixed for 20 min at room temperature in 4% paraformaldehyde (Polysciences, Warrington, PA). Vinculin and paxillin immunostaining was used to locate focal adhesions in T-FGM cells and MEFs as reported previously (Santiago-Josefat et al., 2004). To label actin stress fibers, T-FGM fibroblasts and MEF cells were also stained with rhodamin-phalloidin (Invitrogen) as indicated (Mulero-Navarro et al., 2005). Fluorescence microscopy images were taken using an Axioplan epifluorescence microscope (Carl Zeiss, Jena, Germany) equipped with Plan-Neufluoar lenses with magnifications of 20× (0.50 aperture) and 40× (0.75 aperture). Pictures were also taken using a Plan Apochromat 63× lens (1.40 aperture). Fluorochromes used were Alexa 488, rhodamine, EGFP, and enhanced yellow fluorescent protein (EYFP). Images were captured with a CoolSNAP camera (RS Photometrics, Tucson, AZ) and analyzed using the RS software.
Expression Constructs, Transient Transfections, and RNA Interference
To generate a vector encoding the AhR-EYFP fusion protein, the full-length AhR cDNA was cloned into the pEYFP vector as indicated previously (Santiago-Josefat et al., 2001). To generate a mammalian expression vector for CA-AhR-EYFP, a modified AhR cDNA encoding a constitutively active version of the receptor was amplified by polymerase chain reaction (PCR) from pEFBOS-mAhR-CA and cloned into pEYFP by using previously described protocols (Santiago-Josefat et al., 2001). Transient transfections were performed in T-FGM by using the Lipofectamine Plus reagent (Invitrogen) and 1 μg of AhR-EYFP, CA-AhR-EYFP, or Vav3-EGFP expression vectors (Movilla and Bustelo, 1999). Protein expression was maintained for 24 h, and cultures were analyzed by fluorescence microscopy as indicated below. vav3 siRNA experiments were done in T-FGM cells by transient transfection of either 20 or 100 nM of commercially available siGenome on-Target plus Smart pool (Dharmacon RNA Technologies, Lafayette, CO). Negative control experiments were also performed in parallel by transient transfection of unspecific scramble siRNAs (Dharmacon RNA Technologies).
Reverse Transcription and Real-Time PCR
Total RNA was isolated from T-FGM fibroblasts and MEF cultures by using the RNeasy kit (QIAGEN, Hilden, Germany). Reverse transcription was performed using random hexamers priming and SuperScript II transcriptase as indicated previosly (Gomez-Duran et al., 2006). Real-time PCR was performed to quantify the expression levels of the vav3 and Cyp1a1 mRNAs. Primers for vav3 were 5′-GGAGTGGAGTCAGCCATCTC-3′ (forward) and 5′-ATTGGAACGACCAGCAAATC-3′ (reverse). For Cyp1a1 were 5′-ACAGACAGCCTCATTGAGCA-3′ (forward) and 5′-GGCTCCACGAGATAGCAGTT-3′ (reverse). We used the expression of the β-actin mRNA as normalization control. This cDNA was amplified using the oligonucleotides 5′-GGTCAGAAGGACTCCTATGTGG-3′ (forward) and 5′-TCCCTCTCAGCTGTGGTGGT-3′ (reverse). Reactions were done using SYBR Green I/QTaq DNA Polymerase Mix (BD Biosciences) on an iCycler equipment (Bio-Rad, Hercules, CA), as described previously (Gomez-Duran et al., 2006).
Cloning of the vav3 Promoter, Reporter Gene Analyses, and Chromatin Immunoprecipitation
The mouse vav3 promoter was cloned from the genomic sequence included in the WI1-2862P14 phosmid vector (Wellcome Trust Sanger Institute, Cambridge, United Kingdom). Briefly, the WI1-2862P14 clone was digested with BglII and a 3574-base pair fragment containing the vav3 promoter cloned into the pGL2-Basic vector. The resulting clone was then digested with BglII + BstEII and a 2005-base pair fragment isolated and made blunt ended with Klenow DNA polymerase. The 2005-base pair fragment was finally ligated into the SmaI-digested pGL2-Basic vector. The vav3 promoter sequence was analyzed for potential XRE binding sites by using the consensus element 5′-GCGTG-3′. Correct orientation of the vav3 promoter was confirmed by digestion with a panel of restriction endonucleases. Luciferase reporter gene assays were done as indicated previously (Roman et al., 2008) by using the dual-glow luciferase kit (Promega, Madison, WI). The pRLTK vector was used as normalization control. Firefly and Renilla luciferase activities were determined in a microtiter plate luminometer MLX (Dynex Technologies, Chantilly, VA). Chromatin immunoprecipitation (ChIP) to analyze AhR binding to the vav3 promoter was performed essentially as described previously (Mulero-Navarro et al., 2006; Roman et al., 2008). Oligonucleotides used for PCR were as follows: XRE1 forward 5′-TCATCCGAGGGTTCACG-3′ and reverse 5′-GCACTGGCTTCCGACTG-3′; XRE2 forward 5′-AACCTACTTCTTTCCCTCTAC-3′ and reverse 5′-GGTCCATCCTCAGCCCCTTCC-3′; XRE3-4 forward 5′-TCCTTAGGCTGATTTATTATTGTT-3′ and reverse 5′-GCTGCTGCTGTTTCATTAGAT-3′.
Site-directed Mutagenesis
The XRE1 site in the proximal vav3 promoter was inactivated by site-directed mutagenesis changing the consensus sequence for AhR binding 5′ CACGC 3′ to 5′ CAGGA 3′ by PCR (Gomez-Duran et al., 2008a) (mutated bases underlined). Briefly, the pGL2-Vav3 full-length construct was partially digested with BglI and the linealized DNA further cut with PstI. This fragment was amplified by PCR using the oligonucleotides forward 5′-CCAGCCAGGGCGGCGGGCAGGATCCCACCCG 3′ and reverse 5′ CCGGCCGCCGCTGCAGCAGCGAGTGCC 3′. Amplification was carried out for 30 cycles in 50-μl reaction mixture containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, 0.5 μM each primer, and 2.5 U of Taq polymerase. Cycling conditions were as follows: denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min. This PCR fragment was then digested with BglI and PstI and cloned into the pGL2-Basic vector cut previously with the same restriction endonucleases. The resulting reporter construct (pGL2-Vav3-mutXRE1) was transiently transfected into Hepa 1c1c7 and c2 hepatoma cell lines as described above.
Cell Adhesion and Cell Detachment Assays
Cell adhesion was performed on plastic or fibronectin-coated culture dishes. Twelve-well plates were coated with 0.04, 0.2, 0.7, 1, 4, or 10 μg/ml fibronectin. After washing with phosphate-buffered saline (PBS) and culture medium, T-FGM cells and MEFs were seeded at 3 × 105 cells/well. After an incubation of 15 min (T-FGM cells) or 1 h (MEFs), cells were washed in PBS and fixed overnight at 4°C in 3.7% paraformaldehyde. Cultures were then stained for 15 min with crystal violet (20% methanol solution). After washing and drying, attached and expanded cells were counted de visu by using light microscopy. Cell adhesion on plastic was done as above using untreated culture plates. Cell detachment experiments were also done on plastic using cultures seeded at the same cell density. After treatment for the indicated periods with a solution containing 4 mM EGTA and 1 mM MgCl2, cultures were fixed in ice-cold methanol and stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells that remained attached were counted using the ImageJ software.
RhoA and Rac1 Activation Assays
Pull-down assays with a glutathione transferase (GST) fusion protein containing the RhoA binding domain of rhotekin (rhotekin-GST) or the Rac1 binding domain of PAK-CRIB (PAK-CRIB-GST) were performed essentially as described previously (Mulero-Navarro et al., 2005). Samples were analyzed for activated and total RhoA and Rac1 by Western immunoblotting using specific antibodies.
Microarray Experiments
Two-color cDNA microarrays containing probes for 15,000 mouse genes were used. Total RNA isolation, labeling, hybridization, normalization, and differential expression analyses were made as described previously (Martinez-Delgado et al., 2004).
Statistical Analyses
Data are shown as mean ± SEM. Statistical comparison between experimental conditions was done using GraphPad Prism 4.0 software (GraphPad Software, San Diego, CA). One-way analysis of variance followed by Dunn's test was applied. For the analyses of cell area and minor/major axis ratio, the Mann–Whitney nonparametric median statistical analysis was used.
RESULTS
Dioxin Receptor Deficiency Induces Changes in Cytoskeleton-related Processes in Both Immortalized and Primary Fibroblasts
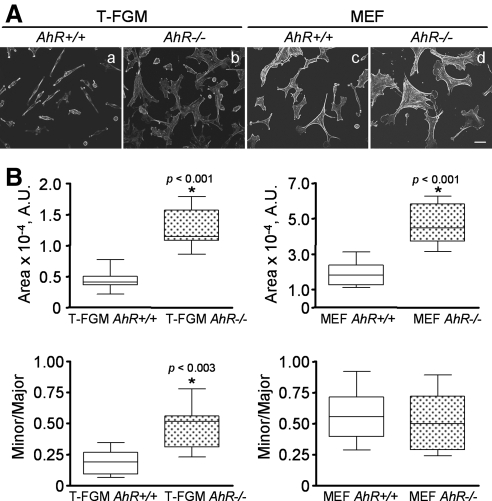
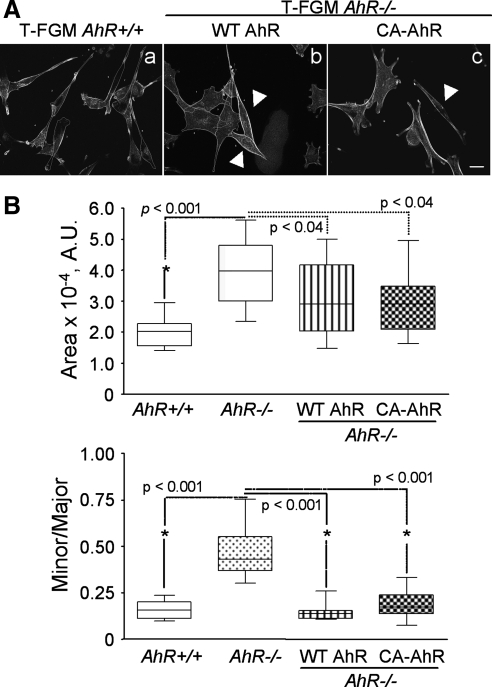
We have shown previously that immortalized mouse fibroblasts lacking AhR expression (T-FGM AhR−/− cells) had impaired migration rates (Mulero-Navarro et al., 2005). To further characterize the influence of AhR deficiency in this biological process, we decided to study in detail the changes in shape and adhesion of those cells. In addition, and to avoid secondary effects derived from the immortalization process, we also conducted these experiments with primary embryonic fibroblasts (MEFs) derived from AhR+/+ or AhR−/− mice. We observed that, irrespective of the cell type used, AhR deficiency always correlated with the generation of cells with enlarged cell area and reduced polarity (Figure 1A). Quantification and statistical comparisons of the minor/major axis ratio and cell areas corroborated these de visu observations (Figure 1B). However, the increase in cell area of AhR−/− cells with respect to AhR+/+ fibroblasts was not the consequence of higher cell volumes as determined by FACS analyses (data not shown). Transfection of AhR-deficient cells with mammalian expression vectors encoding yellow fluorescent proteins (YFPs) fused to either wild-type or constitutively active (lacking the minimal PAS-B domain) versions of AhR restored the typical shape and size of the cells (Figure 2, A and B; and Supplemental Figure S1), confirming that these differences in cell morphology were a direct consequence of the loss of AhR expression. YFP alone did not alter cell shape or size of T-FGM AhR+/+ or T-FGM AhR−/− fibroblasts, further excluding an unspecific effect (Supplemental Figure S2).
Figure 1.
Dioxin receptor expression modulates cell morphology. (A) Immortalized T-FGM cells and MEFs from AhR+/+ and AhR−/− mice were plated at the same cell density for each cell type and starved in the absence of serum for 12 h. After fixation, cultures were stained with rhodamine-phalloidin and the cellular contours defined using the ImageJ software. (B) Cell area was measured for each genotype and cell type, and data represented as median ± SEM (nonparametric Mann–Whitney U test). Minor/major axis ratios were also measured and data analyzed as indicated. At least 15 cells were analyzed for each experimental condition, and the experiments were done in at least two different cultures. The p values for statistical comparison between genotypes are indicated. The calibration bar corresponds to 20 μm. A.U., arbitrary units.
Figure 2.
AhR re-expression changes the AhR-null to the wild-type phenotype. (A) T-FGM AhR−/− cells were plated and transfected with wild-type AhR-EYFP (WT AhR) or constitutively active AhR-EYFP (CA-AhR) expression constructs. Protein expression was allowed for 24 h under fetal bovine serum deprivation. Cells were then stained with rhodamine-phalloidin and analyzed by fluorescence microscopy. The ImageJ software was used to measure YFP-labeled cells in each experimental condition. Arrowheads indicate cells expressing the ectopic proteins. (B) Cell area and minor/major axis ratios were calculated for each experimental condition and data represented as median ± SEM. At least 15 transfected cells were analyzed for each genotype, and the experiments were done in triplicate. The p values for statistical significance are indicated. The calibration bar corresponds to 10 μm. A.U., arbitrary units.
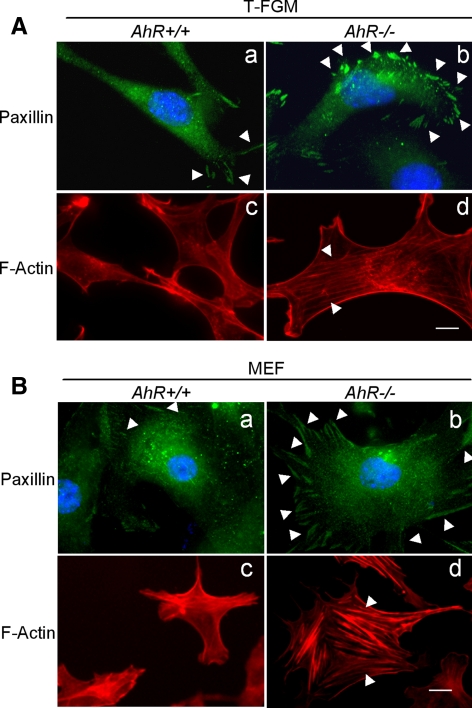
To analyze the cytoskeleton in AhR-deficient fibroblasts, we next stained T-FGM cells and MEFs of the indicated genotypes with antibodies to paxillin and rhodamine-labeled phalloidin to monitor the status of focal adhesions and the F-actin cytoskeleton in the presence or absence of AhR expression, respectively. Anti-paxillin immunofluorescence experiments indicated that the AhR deficiency was associated with an increase in focal adhesions (Figure 3, A and B, a and b) and stress fibers (Figure 3, A and B, c and d) both in T-FGM cells and MEFs. Unlike AhR-positive cells, the focal adhesions present in AhR-deficient cells were usually detected around all the cell perimeter rather than been limited and polarized to areas of membrane protrusions (Figure 3, A and B, a and b). The difference in focal adhesions among AhR+/+ and AhR−/− cells could be also influenced by altered expression of focal adhesion (FA)-localized proteins. Although paxillin expression was similar in AhR+/+ and AhR−/− T-FGM cells and MEFs, vinculin levels seemed moderately elevated in T-FGM and MEF AhR−/− fibroblasts (Supplemental Figure S3). Considering that vinculin stabilizes FAs and thereby decreases cell migration (Ziegler et al., 2006), the contribution of this protein to both increased FA content and lower migration of T-FGM and MEF AhR−/− fibroblast (Mulero-Navarro et al., 2005; Gomez-Duran et al., 2009) deserves further consideration.
Figure 3.
AhR-null cells have unpolarized focal adhesions and increased actin stress fibers. (A) T-FGM AhR+/+ and T-FGM AhR−/− cells were plated, starved for 12 h by fetal bovine serum deprivation and analyzed for focal adhesions content and distribution using anti-paxillin immunostaining and an Alexa 488-labeled secondary antibody (a and b) or for F-actin by using rhodamine-phalloidin staining (c and d). (B) The same experiments were performed in MEF cultures for focal adhesions (a and b) or actin stress fibers (c and d). The experiment was repeated in at least four different cultures of each cell type and genotype with same results. Bar, 5 μm. Arrowheads mark focal adhesions in a and b or actin stress fibers in c and d.
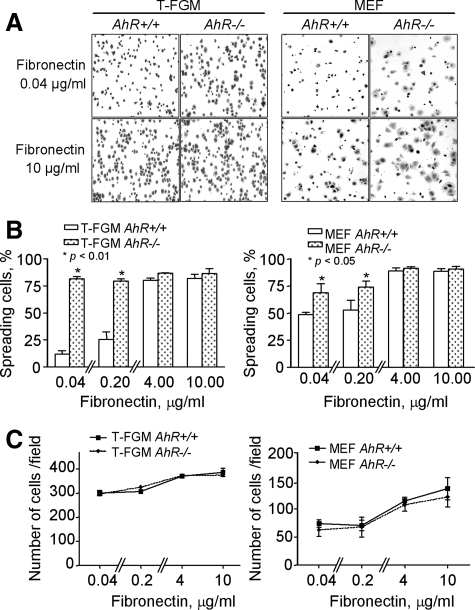
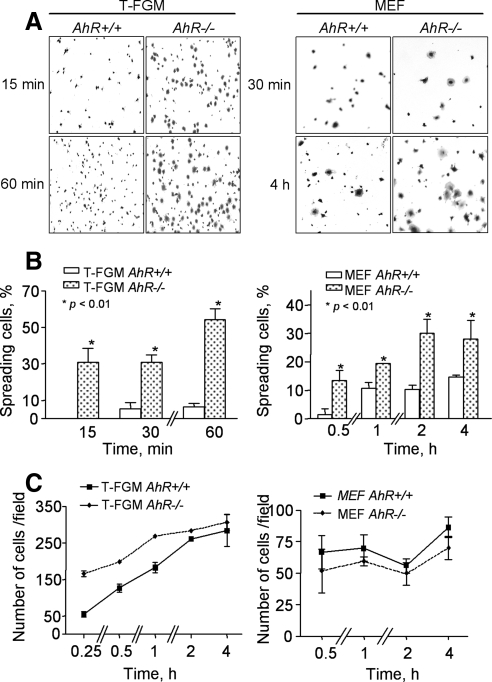
The functional relevance of AhR in cytoskeleton-related events was further analyzed by cell spreading and detachment experiments. When plated on fibronectin (FN)-coated dishes, T-FGM AhR−/− cells displayed higher spreading rates than their wild-type counterparts (Figure 4, A and B, left). However, these differences became less acute when these experiments were repeated with coverslips coated with higher FN concentrations (i.e., 10 μg/ml; Figure 4, A and B, left). Similar results where observed in AhR−/− MEFs, although in this case the difference in spreading with respect to wild-type cells was less pronounced (Figure 4, A and B, right). The lower spreading rates of AhR+/+ T-FGM cells and MEFs was not due to initial defects in substrate attachment, because total cell counts indicate equal numbers of attached cells to the dishes regardless of the genotype used (Figure 4C). The differences in spreading between AhR+/+ and AhR−/− fibroblasts became even more prominent when the plating step was performed on a plastic substratum. Thus, whereas T-FGM AhR+/+ cells showed negligible spreading on plastic after 60 min of being plated, 50% of the T-FGM AhR−/− cells efficiently spread under identical conditions (Figure 5, A and B, left). Likewise, AhR−/− MEFs displayed much higher spreading rates compared with wild-type MEFs (Figure 5, A and B, right). These differences in spreading behavior were detected until at least 4 h after plating (Figure 5B, right). Additional controls indicated again that these differences were not due to initial defects in cell attachment to the plates (Figure 5C).
Figure 4.
The absence of AhR increases cell spreading on fibronectin. (A) T-FGM cells and MEF of the indicated genotypes were cultured on dishes previously coated with increasing concentrations of FN (0.04, 0.2, 4.0, or 10 μg/ml). Representative results obtained at 0.04 and 10 μg/ml are shown for each genotype and cell type. (B) Cell spreading was quantified at 15 min (T-FGM cells) or 60 min (MEFs) by cell counting after crystal violet staining. Cytoplasmic extensions indicative of spreading was determined by blinded analysis. Percentages of spread cells were referred to the total number of cells within the same microscopy field. Between 250 and 300 T-FGM cells or 100 MEFs were counted in six independent fields for each experimental condition. The p values for statistical comparison between genotypes are indicated. (C) The total number of cells attached to the plates was counted for each FN concentration. Data are shown as mean ± SEM from cells counted in at least two different cultures of each genotype and cell type.
Figure 5.
The absence of AhR increases cell spreading on plastic. (A) T-FGM cells and MEFs from AhR+/+ and AhR−/− mice were cultured on untreated plastic dishes for the indicated times and cells stained with crystal violet. (B) Cell spreading was quantified for each experimental condition as indicated in the legend for Figure 4. The p values for statistical comparison between genotypes are indicated. (C) The variation with time of the total number of attached cells was also determined for each cell type and genotype. Data are shown as mean ± SEM from cells counted in at least two different cultures of each genotype and cell type.
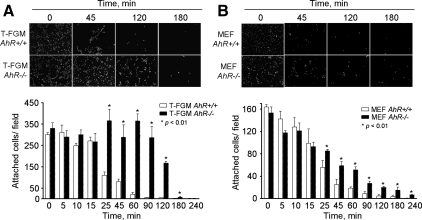
To further verify the implication of AhR in cell–substratum interactions, we resorted to measuring their kinetics of detachment upon Ca2+ chelation by EGTA treatment (see Materials and Methods). As shown in Figure 6A, the kinetic of EGTA-dependent detachment was much slower in T-FGM AhR−/− cells than in their wild-type counterparts. Similar results were obtained when AhR−/− and AhR+/+ MEFs were compared, although, similarly to the cell spreading experiments, the differences with respect to wild-type cells were less pronounced than in the case of the immortalized T-FGM cell line (Figure 6B). Together, these data strongly support a role for AhR in the regulation of cell shape, cytoskeleton organization, and adhesion. These observations are also in good agreement with our previous results indicating that T-FGM AhR−/− fibroblasts show lower migration activity (Mulero-Navarro et al., 2005).
Figure 6.
AhR expression decreases cell adhesion to the substratum. (A) T-FGM AhR+/+ and T-FGM AhR−/− cells were seeded at the same cell density on plastic dishes and then induced to detach by using treatments with EGTA for the indicated times. Nuclei were stained with DAPI, and attached cells counted using the ImageJ software. (B) MEFs of the indicated genotypes were cultured at the same initial cell density and processed and analyzed as described in A. Data are shown as mean ± SEM for measurements made in duplicate in at least two independent cultures for each genotype and cell type. The p values for statistical comparison between genotypes are indicated.
The GTPases Rac1 and RhoA Are Involved in the AhR-dependent Cytoskeleton Changes
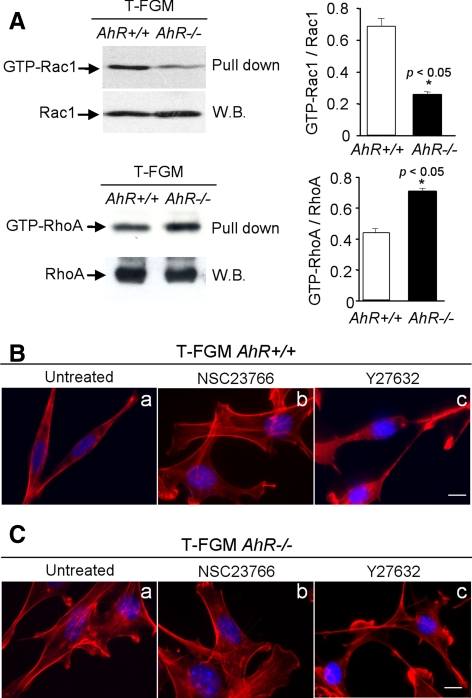
Because cytoskeleton-dependent events such as cell shape and adhesion are regulated by members of the Rho/Rac GTPase subfamily (Nobes and Hall, 1999; Fukata et al., 2003; Bustelo et al., 2007), we next explored whether variations in the expression and/or activity of these GTPases could be the cause of the phenotype detected in our fibroblast cells. Initial experiments indicated that the expression of Rac1, RhoA, and Cdc42 did not change at the mRNA or protein level (Figure 7A; data not shown), suggesting that AhR does not affect the expression of these important cytoskeleton regulators. However, pull-down assays using a PAK-CRIB-GST fusion protein revealed that T-FGM AhR−/− cells had reduced levels of activated GTP-Rac1 compared with wild-type T-FGM fibroblasts (Figure 7A, left). Because it has been reported that Rac1 inhibits RhoA activity in a number of cell systems (Rottner et al., 1999; Sander et al., 1999), we also evaluated whether the increase in stress fiber formation and the reduction in Rac1 activity detected in T-FGM AhR−/− cells could be due to increased activity of the RhoA pathway. To this end, we analyzed the activation status of RhoA by using pull-down assays with a Rhotekin-GST fusion protein. These experiments showed that T-FGM AhR−/− did have higher levels of GTP-bound RhoA than the wild-type cells (Figure 7A, right), indicating that the former cell line has higher levels of RhoA activity than the latter one.
Figure 7.
Rac1 and RhoA contribute to the AhR-dependent control of cell morphology. (A) Levels of Rac1 (GTP-Rac1) and RhoA (GTP-RhoA) activation were measured in T-FGM AhR+/+ and T-FGM AhR−/− cells by pull-down assays by using the PAK-CRIB-GST or Rhotekin-GST target proteins, respectively. Total cellular levels of Rac1 and RhoA were also determined by Western blot (W.B.) using anti-Rac1 and anti-RhoA antibodies. The levels of GTP-RhoA and GTP-Rac1 were quantified and normalized by total RhoA and Rac1 expression, respectively. Data are shown as mean ± SEM from at least two independent cultures. The p values for statistical comparison between genotypes are indicated. (B) T-FGM AhR+/+ and T-FGM AhR−/− cells were plated and treated for 6 h with the Rac1 inhibitor NSC23766 (5 μM) or (C) with the Rock inhibitor Y27632 (10 μM). Cells were then fixed, stained with rhodamine-phalloidin, and photographed. The experiment was done in three different cultures with the same results. Bar, 5 μm.
To verify whether the activity of the Rac1 and RhoA signaling pathways was altered, we treated T-FGM AhR+/+ and T-FGM AhR−/− cells with NSC23766, a drug that blocks the interaction of Rac1 with upstream GDP/GTP exchange factors and, thereby, inhibits its activation in vivo (Gao et al., 2004). NSC23766 efficiently inhibited Rac1 activation in T-FGM AhR+/+ cells (see Figure 10C, bottom) and, interestingly, we observed that the addition of this compound to T-FGM AhR+/+ cells induced a morphologically phenotype similar to that found in T-FGM AhR−/− cells, whereas it did not significantly affect the phenotype of AhR-null cells (Figure 7B, compare a and b). Those changes seen in T-FGM AhR+/+ cells included the prototypical increase in both the cell area and stress fiber formation, a result that suggest that the phenotype induced by AhR deficiency may be mediated by decreased signals from the Rac1 pathway. Next, we treated AhR+/+ and AhR−/− T-FGM cells with Y27632, a specific inhibitor of the serine/threonine kinase Rock (Uehata et al., 1997). This kinase is a downstream element of RhoA that mediates the formation of stress fibers and the actomyosin ring (Riento and Ridley, 2003; Shimokawa and Rashid, 2007). Y27632 induced the rapid disassembly of the stress fibers and, in addition, the acquisition of a T-FGM AhR+/+-like morphology in the AhR-null cells (Figure 7C, compare a and c). These results indicate that the AhR-dependent changes in cell morphology in these fibroblast cell lines rely, at least in part, in the differential engagement of the GTPases Rac1 and RhoA.
Figure 10.
The knockdown of the vav3 mRNA phenocopies the cytoskeleton-related changes observed in T-FGM AhR−/− cells. (A) Vav3 protein expression was determined in T-FGM AhR+/+ and T-FGM AhR−/− cells by Western blot with an anti-Vav3 antibody. In parallel experiments, vav3 siRNAs or unspecific scramble RNAs were transfected in T-FGM cells of both genotypes and Vav3 protein expression determined as described above. The expression of β-actin was used as control. Vav3 protein levels were normalized by β-Actin for each experimental condition and the results obtained represented on the right panel. (B) T-FGM AhR+/+ cells were transfected with vav3 siRNAs or with unspecific scramble RNAs and, 72 h later, stained with rhodamine-phalloidin (Rho-Pha) to visualize the F-actin cytoskeleton (a–c) or used in anti-paxillin immunofluorescence experiments to analyze their pattern of focal adhesions (e–g). T-FGM AhR−/− cells were also stained under the same conditions for comparison (d and h). Actin stress fibers and focal adhesions are indicated by arrowheads in c and d and g and h, respectively. Bar, 5 μm. (C) T-FGM AhR+/+ and T-FGM AhR−/− cells were grown and cell extracts prepared and used in pull-down assays for RhoA and Rac1 by using the rhotekin-GST and the PAK-CRIB-GST fusion proteins as targets, respectively. T-FGM cells of both genotypes were also transfected with vav siRNAs or with unspecific scramble siRNAs as described above and used in pull-down assays as described in B. In additional experiments, T-FGM AhR+/+ and T-FGM AhR−/− cells were treated with NSC23766 as indicated in the legend for Figure 7, and its effect on the levels of Rac1 activation was determined by pull-down assays as described above. Protein levels for RhoA and Rac1 were also determined by Western blotting (W.B.) in the same cell extracts. Levels of GTP-RhoA and GTP-Rac1 were normalized by total RhoA and Rac1 protein, and the values are plotted (right). Data are shown as mean ± SEM from experiments performed in two different cultures of each genotype. The p values for statistical comparison between genotypes under the different experimental conditions are indicated.
AhR Regulates the Constitutive Transcription of Vav3, which Is a Major Intermediate Modulating the AhR-dependent Phenotype
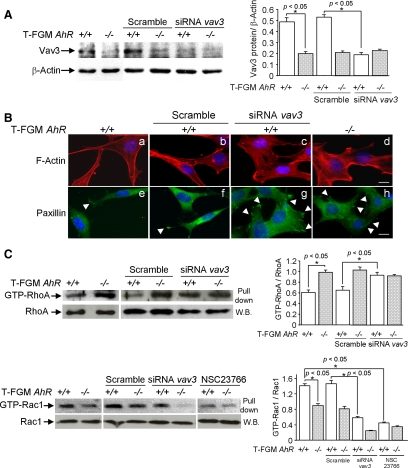
Because the expression levels of Rac1 and RhoA did not change depending on the AhR status of the cell (see above), we speculated that the variations in the activities of their signal transduction pathways could be mediated by alterations in upstream molecules (GEFs), negative regulators (i.e., GAPs), and/or downstream elements. Given the large number of putative AhR targets, we decided to conduct microarray experiments to obtain a global, genome-wide view of the transcriptomal differences present in T-FGM AhR+/+ and T-FGM AhR−/− cells. The functional annotation of the transcriptomal changes indicated that the AhR deficiency induced the down-modulation of a subset of RhoA (Net1) (Alberts and Treisman, 1998), Rac1 (Sos1, Vav3, and Abr) (Chuang et al., 1995; Nimnual et al., 1998; Bustelo, 2000), and Ras (Sos1) (Boguski and McCormick, 1993) GEFs as well as of two Rab family members (Rab1 and Rab9) (Figure 8A). In addition, it up-regulated the expression of two Rho/Rac GAPs (ArhGAP18 and ArhGAP20) and the Rab8 GTPase (Figure 8A). Two main regulators of RhoA and Rac1 GTPases that seemed markedly repressed in T-FGM AhR−/− cells were Vav3 and Net1 (Burridge and Wennerberg, 2004). Given previous results indicating the importance of Vav3 in cytoskeleton organization (Movilla and Bustelo, 1999; Hornstein et al., 2004; Couceiro et al., 2005) and its marked down-regulation in T-FGM AhR−/− cells, we decided to verify whether the modulation of this exchange factor by AhR could be a component of the cytoskeleton-related effects of this transcriptional factor. To this end, we first investigated whether the vav3 proto-oncogene was indeed a potential AhR transcriptional target. Consistent with the microarray results, we observed that the vav3 transcript was remarkably decreased in AhR−/− T-FGM and MEFs cells, as determined by quantitative, real-time PCR analysis (Figure 8B, left and middle). Importantly, unlike other classical AhR targets such as Cyp1a1 (Figure 8B, right) (Hankinson, 1995; Whitlock, 1999; Nebert et al., 2004), the expression of the vav3 mRNA could not be enhanced by the addition of the AhR ligand TCDD to T-FGM AhR+/+ cells (Figure 8B, left and middle). These results indicate that AhR functions as a transcription factor committed to maintain the constitutive expression of vav3, and suggest a relevant role for the physiological levels of Vav3 in the regulation of the AhR-dependent phenotype. Regarding Net1, we have analyzed protein expression by Western immunoblotting and found that, although repressed in our cDNA expression arrays, Net1 had similar protein levels in T-FGM AhR+/+ and T-FGM AhR−/− (Figure 8C). Even though the regulation of Net1 could deserve further analysis, based on its similar protein levels in T-FGM AhR+/+ and T-FGM AhR−/− fibroblasts, we focused our study on Vav3.
Figure 8.
AhR regulates constitutive vav3 mRNA expression. (A) Total RNA purified from T-FGM AhR+/+ and T-FGM AhR−/− cells was analyzed by microarrays as indicated in Materials and Methods. Genes above and below the horizontal dotted line are overexpressed or repressed, respectively. (B) vav3 mRNA expression was determined by real-time PCR in T-FGM cells and MEFs of the indicated genotypes under basal conditions or after a 24-h-long treatment of TCDD (10 nM). As positive control for TCDD-dependent AhR activation, mRNA levels of the target gene Cyp1a1 were determined in parallel. Gene expression was normalized among the different experimental conditions using as comparative control the expression of the β-actin gene. (C) Net1 protein expression was analyzed in T-FGM AhR+/+ and T-FGM AhR−/− by Western immunoblotting using 15 μg total protein and a specific antibody. The expression of β-actin was used as loading control. Data are shown as mean ± SEM from triplicate measurements. The experiments were done in two different cultures of each genotype and cell line. The p values for statistical comparison between genotypes and between untreated and TCDD-treated AhR+/+ T-FGM cells are indicated.
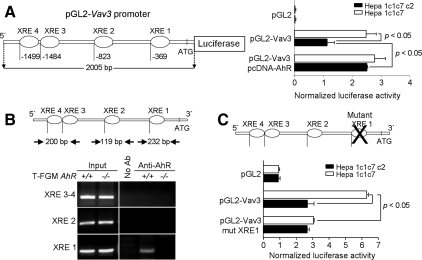
We decided to isolate the mouse vav3 gene promoter and to further characterize its regulation by AhR. A 2005-base pair upstream genomic fragment containing the vav3 promoter was cloned that contained four XRE consensus elements for AhR binding. These were located at −369 (XRE1), −823 (XRE2), −1484 (XRE3), and −1499 (XRE4) with respect to the translation start codon ATG (Figure 9A, top). We then analyzed the functionality of this promoter sequence by luciferase reporter assays using wild-type mouse hepatoma Hepa 1c1c7 cells and its variant c2 clone (expressing <10% wild-type levels of AhR). In agreement with an AhR-dependent process, vav3 promoter activity was higher in wild-type 1c1c7 than in AhR-deficient c2 hepatoma cells (Figure 9A, right). Furthermore, the reexpression of AhR by the pcDNA-AhR expression vector in c2 cells restored vav3 promoter activity to values close to those observed in wild-type 1c1c7 cells (Figure 9A, right). To further demonstrate that AhR contributes to the transcriptional regulation of vav3 expression, ChIP assays were conducted. AhR binding to the XRE1, XRE2, and XRE3-4 elements was analyzed by PCR in AhR immunoprecipitates. The results showed that AhR was recruited to the proximal XRE1 element on the vav3 promoter (Figure 9B), thus supporting the fact that vav3 is an AhR-regulated gene in the mouse. Moreover, the XRE1 element is indeed necessary for vav3 promoter regulation since its inactivation by site-directed mutagenesis abrogated the ability of the vav3 promoter to drive luciferase reporter activity in AhR-expressing wild-type 1c1c7 cells (Figure 9C).
Figure 9.
vav3 is a new transcriptional target of the AhR. (A) A 2005-base pair fragment of upstream genomic sequence containing the vav3 mouse promoter was cloned from the WI1-2862P14 phosmid into the pGL2-Basic vector. The resulting construct was transfected in wild-type Hepa 1c1c7 cells or in the AhR-deficient c2 line (expressing <10% wild-type levels of AhR) and firefly luciferase activity measured and normalized by Renilla luciferase activity. (B) Four XRE consensus elements for AhR binding were located in the vav3 promoter. AhR binding to these XREs was determined by ChIP in T-FGM AhR+/+ and T-FGM AhR−/− cells. The location of the oligonucleotides used to amplify by PCR the regions of the vav3 promoter containing XRE sequences is indicated. Positive controls were performed using the input fractions whereas negative controls were made in the absence of antibody. (C) Site-directed mutagenesis was used to inactivate the XRE1 site in the vav3 promoter. The mutated form of the promoter was cloned into the pGL2-Basic vector and the resulting construct transfected in wild-type Hepa 1c1c7 or in AhR-deficient c2 cells. Mutant vav3 promoter activity was analyzed by luciferase assays as indicated above.
These results prompted us to further analyze the contribution of Vav3 to the AhR-dependent phenotype. We hypothesized that if a reduction in the physiological levels of Vav3 expression in AhR-deficient cells were involved in determining their characteristic morphological phenotype, the knockdown of the vav3 mRNA in wild-type fibroblasts should induce an AhR-null-like phenotype. To address this possibility, we transfected vav3-specific small interfering RNAs (siRNAs) in T-FGM AhR+/+ cells, and, 72 h later, we monitored the morphology, polarity, and cytoskeleton changes in mock- and vav3 siRNA-transfected cells by using microscopy techniques. As an additional control, we used in these analyses untranfected T-FGM AhR−/− cells. We initially determined by Western immunoblotting that T-FGM AhR−/− cells expressed lower Vav3 protein levels and that vav3-specific siRNAs, but not unspecific scramble siRNAs, consistently reduced Vav3 protein expression to ∼30% its basal levels in T-FGM AhR+/+ cells (p < 0.05) (Figure 10A). Notably, vav3 knockdown induced a shift in the morphology of T-FGM cells from the wild-type to the AhR-null-like phenotype (Figure 10B). This change included the increase in cell area (Figure 10B, compare a with c and d and e with g and h), the promotion of stress fiber formation (Figure 10B, compare a with c and d), increased numbers of focal adhesions that localize around the cell edges (Figure 10B, compare e with g and h), and cell depolarization (Figure 10B, compare e with g and h). Transfection of unspecific scramble siRNAs, on the contrary, did not change these phenotypes in T-FGM AhR+/+ cells (Figure 10B, b and f). We then analyzed whether knockdown of Vav3 expression in fact affected its downstream target proteins RhoA and Rac1. Consistent with the role proposed for Vav3 in the regulation of the Rac1 and RhoA signaling pathways, down-modulation of Vav3 protein levels by vav3 siRNAs had an effect on these GTPases, and thus RhoA activation was increased, whereas Rac1 activation was decreased in transfected T-FGM AhR+/+ cells (Figure 10C). As indicated above, transfection of unspecific scramble siRNAs did not affect RhoA or Rac1 activation in T-FGM fibroblasts of either genotype (Figure 10C). We also confirmed that Rac1 has a prominent role in the phenotypic changes induced by NSC23766 in T-FGM AhR+/+ cells (Figure 7B) because this molecule decreased Rac1 activity in our pull-down assays (Figure 10C, bottom right). These results support that AhR, through the control of constitutive Vav3 expression and by modulating the activation of its downstream target proteins RhoA and Rac1, has a relevant role in determining fibroblast cell phenotype. In addition, because Vav3 can be regulated by phosphorylation (Bustelo, 2002; Llorca et al., 2005), we also analyzed whether T-FGM AhR−/− cells had a lower phosphorylation potential that could contribute to a reduction in Vav3 signaling. T-FGM AhR+/+ and T-FGM AhR−/− cells were transfected with a myc-tagged Vav3 expression construct and the levels of pVav3 determined by Western immunoblotting in myc immunoprecipitates. Both cell lines had a similar potential to phosphorylate Vav3 (Supplemental Figure S4), further supporting that decreased Vav3 protein expression is a major determinant in the AhR-dependent phenotype. We also performed overexpression experiments in which T-FGM AhR−/− and T-FGM AhR+/+ fibroblasts were transfected with the wild type or with a constitutively active version of Vav3 (Δ1-184 deletion mutant). We found that Vav3 overexpression produced the same morphological alteration in T-FGM fibroblasts regardless on their AhR status (Supplemental Figure S5), suggesting that conditions in which Vav3 activity largely exceeds its constitutive cellular levels, could abolish the physiological control of cell morphology making the process AhR independent. We used an additional experimental approach to further support that overactivation of Vav3 signaling alters cell morphology independently of AhR. Because Vav3 overactivation should increase the activity of its downstream client protein Rac1, transfection of a constitutively active form of Rac1 should also induce a phenotypic change regardless of AhR expression. T-FGM AhR+/+ and T-FGM AhR−/− cells were transfected with a wild-type or a constitutively active form of Rac1 and their effects on cell morphology analyzed (Supplemental Figure S6). Transfection of wild-type Rac1 did not significantly change the basal morphology of T-FGM AhR+/+ and T-FGM AhR−/− cells (Supplemental Figure S6A). However, the constitutively active version of Rac1 (V12Rac1; del Pozo et al., 1999) induced a change in cell area and polarity that was of similar magnitude in both T-FGM cell lines (Supplemental Figure S6B). Thus, overactivation of Vav3-dependent signaling overrides the influence of AhR in the control of fibroblast cell phenotype. Together, these data support a mechanism in which highly regulated control of constitutive Vav3 expression by the transcriptional activity of AhR has an important role in modulating fibroblast shape and cytoskeleton under physiological conditions.
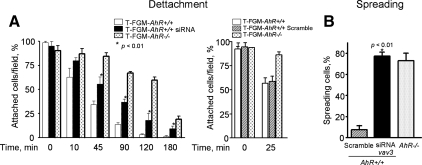
To investigate whether the reduced Vav3 expression also participated in the enhanced adhesion properties of T-FGM AhR−/− cells, we measured the effect of the vav3 knockdown in the detachment and spreading properties of wild-type T-FGM cells. As shown in Figure 11A, the vav3 knockdown increased the attachment efficiency of T-FGM AhR+/+ in the presence of EGTA to values significantly higher than those observed in untransfected wild-type cells. Transfection of unspecific scramble siRNA, on the contrary, did not affect the attachment efficiency of T-FGM AhR+/+ cells (Figure 11A, right). Moreover, vav3 siRNA-transfected T-FGM AhR+/+ cells did gain ability to spread on plastic, reaching values close to those found in T-FGM AhR−/− cells 60 min after plating (Figure 11B). Together, these data indicate that Vav3 is a bonafide AhR target implicated in the cytoskeleton-dependent effects induced by this transcriptional factor.
Figure 11.
Vav3 down-regulation increases the spreading and attachment efficiency of T-FGM AhR+/+ cells. (A) vav3 siRNAs- or unspecific scramble siRNAs-transfected T-FGM AhR+/+ fibroblasts were used in EGTA-induced detachment experiments. At the indicated times, nuclei were stained and counted as indicated in Figure 6. The p values for statistical comparison between vav3 siRNA-transfected T-FGM AhR+/+ and untransfected wild-type cells are indicated. (B) T-FGM AhR+/+ cells were transfected with vav3 siRNAs and their spreading efficiency determined after crystal violet staining. Between 250 and 300 T-FGM cells were counted in six independent fields for each experimental condition. The p values for statistical comparison in the increase of cell spreading with time in T-FGM AhR+/+ cells are indicated. Data are shown as mean ± SEM from duplicate experiments in two different cultures.
Vav3 Null MEFs Had Increased Cell Area and Enhanced Actin Stress Fibers Content
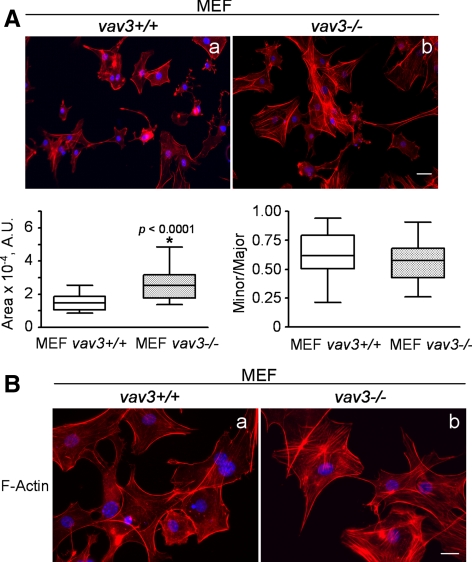
To provide even further support to our hypothesis that Vav3 has an important contribution to the AhR-dependent phenotype in fibroblast cells, vav3−/− MEFs were isolated and their morphology and ability to generate actin stress fibers was analyzed. Cell area and minor/mayor axis ratio were determined as indicated in Figure 1. vav3−/− MEFs had a significant increase in cell area without a change in minor/mayor axis ratio (Figure 12A), a phenotype closely mimicking that found in AhR−/− MEFs (see Figure 1). F-Actin staining with rhodamine-labeled phalloidin was used to analyze actin stress fibers content. Loss of Vav3 expression produced an increase in stress fibers with respect to vav+/+ MEFs (Figure 1, A and B, b) that resembled the effects described in AhR-null MEF cells (Figures 3, 7, and 10). The fact that genetically modified vav3−/− MEFs share common phenotypes regarding cell morphology and actin stress fibers with AhR−/− MEFs, strongly support our premise that Vav3 is a downstream target of AhR and that both proteins have a role in determining the phenotype of fibroblastic cells.
Figure 12.
vav3−/− MEFs have increased cell area and enhanced actin stress fibers content. (A) MEF cells were isolated from vav3+/+ and vav3−/− mice and analyzed for cell size and polarity (minor/major axis ratio) as indicated in the legend for Figure 1. (B) The F-actin stress fibers were revealed by rhodamin-labeled phalloidin as indicated in the legend for Figure 3. At least 20 cells were analyzed for each experimental condition, and the experiments were done in two different MEF cultures. The p values for statistical comparison between genotypes are indicated. Bar, 10 μm (A) and 5 μm (B). A.U., arbitrary units.
DISCUSSION
AhR is a critical intermediate in xenobiotic-induced toxicity and carcinogenesis (Fernandez-Salguero et al., 1996; Peters et al., 1999; Shimizu et al., 2000) and many studies have analyzed its role in cell cycle, differentiation, apoptosis, and genome integrity (Lahvis et al., 2005; Barouki et al., 2007; Furness et al., 2007; McMillan and Bradfield, 2007). Despite these known functions, emerging evidence indicates that AhR may be also involved in cytoskeleton-related processes. Thus, it has been shown that cell detachment leads to the transcriptional activation of AhR in several adherent cells such as keratinocytes (Sadek and Allen-Hoffmann, 1994a), hepatoma Hepa I cells (Sadek and Allen-Hoffmann, 1994b), and C3H10T1/2 fibroblasts (Cho et al., 2004). Moreover, it has been observed that AhR has a predominantly nuclear distribution in cells located at the migrating edge in wound healing experiments (Ikuta and Kawajiri, 2006). It was also reported that both the ligand-dependent and the constitutive activation of AhR in the immune system leads to enhanced migration of CD4−CD8− double-negative lymphocytes from the thymus (Esser et al., 2004; Temchura et al., 2005). Finally, a recent work has shown that either dioxin or a constitutively active form of the AhR induce morphological changes that affect the F-actin cytoskeleton, cell–cell interactions, and the motility of breast tumor cells (Diry et al., 2006). Despite this knowledge, the contribution of AhR to cell morphology, adhesion, and migration under physiological, xenobiotic-free conditions, remains largely unknown.
Consistent with a link between AhR and cytoskeleton-related processes, we report here that AhR status affects cell shape, F-actin cytoskeleton, adhesion, and spreading of murine fibroblasts. A first indication of the mechanism through which AhR can modulate cell morphology and adhesion comes from our results with the constitutively active AhR protein. The expression of constitutively active AhR in AhR−/− fibroblasts rescues the morphological phenotype of these cells, suggesting that the process requires the transcriptional activity of the receptor. The spread morphology of AhR−/− cells was associated to prominent actin stress fibers and to increased numbers of focal adhesions with a depolarized distribution around the cell area. Focal adhesions are major structures for cell–substratum interaction (Sieg et al., 1999; Schaller, 2001) and, indeed, spreading and attachment were significantly enhanced in AhR-null with respect to wild-type cells. These are relevant data that could help interpreting the lower migration rates observed in T-FGM AhR−/− fibroblasts (Mulero-Navarro et al., 2005) and MEF AhR−/− cells (Carvajal-Gonzalez, unpublished data).
Rac1 and RhoA play crucial roles in cell migration, polarity, adhesion, and cytoskeleton reorganization (Nobes and Hall, 1999; Bishop and Hall, 2000). The importance of these GTPases in determining AhR-dependent morphology was confirmed by two facts: 1) more spread and highly attached T-FGM AhR−/− cells had higher RhoA and lower Rac1 activation than wild-type fibroblasts and 2) the Rac1 inhibitor NSC23766 changed the morphology of AhR+/+ fibroblasts to that of AhR−/− cells, whereas the RhoA kinase inhibitor Y27632 did the opposite and transformed AhR-null cells into the wild-type phenotype. Moreover, these results are in close agreement with previous data indicating that Rac1 and RhoA have antagonistic effects in different cell types (Leeuwen et al., 1997; Sander et al., 1999). Interestingly, however, protein expression for Rac1 and RhoA was similar between T-FGM AhR+/+ and T-FGM AhR−/− cells, suggesting that AhR regulated these GTPases at the level of their activation and/or downstream elements. To find possible targets, we carried out expression microarrays in T-FGM AhR+/+ and T-FGM AhR−/− cells and searched for GTPases and/or GTPase regulators. This analysis indicated that the phosphorylation-dependent exchange factors Vav3 and Net 1 (Movilla and Bustelo, 1999; Hornstein et al., 2004) were significantly down-regulated in AhR−/− cells. We focused on Vav3 because Net1 protein levels did not significantly differ between T-FGM AhR+/+ and T-FGM AhR−/− cells. Cloning of the murine vav3 promoter revealed that AhR expression increases vav3 promoter activity and that AhR is recruited to a proximal XRE element in the vav3 promoter. Interestingly, the high-affinity AhR ligand TCDD did not increase the AhR-dependent vav3 expression, suggesting not only that AhR is committed to regulate constitutive gene transcription of vav3 but also that the physiological levels of Vav3 could be relevant to mediate the effects of AhR on cell phenotype.
The constitutive transcription of Vav3 has been related to the reorganization of actin stress fibers and to the formation of lamellipodia and membrane ruffles (Movilla and Bustelo, 1999; Bustelo, 2000) and, as such, represents a candidate target gene whose regulation by AhR could modulate cell morphology, adhesion, and spreading. We hypothesized that the down-regulation of vav3 mRNA and protein in fibroblasts could account for the cytoskeleton-dependent changes observed in AhR-deficient cells. Indeed, siRNA-based experiments, which down-regulate the physiological levels of target genes, demonstrated that this exchange factor represents an important intermediary in the AhR-dependent signaling pathways determining cell morphology. Thus, we observed that down-regulation of Vav3 in T-FGM AhR+/+ cells induced a T-FGM AhR-null-like phenotype regarding cell shape, cytoskeleton organization, polarity, adhesion, and spreading that was consistent with the changes observed in the activation of the Vav3 downstream GTPases Rac1 and RhoA. The fact that overexpression of the wild-type or a constitutively active Vav3 or a constitutively active form of Rac1 changed cell morphology independently on AhR expression, further support the hypothesis that the physiological level of Vav3 is a highly regulated parameter in the AhR-dependent modulation of cellular morphology. Indeed, vav3−/− MEFs mimicked the AhR−/− phenotype with respect to increased cell area and enhanced actin stress fibers content, strongly indicating not only that Vav3 is a downstream target of AhR but also that constitutive Vav3 levels are required to maintain fibroblast morphology and cytoskeleton. Therefore, the AhR-dependent phenotype for cell morphology, adhesion, and spreading is controlled, at least in part, by the precise regulation of the physiological levels of the exchange factor Vav3. This is a relevant example of how AhR can modulate physiologically defined cellular properties through the transcriptional regulation of an endogenous target gene that controls migration-related signaling pathways. Although these data attribute a relevant role for the transcriptional activity of AhR in determining cell phenotype, additional, transcription-independent mechanisms, cannot yet be excluded. For example, as a cullin 4B ubiquitin ligase (Ohtake et al., 2007), the liganded AhR could regulate the levels of migration-related proteins through the proteasome by a mechanism similar to that demonstrated previously for sex steroid receptors (Ohtake et al., 2007).
It is worth noting that our data do not exclude the participation of other signaling molecules in the AhR-mediated cytoskeleton effects in fibroblasts. For example, scaffolding proteins such as vinculin or other GEFs and GAPs are also dependent on the presence of AhR, at least in the case of fibroblasts. It is therefore possible that these additional proteins could cooperate with, or participate in parallel pathways, to those already engaged by Vav3. Further work in this area will address this possibility and unveil the level of cross-talk and cooperativity between these pathways. Given the important role of Rho/Rac pathways in pathologies such as cancer, immunodeficiencies or cardiovascular disease, it will be also interesting to see whether AhR may contribute to the regulation of those pathways in the context of these pathological states.
Supplementary Material
ACKNOWLEDGMENTS
The plasmid vector expressing constitutively active AhR (CA-AhR) was a generous gift from Dr. Yoshiaki Fujii-Kuriyama. We thank Drs. Pedro Casero and Ilda Casimiro for support with fluorescence microscope. This work was supported by grants to P.M.F.-S. from the Spanish Ministry of Education and Sciences (SMES) (SAF2005-00130, SAF2008-0462), the Junta de Extremadura (2PR04A060) and the Red Tem‡tica de Investigación Cooperativa en Cáncer (RTICC) (RD06/0020/1016, Fondo de Investigaciones Sanitarias (FIS), Carlos III Institute, Spanish Ministry of Health) and by grants to X.R.B. from the US National Cancer Institute/NIH (5R01-CA73735-11), the Spanish Ministry of Education and Science (MES) (SAF2006-01789), the Castilla-León Autonomous Government (SA053A05), and the Red Tem‡tica de Investigación Cooperativa en Cáncer (RTICC) (RD06/0020/0001, Fondo de Investigaciones Sanitarias (FIS), Carlos III Institute, Spanish Ministry of Health). J.M.C.-G. and A.C.R. were supported by fellowships from the Junta de Extremadura and the SMES, respectively. V. S. has been partially supported by the SMES Juan de la Cierva program and the National Institutes of Health. All Spanish funding is cosponsored by the European Union FEDER program.
Glossary
Abbreviations used:
- AhR
aryl hydrocarbon (dioxin) receptor
- FGM
immortalized mouse fibroblasts
- FN
fibronectin
- GAP
GTPase-activating protein
- GEF
guanosine diphosphate/guanosine triphosphate exchange factor
- MEF
mouse embryonic fibroblast
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0451) on January 21, 2009.
REFERENCES
- Alberts A. S., Treisman R. Activation of RhoA and SAPK/JNK signalling pathways by the RhoA-specific exchange factor mNET1. EMBO J. 1998;17:4075–4085. doi: 10.1093/emboj/17.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouki R., Coumoul X., Fernandez-Salguero P. M. The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett. 2007;581:3608–3615. doi: 10.1016/j.febslet.2007.03.046. [DOI] [PubMed] [Google Scholar]
- Bishop A. L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- Boguski M. S., McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- Bustelo X. R. Regulatory and signaling properties of the Vav family. Mol. Cell Biol. 2000;20:1461–1477. doi: 10.1128/mcb.20.5.1461-1477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo X. R. Regulation of Vav proteins by intramolecular events. Front. Biosci. 2002;7:d24–d30. doi: 10.2741/A766. [DOI] [PubMed] [Google Scholar]
- Bustelo X. R., Sauzeau V., Berenjeno I. M. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E. A., King W. G., Brugge J. S., Symons M., Hynes R. O. Integrin-mediated signals regulated by members of the rho family of GTPases. J. Cell Biol. 1998;142:573–586. doi: 10.1083/jcb.142.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couceiro J. R., Martin-Bermudo M. D., Bustelo X. R. Phylogenetic conservation of the regulatory and functional properties of the Vav oncoprotein family. Exp. Cell Res. 2005;308:364–380. doi: 10.1016/j.yexcr.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X., Fan Y., Karyala S., Schwemberger S., Tomlinson C. R., Sartor M. A., Puga A. Ligand-independent regulation of transforming growth factor beta1 expression and cell cycle progression by the aryl hydrocarbon receptor. Mol. Cell Biol. 2007;27:6127–6139. doi: 10.1128/MCB.00323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. H., Tzen J. T., Bresnick A. R., Chen H. C. Roles of Rho-associated kinase and myosin light chain kinase in morphological and migratory defects of focal adhesion kinase-null cells. J. Biol. Chem. 2002;277:33857–33863. doi: 10.1074/jbc.M204429200. [DOI] [PubMed] [Google Scholar]
- Cho Y. C., Zheng W., Jefcoate C. R. Disruption of cell-cell contact maximally but transiently activates AhR-mediated transcription in 10T1/2 fibroblasts. Toxicol. Appl. Pharmacol. 2004;199:220–238. doi: 10.1016/j.taap.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Chuang T. H., Xu X., Kaartinen V., Heisterkamp N., Groffen J., Bokoch G. M. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein family. Proc. Natl. Acad. Sci. USA. 1995;92:10282–10286. doi: 10.1073/pnas.92.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo M. A., Vicente-Manzanares M., Tejedor R., Serrador J. M., Sanchez-Madrid F. Rho GTPases control migration and polarization of adhesion molecules and cytoskeletal ERM components in T lymphocytes. Eur. J. Immunol. 1999;29:3609–3620. doi: 10.1002/(SICI)1521-4141(199911)29:11<3609::AID-IMMU3609>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Diry M., Tomkiewicz C., Koehle C., Coumoul X., Bock K. W., Barouki R., Transy C. Activation of the dioxin/aryl hydrocarbon receptor (AhR) modulates cell plasticity through a JNK-dependent mechanism. Oncogene. 2006;25:5570–5574. doi: 10.1038/sj.onc.1209553. [DOI] [PubMed] [Google Scholar]
- Disanza A., et al. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat. Cell Biol. 2006;8:1337–1347. doi: 10.1038/ncb1502. [DOI] [PubMed] [Google Scholar]
- Esser C., Temchura V., Majora M., Hundeiker C., Schwarzler C., Gunthert U. Signaling via the AHR leads to enhanced usage of CD44v10 by murine fetal thymic emigrants: possible role for CD44 in emigration. Int. Immunopharmacol. 2004;4:805–818. doi: 10.1016/j.intimp.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero P., Pineau T., Hilbert D. M., McPhail T., Lee S. S., Kimura S., Nebert D. W., Rudikoff S., Ward J. M., Gonzalez F. J. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero P. M., Hilbert D. M., Rudikoff S., Ward J. M., Gonzalez F. J. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- Frericks M., Meissner M., Esser C. Microarray analysis of the AHR system: tissue-specific flexibility in signal and target genes. Toxicol. Appl. Pharmacol. 2007;220:320–332. doi: 10.1016/j.taap.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Fukata M., Nakagawa M., Kaibuchi K. Roles of Rho-family GTPases in cell polarisation and directional migration. Curr. Opin. Cell Biol. 2003;15:590–597. doi: 10.1016/s0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- Furness S. G., Lees M. J., Whitelaw M. L. The dioxin (aryl hydrocarbon) receptor as a model for adaptive responses of bHLH/PAS transcription factors. FEBS Lett. 2007;581:3616–3625. doi: 10.1016/j.febslet.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Gao Y., Dickerson J. B., Guo F., Zheng J., Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Duran A., Ballestar E., Carvajal-Gonzalez J. M., Marlowe J. L., Puga A., Esteller M., Fernandez-Salguero P. M. Recruitment of CREB1 and histone deacetylase 2 (HDAC2) to the mouse Ltbp-1 promoter regulates its constitutive expression in a dioxin receptor-dependent manner. J. Mol. Biol. 2008a;380:1–16. doi: 10.1016/j.jmb.2008.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Duran A., Carvajal-Gonzalez J. M., Mulero-Navarro S., Santiago-Josefat B., Puga A., Fernandez-Salguero P. M. Fitting a xenobiotic receptor into cell homeostasis: how the dioxin receptor interacts with TGFbeta signaling. Biochem. Pharmacol. 2009;77:700–712. doi: 10.1016/j.bcp.2008.08.032. [DOI] [PubMed] [Google Scholar]
- Gomez-Duran A., Mulero-Navarro S., Chang X., Fernandez-Salguero P. M. LTBP-1 blockade in dioxin receptor-null mouse embryo fibroblasts decreases TGF-beta activity: Role of extracellular proteases plasmin and elastase. J. Cell Biochem. 2006;97:380–392. doi: 10.1002/jcb.20637. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hornstein I., Alcover A., Katzav S. Vav proteins, masters of the world of cytoskeleton organization. Cell Signal. 2004;16:1–11. doi: 10.1016/s0898-6568(03)00110-4. [DOI] [PubMed] [Google Scholar]
- Ikuta T., Kawajiri K. Zinc finger transcription factor Slug is a novel target gene of aryl hydrocarbon receptor. Exp. Cell Res. 2006;312:3585–3594. doi: 10.1016/j.yexcr.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Ito T., et al. A constitutively active arylhydrocarbon receptor induces growth inhibition of jurkat T cells through changes in the expression of genes related to apoptosis and cell cycle arrest. J. Biol. Chem. 2004;279:25204–25210. doi: 10.1074/jbc.M402143200. [DOI] [PubMed] [Google Scholar]
- Kolluri S. K., Balduf C., Hofmann M., Gottlicher M. Novel target genes of the Ah (dioxin) receptor: transcriptional induction of N-myristoyltransferase 2. Cancer Res. 2001;61:8534–8539. [PubMed] [Google Scholar]
- Lahvis G. P., Pyzalski R. W., Glover E., Pitot H. C., McElwee M. K., Bradfield C. A. The aryl hydrocarbon receptor is required for developmental closure of the ductus venosus in the neonatal mouse. Mol. Pharmacol. 2005;67:714–720. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- Leeuwen F. N., Kain H. E., Kammen R. A., Michiels F., Kranenburg O. W., Collard J. G. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J. Cell Biol. 1997;139:797–807. doi: 10.1083/jcb.139.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Guan J. L., Chien S. Biochemistry and biomechanics of cell motility. Annu. Rev. Biomed. Eng. 2005;7:105–150. doi: 10.1146/annurev.bioeng.7.060804.100340. [DOI] [PubMed] [Google Scholar]
- Llorca O., Arias-Palomo E., Zugaza J. L., Bustelo X. R. Global conformational rearrangements during the activation of the GDP/GTP exchange factor Vav3. EMBO J. 2005;24:1330–1340. doi: 10.1038/sj.emboj.7600617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe J. L., Puga A. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell Biochem. 2005;96:1174–1184. doi: 10.1002/jcb.20656. [DOI] [PubMed] [Google Scholar]
- Martinez-Delgado B., et al. Expression profiling of T-cell lymphomas differentiates peripheral and lymphoblastic lymphomas and defines survival related genes. Clin. Cancer Res. 2004;10:4971–4982. doi: 10.1158/1078-0432.CCR-04-0269. [DOI] [PubMed] [Google Scholar]
- Matikainen T., et al. Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat. Genet. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- McMillan B. J., Bradfield C. A. The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol. Pharmacol. 2007;72:487–498. doi: 10.1124/mol.107.037259. [DOI] [PubMed] [Google Scholar]
- Mimura J., et al. Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells. 1997;2:645–654. doi: 10.1046/j.1365-2443.1997.1490345.x. [DOI] [PubMed] [Google Scholar]
- Mitra S. K., Hanson D. A., Schlaepfer D. D. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- Movilla N., Bustelo X. R. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol. Cell Biol. 1999;19:7870–7885. doi: 10.1128/mcb.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Navarro S., Carvajal-Gonzalez J. M., Herranz M., Ballestar E., Fraga M. F., Ropero S., Esteller M., Fernandez-Salguero P. M. The dioxin receptor is silenced by promoter hypermethylation in human acute lymphoblastic leukemia through inhibition of Sp1 binding. Carcinogenesis. 2006;27:1099–1104. doi: 10.1093/carcin/bgi344. [DOI] [PubMed] [Google Scholar]
- Mulero-Navarro S., et al. Immortalized mouse mammary fibroblasts lacking dioxin receptor have impaired tumorigenicity in a subcutaneous mouse xenograft model. J. Biol. Chem. 2005;280:28731–28741. doi: 10.1074/jbc.M504538200. [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Dalton T. P., Okey A. B., Gonzalez F. J. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Nimnual A. S., Yatsula B. A., Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- Nobes C. D., Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T., Mimura J., Hikida M., Fujimoto H., Fujii-Kuriyama Y., Ohmori H. Expression of human and mouse genes encoding polkappa: testis-specific developmental regulation and AhR-dependent inducible transcription. Genes Cells. 2001;6:943–953. doi: 10.1046/j.1365-2443.2001.00478.x. [DOI] [PubMed] [Google Scholar]
- Ohtake F., et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- Peters J. M., Narotsky M. G., Elizondo G., Fernandez-Salguero P. M., Gonzalez F. J., Abbott B. D. Amelioration of TCDD-induced teratogenesis in aryl hydrocarbon receptor (AhR)-null mice. Toxicol. Sci. 1999;47:86–92. doi: 10.1093/toxsci/47.1.86. [DOI] [PubMed] [Google Scholar]
- Qin H., Powell-Coffman J. A. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev. Biol. 2004;270:64–75. doi: 10.1016/j.ydbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Riento K., Ridley A. J. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- Roman A. C., Benitez D. A., Carvajal-Gonzalez J. M., Fernandez-Salguero P. M. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:1632–1637. doi: 10.1073/pnas.0708366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K., Hall A., Small J. V. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr. Biol. 1999;9:640–648. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- Sadek C. M., Allen-Hoffmann B. L. Cytochrome P450IA1 is rapidly induced in normal human keratinocytes in the absence of xenobiotics. J. Biol. Chem. 1994a;269:16067–16074. [PubMed] [Google Scholar]
- Sadek C. M., Allen-Hoffmann B. L. Suspension-mediated induction of Hepa 1c1c7 Cyp1a-1 expression is dependent on the Ah receptor signal transduction pathway. J. Biol. Chem. 1994b;269:31505–31509. [PubMed] [Google Scholar]
- Safe S. Molecular biology of the Ah receptor and its role in carcinogenesis. Toxicol. Lett. 2001;120:1–7. doi: 10.1016/s0378-4274(01)00301-0. [DOI] [PubMed] [Google Scholar]
- Sander E. E., ten Klooster J. P., van Delft S., van der Kammen R. A., Collard J. G. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Josefat B., Mulero-Navarro S., Dallas S. L., Fernandez-Salguero P. M. Overexpression of latent transforming growth factor-{beta} binding protein 1 (LTBP-1) in dioxin receptor-null mouse embryo fibroblasts. J. Cell Sci. 2004;117:849–859. doi: 10.1242/jcs.00932. [DOI] [PubMed] [Google Scholar]
- Santiago-Josefat B., Pozo-Guisado E., Mulero-Navarro S., Fernandez-Salguero P. M. Proteasome inhibition induces nuclear translocation and transcriptional activation of the dioxin receptor in mouse embryo primary fibroblasts in the absence of xenobiotics. Mol. Cell Biol. 2001;21:1700–1709. doi: 10.1128/MCB.21.5.1700-1709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauzeau V., Sevilla M. A., Rivas-Elena J. V., de Alava E., Montero M. J., Lopez-Novoa J. M., Bustelo X. R. Vav3 proto-oncogene deficiency leads to sympathetic hyperactivity and cardiovascular dysfunction. Nat. Med. 2006;12:841–845. doi: 10.1038/nm1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M. D. Biochemical signals and biological responses elicited by the focal adhesion kinase. Biochim. Biophys. Acta. 2001;1540:1–21. doi: 10.1016/s0167-4889(01)00123-9. [DOI] [PubMed] [Google Scholar]
- Schmidt J. V., Su G. H., Reddy J. K., Simon M. C., Bradfield C. A. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. USA. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Nakatsuru Y., Ichinose M., Takahashi Y., Kume H., Mimura J., Fujii-Kuriyama Y., Ishikawa T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA. 2000;97:779–782. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa H., Rashid M. Development of Rho-kinase inhibitors for cardiovascular medicine. Trends Pharmacol. Sci. 2007;28:296–302. doi: 10.1016/j.tips.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Sieg D. J., Hauck C. R., Schlaepfer D. D. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J. Cell Sci. 1999;112:2677–2691. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- Temchura V. V., Frericks M., Nacken W., Esser C. Role of the aryl hydrocarbon receptor in thymocyte emigration in vivo. Eur. J. Immunol. 2005;35:2738–2747. doi: 10.1002/eji.200425641. [DOI] [PubMed] [Google Scholar]
- Uehata M., et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr. Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- Ziegler W. H., Liddington R. C., Critchley D. R. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.