Abstract
Cilia and flagella are structurally and functionally conserved organelles present in basal as well as higher eukaryotes. The assembly of cilia requires a microtubule based scaffold called a basal body. The ninefold symmetry characteristic of basal bodies and the structurally similar centriole is organized around a hub and spoke structure termed the cartwheel. To date, SAS-6 is one of the two clearly conserved components of the cartwheel. In some organisms, overexpression of SAS-6 causes the formation of supernumerary centrioles. We questioned whether the centriole assembly initiation capacity of SAS-6 is separate from or directly related to its structural role at the cartwheel. To address this question we used Tetrahymena thermophila, which expresses two SAS-6 homologues, TtSAS6a and TtSAS6b. Cells lacking either TtSAS6a or TtSAS6b are defective in new basal body assembly. TtSas6a localizes to all basal bodies equally, whereas TtSas6b is enriched at unciliated and assembling basal bodies. Interestingly, overexpression of TtSAS6b but not TtSAS6a, led to the assembly of clusters of new basal bodies in abnormal locations. Our data suggest a model where TtSAS6a and TtSAS6b have diverged such that TtSAS6a acts as a structural component of basal bodies, whereas TtSAS6b influences the location of new basal body assembly.
INTRODUCTION
Cilia and flagella are microtubule based cellular appendages used for locomotion, the movement of fluids over an epithelial layer, and sensing the environment (Fliegauf et al., 2007). In humans and other metazoans, cilia function in all of these capacities (Fliegauf et al., 2007). Based on the presence of these organelles in basal eukaryotes as well as more complex organisms such as humans, it is believed that these structures have an ancient origin (Cavalier-Smith, 2002; Satir et al., 2007). Cilia are responsible for many important developmental, cell signaling, and physiological processes; therefore, it is not surprising to find that a growing number of genetic diseases with pleiotropic symptoms are caused by mutations in genes encoding components of cilia or basal bodies (Badano et al., 2006). Thus it is of significant interest to understand how these organelles are normally assembled and maintained.
Cilia are assembled from a structure called the basal body, which in mammals and other species is derived from a centriole (Sorokin, 1962) or by de novo synthesis in ciliated epithelia (Dirksen, 1971). Centrioles and basal bodies contain a barrel-shaped ninefold symmetric array of microtubules. In humans and most other species in which centrioles or basal bodies have been examined, the microtubules of the basal body are arranged in triplet bundles. However, the centrioles of Caenorhabditis elegans are composed of a ninefold symmetric array of microtubule singlets. Elegant electron microscopy studies of centriole assembly steps have shown that the microtubule array is organized around a hub-and-spoke structure termed the cartwheel in organisms as diverse as ciliates, Chlamydomonas, Drosophila, and mammalian species (Dippell, 1968; Allen, 1969; Cavalier-Smith, 1974; Vorobjev and Chentsov Yu, 1982; Callaini et al., 1997) or around a central tube in the case of C. elegans (Pelletier et al., 2006).
Here we use the ciliated protozoan Tetrahymena thermophila to study basal body biogenesis. Tetrahymena have hundreds of basal bodies per cell, and before cell division many new basal bodies assemble in specific regions of the cell (Allen, 1969). Furthermore, the basal bodies of Tetrahymena are similar in morphology and protein composition to the basal bodies of humans (Allen, 1969; Kilburn et al., 2007). From a structural standpoint, the basal body assembly process in Tetrahymena was meticulously documented (Allen, 1969), but few investigations have analyzed specific components of Tetrahymena basal bodies and their role in the assembly of this complex structure. Recently, several studies have analyzed a number of widely conserved genes that are important for centriole assembly and developed an assembly pathway. Within this group, SAS-6 has a relatively early role in assembly (Dammermann et al., 2004; Habedanck et al., 2005; Leidel et al., 2005; Delattre et al., 2006; Pelletier et al., 2006; Kleylein-Sohn et al., 2007; Nakazawa et al., 2007; Peel et al., 2007; Rodrigues-Martins et al., 2007a,b; Strnad et al., 2007). SAS-6 is a component of the central tube in C. elegans (Pelletier et al., 2006) and the cartwheel in Chlamydomonas (Nakazawa et al., 2007). In C. elegans, the nine microtubule singlets of the centriole cannot be organized into a centriole without SAS-6 (Pelletier et al., 2006). Similarly Chlamydomonas or Drosophila SAS-6 mutants display aberrant numbers of microtubule triplets in their basal bodies (Nakazawa et al., 2007; Rodrigues-Martins et al., 2007a). These studies demonstrate that SAS-6 has a role in cartwheel formation. Additionally, overexpression of SAS-6 in some organisms leads to the formation of supernumerary centrioles (Leidel et al., 2005; Peel et al., 2007; Strnad et al., 2007). These observations led us to question whether the centriole assembly initiation activity of SAS-6 is separate from or linked to its structural role at the cartwheel. If these activities are in fact separate functions of the SAS-6 gene, then a system that possesses two SAS-6 homologues may be able to help answer this question. Tetrahymena expresses two SAS-6 homologues, which provided us with a potential model system that we could use to investigate this possibility.
MATERIALS AND METHODS
Strains and Culture Conditions
Tetrahymena thermophila strains B2086, CU428, and SB1969 (gifts from Peter Bruns, Cornell University, and Eduardo Orias, University of California Santa Barbara) were used for the generation of transgenic cell lines. Cells were grown in SPP media consisting of 2% proteose peptone, 0.1% yeast extract, 0.2% glucose, and 0.003% FeEDTA at 30°C. Cells were starved in 10 mM Tris, pH 7.4. Matings between cells were performed as described in Bruns and Cassidy-Hanley (2000). SAS6a shutoff cells were grown in 1% SPP with 100 ng/ml CdCl2 to activate expression of SAS6a or with 100 μM EDTA to inactivate expression of the gene. In SAS6b overexpression experiments, cells were grown in 2% SPP with 400 ng/ml CdCl2 and 15 μg/ml cyclohexamide.
Identification of Tetrahymena SAS-6 Homologues
Possible SAS-6 homologues were identified by a BLAST search of the Tetrahymena thermophila predicted proteome (Eisen et al., 2006) from Tetrahymena Genome Database (TGD) using published sequences (Dammermann et al., 2004; Leidel et al., 2005). We identified two predicted open reading frame (ORFs): TTHERM_00388200 corresponds to TtSAS6a and TTHERM_00137600 corresponds to TtSAS6b.
Putative SAS-6 PISA domain amino acid sequences from organisms listed in Figure 1a were aligned and used to generate a distance matrix using PROTDIST (Jones-Taylor-Thornton matrix). This distance matrix was used in the construction of a tree using NEIGHBORS and DRAWTREE (Felsenstein, 1997).
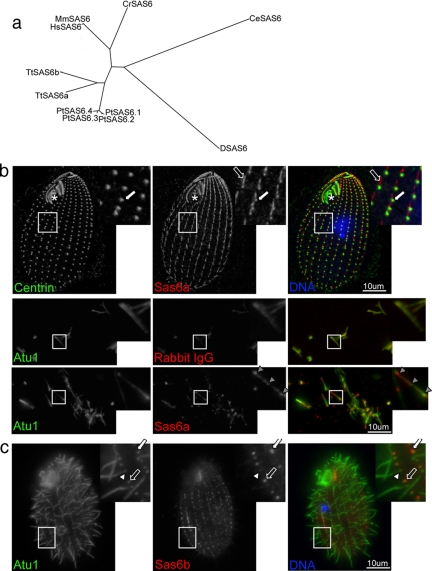
Figure 1.
Conservation and localization of Tetrahymena Sas6 proteins. (a) Dendrogram showing relative distances between SAS-6 genes based on PISA domain alignment. Tt, Tetrahymena thermophila TTHERM_00388200 (TtSAS6a) and TTHERM_00137600 (TtSAS6b); Pt, Paramecium tetraurelia GSPATP00011824001 (PtSAS6.1); GSPATP00008149001 (PtSAS6.2), GSPATP00005603001 (PtSAS6.3), and GSPATP00024268001 (PtSAS6.4); Hs, Homo sapiens GI:35038001; Mm, Mus musculus GI:62511043; Cr. Chlamydomonas reinhardtii GI:161727419; Ce. C. elegans GI:1754348; D, Drosophila melanogaster GI:62511095. (b) Sas6a localizes to basal bodies and kinetodesmal fibers in whole cells and to foci in isolated cilia preparations. Cells are oriented in this and all fluorescent images with anterior side up. Fixed wild-type Tetrahymena cells were labeled with antibodies to centrin (left panel, green in merge) and Sas6a (middle panel, red in merge) as well as DAPI stain to mark DNA (blue in merge). The asterisk indicates the oral apparatus, the white arrow shows centrin and Sas6a basal body localization, and the unfilled arrow shows Sas6a kinetodesmal fiber localization. In cilia fluorescent images, fixed isolated cilia preparations were labeled with antibodies to α-tubulin (Atu1, left panel, green in merge) and rabbit IgG or Sas6a (middle panels, red in merge). The gray arrowheads show Sas6a foci present along the length of the cilia. (c) Sas6b is enriched at unciliated and assembling basal bodies. Fixed wild-type Tetrahymena cells were labeled with antibodies to Atu1 (left panel, green in merge), Sas6b (middle panel, red in merge) as well as DAPI stain to mark nuclei. The white arrow shows an example of an assembling basal body, the unfilled arrow shows an unciliated basal body, and the white arrowhead shows a ciliated basal body.
Generation of Polyclonal Antibodies to Sas6a and Sas6b
DNA encoding the first 160 amino acids of both SAS6a and SAS6b and optimized for expression in Escherichia coli was synthesized (Integrated DNA Technologies, Coralville, IA). This region was chosen because it contains the conserved PISA domain. These DNA fragments were cloned into N-terminal glutathione S-transferase (GST) fusion expression vector, pGEX-6P1 (Amersham, Buckinghamshire, United Kingdom), using BamHI and XhoI sites and were transformed into E. coli strain BL21 for expression. Recombinant Sas6a and Sas6b protein fragments were cleaved from the GST affinity tag using HRV3C protease and eluted off beads with few contaminants (Supplemental Figure S1b). Two milligrams of Sas6a- or Sas6b-enriched samples were injected into rabbits for generation of polyclonal antibodies (Affinity BioReagents, Golden, CO). Antibodies were affinity-purified against agarose-bound recombinant Sas6a and Sas6b fragments using instructions from the manufacturer (Pierce, Rockford, IL) and depleted for cross-reactivity by binding affinity-purified anti-Sas6a antibodies to the Sas6b column and vice versa. In this manner we generated antibodies specific to Sas6a and Sas6b with minimal cross-reactivity to one another. To control for the specificity of the Sas6a antibody, we performed Western blot analysis using this antibody on cells expressing green fluorescent protein (GFP)-SAS6a with or without inclusion of a 50-fold molar excess of Sas6a or Sas6b antigens. Supplemental Figure S1c shows three different cellular fractions together comprising total cellular protein. When these fractions were blotted for Sas6a, two bands representing the predicted molecular weights of GFP-Sas6a and full-length Sas6a were present at approximately 110 and 82 kDa, respectively (Supplemental Figure S1c, left). None of the anti-Sas6a reactive bands were eliminated by the addition of Sas6b antigen (Supplemental Figure S1c, middle). In contrast to this, all of the bands were eliminated or severely reduced by the addition of Sas6a antigen (Supplemental Figure S1c, right). To control for the specificity of the Sas6b antibody, fixed wild-type cells were incubated with Sas6b antibody alone or with a 50-fold molar excess of the Sas6a or Sas6b antigens (Supplemental Figure S1d, left). Inclusion of Sas6a antigen had no effect on the labeling pattern of the anti-Sas6b antibody (Supplemental Figure S1d, middle), but in the presence of Sas6b antigen the basal body signal was completely eliminated (Supplemental Figure S1d, right). These data show that the Sas6a antibody is not cross-reactive to Sas6b and vice versa.
Tetrahymena Cellular Fractionation
Tetrahymena strain SB255 (mucocyst deficient, gift of Eduardo Orias, University of California Santa Barbara) was deciliated using the protocol described in Thompson et al. (1974). Fraction 1 is represented as 0.2% of the total cilia-containing fraction. Pellicles (fractions 3 and 4) and basal body/kinetodesmal fibers (fractions 5 and 6) were made from these deciliated cells according to the method described by Hyams and King (1985). Triton-soluble protein (25 μg at 1%) from the generation of pellicles and kinetodesmal fibers was loaded (fraction 2). Proteins were solubilized from pellicles and basal body/kinetodesmal fiber fractions in HpHS buffer (50 mM Tris, pH 9.0, and 500 mM NaCl). HpHS-soluble pellicle extract, 8 μg, is represented in fraction 3, whereas an equal volume of the insoluble pellet from this extraction was loaded as fraction 4. HpHS-soluble kinetodesmal fiber protein, 2 μg, is represented as fraction 5, and an equal volume of the insoluble pellet is represented as fraction 6.
Western Blot Analysis
Blots were blocked in 5% milk TBST (50 mM Tris, pH 8.0, 150 mM NaCl, and 0.2% Tween-20) for 40 min at room temperature. Affinity-purified anti-Sas6a was used at 1:2500 in 5% milk TBST. Monoclonal anti-Atu1 antibody (12G10) was used at 1:5000 in 5% milk TBST. All incubations were performed for 1 h at room temperature or at 4°C overnight. Blots were washed three to four times for 5 min with TBST after incubation in antibody-containing solution. Western blots were scanned on a LI-COR Odyssey (Li-Cor, Lincoln, NE) using appropriate secondary antibodies (Invitrogen, Carlsbad, CA, and Rockland Biosciences, Gilbertsville, PA).
Immunofluorescence Microscopy
Cells were fixed as described in Stuart and Cole (2000), except formaldehyde fixation lasted 25 min. Centrin was labeled using 20H5 (gift from J. Salisbury, Mayo Clinic, Rochester, MN) at 1:2500 in phosphate-buffered saline (PBS), pH 6.9, 1% BSA. Sas6a antibody was used at 1:2500 in PBS, 1% BSA. Sas6b antibody was used at 1:2500 in PBS, 1% BSA. Atu1 was labeled using 12G10 (gift from Joseph Frankel, University of Iowa, Iowa City, IA) at 1:75 in PBS, 1% BSA. Cen1 antibody (Stemm-Wolf et al., 2005) was used at 1:1500 in PBS, 1% BSA. GFP antibody (Rockland, Gilbertsville, PA) was used at 1:500 in PBS, 1% BSA. All incubations were performed at room temperature for 1 h or at 4°C overnight. Alexa fluor and Texas Red (Invitrogen and Jackson ImmunoResearch, West Grove, PA) secondary antibodies were used according to manufacturer's instructions. Images were acquired on a Leica DMRXA/RF4/V automated microscope (Leica Microsystems, Bannockburn, IL) with a Photometrics COOL SNAP HQ2 digital camera (Photometrics, Tucson, AZ). Images were collected and projection images generated of one hemisphere of the cell using the Metamorph imaging software (Molecular Devices, Sunnyvale, CA).
Plasmid Construction and Molecular Techniques
Total RNA was extracted from log-phase cells using TRI Reagent (Molecular Research Center, Cincinnati, OH). Total cDNA was generated using oligo dT primed Moloney murine leukemia virus (MMLV) reverse transcriptase reactions (Invitrogen). We were able to amplify cDNAs for SAS6a and SAS6b of the size and sequence predicted by TGD. GFP-SAS6a and GFP-SAS6b constructs were built by cloning SAS6a or SAS6b cDNAs into pIGF vector using XhoI and ApaI sites (Doug Chalker, Washington University, St. Louis, MO). MTT-GFP-SAS6b was cloned into pBSMTTGFP using the Gateway system (Invitrogen and Doug Chalker, Washington University, St. Louis, MO). pMTT 6xHisSAS6a was built by removing the Neo1 cassette from p4T2-1 (gift from Jacek Gaertig, University of Georgia, Athens, GA) using EcoRV and SmaI. This was inserted into pMTT-G1 (gift of Martin Gorovsky, University of Rochester) using a blunted BglII site just 5′ of the MTT promoter. SalI and XhoI sites were destroyed in pMTT-G1 + Neo by cutting these sites and filling in the gap. This plasmid was then cut with HindIII and BamHI to remove the IchG1 coding region. An MCS was then cloned into this site containing PmeI and ApaI. Annealed doubled-stranded oligonucleotides coding for RGSHHHHHH and a HRV3C cut site was then ligated into PmeI and XhoI sites. In this manner an inducible RGS6xHis-SAS6a construct was generated. A SAS6a knockout construct was built into the pMNBL plasmid. Six hundred base pairs 5′ of the start codon was used for the 5′ arm and 1.6 kb of DNA flanking the stop codon was used for the 3′ arm. The 5′ arm was cloned in using ApaI and XhoI. The 3′ arm was cloned into BamHI and XbaI sites. An alternative version of pMNBL-SAS6a was built by removing the Neo3 cassette and replacing it with the Bsr cassette (gift from Jacek Gaertig, University of Georgia, Athens, GA) by cloning into XhoI and SmaI sites. This knockout construct was used for the generation of the SAS6a shutoff. The SAS6b knockout construct was built by cloning into p4T2-1. One kilobase of DNA flanking the start codon was used for the 5′ arm, and 2 kb of DNA flanking the stop codon was used for the 3′ arm. The 5′ arm was cloned into ApaI and XhoI sites, and the 3′ arm was cloned into BamHI and NotI sites. pGEX-SAS6a was built by cloning the SAS6a minigene (Integrated DNA Technologies) into BamHI and XhoI sites present in pGEX-6P1 (Amersham). pGEX-SAS6b was built by cloning the SAS6b minigene into BamHI and XhoI sites present in pGEX-6P1. Genomic DNA was isolated from cells using the method of Gaertig et al. (1994). Southern blot for SAS6a and SAS6b knockouts was performed according to standard techniques. Briefly for the SAS6a knockout, 30 μg of genomic DNA from CU428 (WT), SAS6a KO cells without 6xHis-SAS6a rescue, and SAS6a KO cells with 6xHis-SAS6a rescue digested with SphI and XmnI were loaded on a 0.7% agarose gel. The blot was transferred by the capillary method. The same fragment of DNA used to build the 5′ arm of the knockout construct was used as a radioactive probe of the blot. For the SAS6b knockout, 30 μg of genomic DNA from WT, SAS6b macronuclear knockout, and SAS6b micronuclear knockout was digested with NheI and XhoI and done as above. The 5′ arm of p4T2-1 SAS6b was used as probe in this blot. Northern blot analysis was performed on 30 μg of total RNA from actively dividing cells. SAS6a or SAS6b cDNA, 1 ng, was used for normalization of blots. Hybridization and washing were done according to standard procedures using a formamide based buffer. Blots were scanned on a STORM PhosphorImager (GE Healthcare, Pittsburgh, PA) and analyzed using the Image Quant software (GE Healthcare).
Oligonucleotides
SAS6a ORF: forward, ATGGATAGTTTATCTTAAAAGTCTGGAAGAAGCTAGC; reverse, TCACTAATTTTTTGTTGGATCACGATATTTTATTGGAAC; SAS6b ORF: forward, ATGGCTGAGTACTAACCTTCATACAGAAAG; reverse, TCAGTTTTAATTTGGATTTTTTGGTTAGCGG; SAS6a 5′ KO arm: forward, GTTTATAAACTGACATGCCTAAACAAATGC; reverse, CTTATTAACTCTACTATTATATATGATATGCGAGG; SAS6a 3′ KO arm: forward, CCAATAAAATATCGTGATCCAACAAAAAATTAG; reverse, CACTACAAGTATAGAAGATGGTCTCCGAGC; SAS6b 5′ KO arm: forward, GCATATTATTCAGCTTTCAGCAGAAGAG; reverse, CTTTATAAGTTTGTATGCTTATTAAACTTG; and SAS6b 3′ KO arm: forward, CCGCTAACCAAAAAATCCAAATTAAAACTGA; reverse, CTTTATCATAACATTCCAAATAAATGGC.
Tetrahymena Strain Construction
Tetrahymena micro- and macronuclear integrants were transformed by biolistics (Bio-Rad, Hercules, CA) as described in (Bruns and Cassidy-Hanley, 2000). The SAS6a shutoff was generated by transforming pMTT-6xHis-SAS6a into the B2086 background. This inducible allele integrates at one of the two β-tubulin (BTU1) loci in Tetrahymena. This strain was then transformed with the blasticidin resistance variant of the pMNBL-SAS6a plasmid, pBLAST-SAS6a. 6xHis-Sas6a localizes to basal bodies and was considered to be functional based on this result (data not shown). The knockout allele was then selected for by growing cells in increasing concentrations of blasticidin (Invitrogen, Carlsbad, CA) in the presence or absence of induction of 6xHis-SAS6a. 6xHis-SAS6a was induced using 500 ng/ml CdCl2. When a concentration of blasticidin was reached at which cells were unable to divide anymore (5 mg/ml), then selection was halted, but these cells continued to be grown with or without induction of 6xHis-SAS6a. A flow chart for how this strain was generated is shown in Supplemental Figure S2a. Only when expression of the SAS6a allele at the BTU1 locus was induced were we able to completely eliminate endogenous SAS6a from the macronucleus (Supplemental Figure S2b). This experiment demonstrated that SAS6a is an essential gene and that we had generated a conditional null allele of SAS6a. We were now able to effectively repress transcription of the sole source of SAS6a by omitting CdCl2 from the media or chelating the available CdCl2 from the media using EDTA (Shang et al., 2002b). The SAS6b knockout was generated by transforming a mating culture of strains B2086 and CU428 with p4T2-1 SAS6b. Micronuclear integrants were homozygosed by mating to B*VI. SAS6b knockout heterokaryon cell line was outcrossed to CU428. Different mating types of heterozygous paromomycin-sensitive clones were homozygoused for the SAS6b knockout allele by mating to B*VI. SAS6b knockout heterokaryon cells of different mating types were mated to each other and mated cells were selected based on resistance to paromomycin. SAS6b was absent from these cells based on immunofluorescence microscopy, PCR, and Southern blot. SAS6b macronulcear knockouts were generated by transforming starved B2086 cells with p4T2-1 SAS6b. Increasing copies of the knockout allele were selected for by growing cells in increasing concentrations of paromomycin until cells could no longer survive (4 mg/ml). Cells were taken from next lowest concentration and assayed for the presence of SAS6b by immunofluorescence and Southern blot.
Cellular Measurements
Basal bodies were counted based on the number of foci of colocalized centrin and α-tubulin present along a cortical row. Three to six rows in 40–100 cells were counted for each condition. The number of Cen1/Atu1 spots present was divided by the length of the ciliary row (μm) on which they were present. This number was multiplied by 10 and rounded to the nearest tenth to yield the number of basal bodies per 10 μm. The graphs represent the percentage of the population exhibiting the given density range of basal bodies was observed. Cilia were measured from the basal body to tip. Cilia from at least 20 cells (n = 140) were counted for each condition and were measured only if the signal was longer than the centrin signal to which it associated. Cilia lengths were rounded to the nearest tenth and grouped as shown in the figure. The ratio of α-tubulin foci associated with Cen1 foci was determined by performing linescans along a ciliary row and counting the number of times a Cen1 signal coincided with an α-tubulin signal from projection images of Tetrahymena cells in the indicated conditions. These numbers were converted to a ratio for each ciliary row examined. This ratio was rounded to the nearest tenth, and the number of times this ratio was observed was quantified in terms of percentage. All measurements were performed using the Metamorph imaging program (Molecular Devices). Microsoft Excel (Redmond, WA) was used for all of the calculations and statistical analysis.
Protein Extractions
One milliliter buffer/1 × 106 Tetrahymena cells was used to solubilize cells in indicated conditions. Cells were resuspended in an appropriate amount of HBS (50 mM HEPES, pH 7.4, 110 mM KOAc, 150 mM NaCl, 2 mM MgCl2, and 0.1% Tween-20) or HBST buffer (HBS with 1% Triton X-100), sonicated briefly, and incubated at 4°C with gentle agitation for 10 min. These extracts were spun down for 10 min at 6000 × g at 4°C, and the supernatant was removed. The HBST supernatant is represented as fraction 1 in Supplemental Figure S1c. The pellet from this step was resuspended in an appropriate amount of HpHS buffer (Tris, pH 9.0, and 500 mM NaCl), sonicated, incubated, and spun down as above. The supernatant is represented as fraction 2 in Supplemental Figure S1c, and the pellet from this fraction is represented as fraction 3 in Supplemental Figure S1c. In the SAS6a knockout experiments, 8 μg of HpHS-soluble protein and an equivalent volume of the pellet are shown in this blot. Sas6a was not present in the HBST fraction in either condition. Extractions were carried out in the presence of leupeptin and pepstatin (10 and 1 μM final concentrations, respectively).
Electron Microscopy
Tetrahymena cells were either chemically fixed (Allen 1969) or high-pressure frozen (Stemm-Wolf et al., 2005). Briefly, Tetrahymena cells were pelleted, fixed in 3% glutaraldehyde, postfixed in 1%OsO4, and embedded in Epon resin. A BAL-TEC HPM-010 high-pressure freezer (Boeckeler Instruments, Tucson, AZ) was used to freeze pelleted cells. These cells were freeze-substituted in 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone and embedded in Lowicryl HM20 resin. Sections at 80 nm were cut, stained with uranyl acetate and lead citrate, and viewed in a Philips CM10 electron microscope (Mahwah, NJ). For immunoelectron microscopy, 55-nm serial sections were produced and labeled with either anti-Sas6a (1:50), anti-Sas6b (1:50), or anti Cen1 (1:200) primary antibodies (described above) after blocking with 1% nonfat milk PBS/Tween 20. Twenty-five-nanometer gold-conjugated goat anti-rabbit secondary antibody (Ted Pella, Redding, CA) was used to visualize the antigens. Grids were stained with uranyl acetate and lead citrate.
RESULTS
Identification, Cloning, and Expression of Tetrahymena SAS-6 Homologues
Two possible Tetrahymena thermophila SAS-6 homologues were identified by BLAST search of the TGD (Eisen et al., 2006) using published and predicted SAS-6 sequences (Leidel et al., 2005). We named the two putative ORFs, TtSAS6a and TtSAS6b, and reported the identity of TtSAS6a in the Tetrahymena basal body proteome (Kilburn et al., 2007). Hereafter we will refer to TtSAS6a as SAS6a and TtSAS6b as SAS6b. Members of the SAS-6 gene family have two common features: a central coiled-coil domain and an N-terminal PISA domain (Leidel et al., 2005), both SAS6a and SAS6b have these sequence motifs. Although the function of the PISA domain is unknown, it is the region of highest sequence similarity and identity among SAS-6 family members (Leidel et al., 2005). On the basis of this, we used an alignment of the PISA domains of known and putative SAS-6 homologues to construct the phylogenetic tree in Figure 1a. Organisms with similar basal body morphologies, i.e., triplet microtubule bundles, appear to be more closely related to each other on this dendrogram, whereas those organisms with centriole and basal body morphologies that are less similar, i.e., microtubule singlets or doublets, have higher degrees of sequence divergence. Interestingly, Paramecium also has more than one SAS-6 homolog, which points to a gene duplication event somewhere in the ciliate lineage. Although the Tetrahymena SAS-6 homologues are most closely related to one another, they are only 59% identical across the 53-amino acid PISA domain and 34% identical across the entire protein. The magnitude of the amino acid differences between Sas6a and Sas6b lead us to speculate that these proteins may have functionally diverged as well.
To verify that these are indeed functional genes, we isolated RNA from actively dividing cells and checked for the presence of SAS6a and SAS6b transcripts by RT-PCR. We were able to amplify cDNAs, as predicted by TGD, for both SAS6a and SAS6b. Northern blot analysis of these same total RNA pools showed that SAS6a is expressed at ∼1.3 times the level of SAS6b (data not shown).
Sas6a and Sas6b Share Basal Body Localization, But Differ in Other Subcellular Distributions
We have previously reported that GFP-Sas6a localized to basal bodies (Kilburn et al., 2007) and found GFP-Sas6b to have a similar, but not identical localization pattern (Supplemental Figure S1a). However, we were interested in confirming the expression and localization of both proteins at endogenous levels. For this reason we generated polyclonal antibodies to fragments of Sas6a and Sas6b with no detectable cross-reactivity to each other (see Materials and Methods and Supplemental Figure 1, c and d). Using centrin as a marker for basal bodies, Sas6a localizes to all three major sites of basal bodies in Tetrahymena: the cortical row basal bodies (Figure 1b, arrows inset), the oral apparatus basal bodies (Figure 1b, asterisk), and the developing oral apparatus basal bodies (data not shown). Sas6a was also found at kinetodesmal fibers, observed as discontinuous lines closely associated with basal bodies (Figure 1b, unfilled arrow, and Figure 2d, white arrows). Interestingly, GFP-SAS6a does not localize to kinetodesmal fibers (Supplemental Figure S1a). Kinetodesmal fibers are non-microtubule-based fibers attached to the anterior side of cortical row basal bodies only after they have been inserted into the plasma membrane and as such are markers of the mature organelle (Allen, 1969). In addition to these sites, Sas6a also localized to foci in isolated cilia preparations (Figures 1b, gray arrowheads, and 2e). Western blot analysis confirmed the presence of Sas6a in this cilia preparation (Supplemental Figure S1e, fraction 1).
Figure 2.
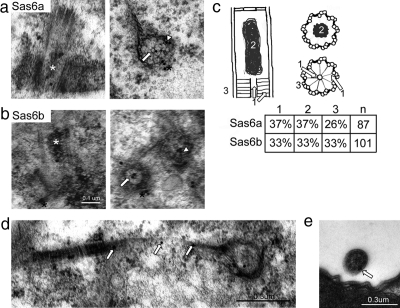
Sas6a and Sas6b localize to all of the same structural domains within the basal body. (a) Sas6a localizes to the cartwheel hub, spokes, electron-dense lumen, and periphery of the basal body at the proximal end. Immunoelectron micrographs showing examples of Sas6a localization observed in whole cell preparations. The Sas6a antibody was visualized using a 25-nm gold–conjugated secondary antibody. The white asterisk indicates the electron-dense lumen, the arrow points to the hub of the cartwheel, the arrowhead shows cartwheel spoke to “A” tubule attachment point, and the black asterisk shows the basal body periphery associated Sas6a. (b) Sas6b shares all of the same localizations as Sas6a. The Sas6b antibody was visualized using a 25-nm gold–conjugated secondary antibody. Immunoelectron micrographs of whole cell preparations showing examples of Sas6b localizations, the basal body structural domains are indicated as in panel a. (c) Sas6a and Sas6b localize at nearly the same frequency to the same structural domains within the basal body. The schematic shows a basal body in longitudinal section (left drawing) as well as a view of the electron-dense lumen in cross section (right, top drawing) and the cartwheel (right, bottom drawing). Structural domains were counted as indicated in the drawings (1, cartwheel hub and spokes; 2, electron-dense lumen; 3, periphery of the basal body). The chart shows the frequency (n) gold particles were observed for each antibody at each position. (d) Sas6a localizes to the kinetodesmal fiber. The white arrows indicate anti-Sas6a particles found along the kinetodesmal fiber associated with a basal body in cross section (right). (e) Sas6a localizes to the periphery of cilia. The arrow indicates the position of a 25-nm gold particle.
Like Sas6a and centrin, Sas6b localized to all basal bodies present in Tetrahymena, but with varying intensity compared with the more uniform intensity of Sas6a (Figure 1c, inset). Using an antibody to α-tubulin (Atu1) to label cilia and basal bodies, we found Sas6b to be specifically enriched at actively assembling basal bodies (Figure 1c, white arrow) and at unciliated basal bodies (Figure 1c, unfilled arrow), compared with mature ciliated basal bodies (Figure 1c, arrowhead). Tetrahymena cortical row basal bodies assemble near the anterior side of an existing basal body (Allen, 1969; Nanney, 1975), therefore the more intense Sas6b signal in the inset of Figure 1c (white arrow) likely represents an actively assembling or a recently assembled basal body. The differences of localization patterns between Sas6a and Sas6b, suggests that the two proteins do not function completely redundantly in Tetrahymena.
Sas6a and Sas6b Localize to the Cartwheel Hub and Spoke Tips, Electron-dense Lumen, and Site of New Assembly
Sas6a and Sas6b were both found at basal bodies, but the resolution of light microscopy was insufficient to determine which specific structure(s) within the basal body contain these two proteins. We therefore used immunoelectron microscopy of whole cells to determine the high-resolution localization of both proteins at basal bodies. All mature Tetrahymena basal bodies contain three recognizable structures in the lumen of the basal body: the cartwheel (most proximal), the electron-dense material (midpoint), and the transition zone (most distal; Allen, 1969; Kilburn et al., 2007). Previous work from our lab showed that overexpressed GFP-Sas6a localized to the region of the basal body occupied by the hub of the cartwheel (Kilburn et al., 2007). Additionally, a recent report on SAS-6 in Chlamydomonas showed an identical ultrastructural localization pattern (Nakazawa et al., 2007). We found that Sas6a, which like its GFP-tagged counterpart localized to the cartwheel hub (Figure 2a, arrow) but surprisingly also to the spoke attachment points to the “A” tubule of the microtubule triplets (Figure 2a, arrowhead). Sas6a was also found in the electron-dense material in the basal body lumen (Figure 2a, white asterisk) and on the periphery near the base (Figure 2a, black asterisk). The frequency of these basal body localizations are quantified in Figure 2c (1: cartwheel localization, 2: electron-dense lumen, 3: area surrounding proximal end of basal body).
Sas6b shared all of the same basal body localizations with Sas6a: the cartwheel hub (Figure 2b, arrow) and spoke attachment points (Figure 2b, arrowhead), the electron-dense luminal material (Figure 2b, white asterisk), and the area surrounding the proximal end of the basal body (Figure 2b, black asterisk). Both proteins were observed in the same locations in the basal body at approximately equal frequencies (Figure 2c). These data combined with our fluorescence data suggest that SAS6a and SAS6b may have evolved overlapping as well as distinct roles in basal body assembly and function.
SAS6a Is Required for Generation of New Basal Bodies and Maintenance of Cilia
To test the hypothesis that SAS6a has evolved an essential function separate from that of SAS6b, we designed an experiment to test the requirements of SAS6a for cell viability and basal body assembly (see Materials and Methods and Supplemental Figure S2a). We could only generate a complete assortment of a SAS6a deletion allele when an inducible 6xHis-tagged version of SAS6a was expressed from a different locus (Supplemental Figure S2b). This experiment demonstrated that SAS6a is required for cell viability and that we had constructed a strain in which we could control expression of SAS6a. It also showed that SAS6b alone is not sufficient for viability. Under conditions where SAS6a expression was repressed, 86% of cells divided three times or less compared with 6% of wild-type cells under identical conditions (Supplemental Figure S2c). Based on these data, SAS6a is essential for cell division, and the SAS-6 genes in Tetrahymena are not redundant for this essential function.
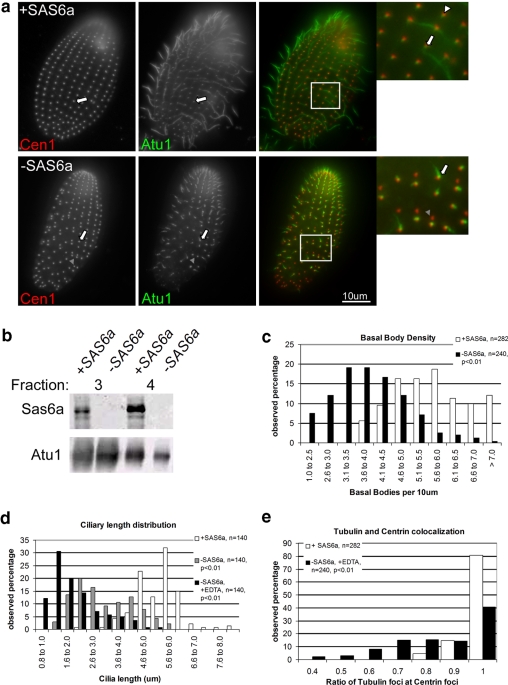
SAS-6 is essential for centriole and basal body assembly in C. elegans, Chlamydomonas, Drosophila, and mammalian cell culture (Dammermann et al., 2004; Leidel et al., 2005; Nakazawa et al., 2007; Rodrigues-Martins et al., 2007a; Vladar and Stearns, 2007). We therefore predicted that SAS6a would be essential for basal body duplication in Tetrahymena. Tetrahymena cells assemble new basal bodies anterior to existing ones and at semiregular intervals along the anterior-posterior axis of the cell (Allen, 1969; Nanney, 1975). Before cell division, the basal bodies along the middle and posterior of the cell duplicate at a higher frequency than the basal bodies at the cell anterior (Nanney, 1975). Tetrahymena divide along the cell equator, and thus both daughter cells possess basal bodies and their associated cilia from the parent. A defect in new basal body assembly would result in larger than normal gaps between existing basal bodies as well as fewer basal bodies at the anterior or posterior end of a cell that had just divided. Alternatively, if SAS6a were required for maintenance of existing basal bodies, then we may expect to see all basal bodies disappear as was observed in γ-tubulin (GTU1) and centrin (CEN1) null mutants (Shang et al., 2002a; Stemm-Wolf et al., 2005). Figure 3a shows cells labeled for centrin (Cen1) and α-tubulin (Atu1) to mark basal bodies, in which the SAS6a gene has been induced or repressed for 2 d. In cases in which SAS6a has been induced (+SAS6a), we found that all of the cells were ciliated (Figure 3a, arrow inset) and that Cen1 and Atu1 colocalized at basal bodies as was expected with this control (Figure 3a, arrowhead). Instances of Cen1 and Atu1 colocalization indicate either fully formed basal bodies or basal bodies that have developed beyond the early intermediates, regardless of the presence of a cilium. When the SAS6a gene was repressed using EDTA (−SAS6a) cilia were severely truncated or absent (Figure 3a, inset arrow). In addition to the short cilia observed upon repression of SAS6a, Cen1 labeling did not always colocalize with Atu1 labeling (Figure 3a, gray arrowhead). The lack of Atu1 signal at Cen1-positive locations indicates there were fewer bona fide basal bodies per cell than Cen1 signal alone indicated. Western blot analysis confirmed that cells grown in EDTA lacked detectable Sas6a (Figure 3b). We used instances of Cen1 and Atu1 colocalization to quantify the number of basal bodies present in each condition. On average, cells expressing SAS6a (+SAS6a) had 5.7 ± 1.1 basal bodies per 10 μm (n = 282) and cilia 5.5 ± 1 μm long (n = 140), whereas cells with repressed SAS6a expression (−SAS6a, + EDTA) had an average of 3.9 ± 1 basal bodies per 10 μm (Figure 3c, n = 240, p <0.01) and cilia 2 ± 1 μm long (Figure 3d, n = 140, p < 0.001). As evidence that the presence of EDTA in the media suppresses expression of SAS6a more so than absence of CdCl2 from the media, cells grown in media lacking CdCl2 (−SAS6a) displayed cilia of intermediate length, 3.2 ± 1.1 μm long (Figure 3d, n = 140, p < 0.001).
Figure 3.
SAS6a is required for new basal body assembly. (a) SAS6a is required for microtubule incorporation into basal bodies and for maintenance of cilia length. Fixed whole cells either grown in the presence (+SAS6a) or absence (−SAS6a) of SAS6a were labeled with antibodies to Cen1 (left panels, red in merge) and Atu1 (middle panels, green in merge). The white arrows show examples of a ciliated basal body, the white arrowhead shows an unciliated basal body, and the gray arrowhead shows a putative incomplete basal body: Cen1 signal is present, but Atu1 signal is absent. (b) SAS6a expression is effectively repressed by inclusion of 100 μM EDTA in the media of the SAS6a conditional null strain. Western blot showing levels of Sas6a and Atu1 in the indicated fractions for the indicated conditions. Equal total protein was loaded in this blot. Fraction 3, protein soluble in HpHS buffer (see Materials and Methods), and fraction 4, the insoluble pellet from this treatment. Sas6a was not found in fractions 1 and 2 in either condition (data not shown). (c) Graphical representation of the basal body assembly defect seen upon activation or repression of SAS6a. Basal bodies were counted based on instances of Cen1 and Atu1 colocalization. Graph shows the frequency of basal body density observed in a population of cells grown with (+SAS6a) or without (−SAS6a) SAS6a. −SAS6a cells have significantly fewer basal bodies per 10 μm than +SAS6a cells. (d) Cells grown without induction of SAS6a display shorter cilia than cells grown with induction of SAS6a. Graphical representation of the distribution of cilium length seen in a population of cells grown with active repression of SAS6a (−SAS6a, +EDTA), incomplete repression of SAS6a (−SAS6a), or with expression of SAS6a (+SAS6a). Cilia were measured from the base to the tip and plotted as the frequency of range of lengths observed in the population. (e) Loss of SAS6a results in a significant increase of aberrant basal bodies. Graphical representation of the frequency at which Cen1 foci were observed with Atu1 foci shows that cells grown in the absence of SAS6a (−SAS6a) had fewer instances of Cen1 and Atu1 colocalization than cells grown with SAS6a (+SAS6a). The number of Cen1 foci colocalizing with Atu1 foci were counted along individual ciliary rows for these calculations.
To quantify the frequency that we observed Cen1 foci lacking Atu1 signal, we counted the number of Cen1 foci and the number of Atu1 foci that colocalized with these spots. These numbers were converted to a ratio for each ciliary row counted. Cells expressing SAS6a had a tubulin-to-centrin ratio of 1 ± 0.05 (n = 282), indicating that the vast majority of Cen1-positive foci represented fully formed basal bodies. In contrast to this, repression of SAS6a resulted in an average tubulin-to-centrin ratio of 0.85 ± 0.17 (Figure 3e, n = 240, p < 0.001), indicating that a significant portion of Cen1-positive locations likely did not represent fully formed basal bodies.
We next tested whether loss of SAS6a affected the localization of Sas6b. In a manner indistinguishable from WT cells, Sas6b and Atu1 colocalized at unciliated and most ciliated basal bodies when SAS6a was expressed (+SAS6a, Figure 4a). When relative fluorescence intensity was quantified, we found that 2.9-fold more Sas6b signal was present at unciliated and assembling basal bodies than at ciliated basal bodies (n = 149). In contrast to this, cells in which SAS6a was repressed (−SAS6a) had only 1.2-fold more Sas6b fluorescence signal at unciliated basal bodies than was present at ciliated basal bodies (n = 151). These cells also more frequently displayed Sas6b foci in the absence of an Atu1 signal (Figure 4a, arrowheads). +SAS6a cells usually had Sas6b present at Atu1 foci, with an average Atu1 foci-to-Sas6b foci ratio of 0.99 ± 0.13 (n = 107). In contrast to this, −SAS6a cells had significantly more Sas6b foci per ciliary row than Atu1 foci, with an average ratio of 0.73 ± 0.12 (Supplemental Figure S3b, n = 120, p < 0.001). On the basis of the observation that Sas6b localizes to presumptive sites of new assembly independently of Sas6a and our differential localization data, we suggest that Sas6b may in fact act upstream of or parallel to Sas6a in the basal body assembly pathway.
Figure 4.
SAS6a is required for the maintenance of the cartwheel structure. (a) Fixed cells grown in the presence (+SAS6a) or absence (−SAS6a) were labeled with antibodies to Atu1 (left panels, green in merge) and Sas6b (middle panels, red in merge). Sas6b signal at unciliated or assembling basal bodies (arrowheads) was on average 2.9-fold greater than Sas6b signal at ciliated basal bodies (arrows) when SAS6a was expressed (n = 149). When SAS6a expression was repressed (−SAS6a), this ratio decreased to 1.2 (n = 151). (b) Repression of SAS6a is correlated with the loss of the cartwheel from mature basal bodies and the appearance of ribosomes in lumen of the basal body. Electron micrographs of whole cell preparations showing basal bodies in longitudinal section (left panels) or cross section (right panels) grown with (+SAS6a) or without (−SAS6a). +SAS6a basal bodies have the cartwheel (unfilled arrows), the electron-dense lumen (white arrowhead), and the markers of mature basal bodies: the postciliary microtubules (PC) and kinetodesmal fibers (KF). Ribosomes (white arrows) are excluded from the basal body lumen. −SAS6a basal bodies often lacked a complete cartwheel (unfilled arrows), and loss of the cartwheel was correlated with the appearance of ribosomes in the lumen of the basal body. The percentages indicate the frequency at which basal bodies like the ones in the figure were observed (see Materials and Methods). Examination of serial sections of two basal bodies in cross section (1, most proximal; 4, most distal) demonstrates this. Structural features are indicated as in +SAS6a condition.
Although Western blot analysis showed undetectable levels of Sas6a when expression was repressed (Figure 3b), we wanted to determine whether Sas6a could be detected by immunofluorescence. In cells in which SAS6a expression was induced (+SAS6a) the presence of an Atu1 signal correlated with the presence of Sas6a signal (Supplemental Figure S3a, arrow). Not surprisingly, cells in which SAS6a expression was repressed (−SAS6a) showed significantly reduced levels of Sas6a (Supplemental Figure S3a). These cells also displayed basal bodies where Sas6a was absent (Supplemental Figure S3a, arrowhead). Furthermore, cells expressing SAS6a (+SAS6a) had an Sas6a signal associated with centrin signal (Supplemental Figure S2d, arrow inset). However, cells where SAS6a expression was repressed (−SAS6a) often displayed centrin only foci (Supplemental Figure S2d, arrowheads inset) in addition to the normal basal body associated Sas6a (Supplemental Figure S2d, arrow inset). The lingering presence of Sas6a at some basal bodies indicates that either some Sas6a is being expressed in these cells or that some fraction of the protein remains stably associated with basal bodies. Although Sas6a signal is absent from most basal bodies, it remains at the basal bodies found at the anterior end of the cell. These basal bodies are among the oldest present in the cell (Nanney, 1975). Our data shows that SAS6a is required for new basal body assembly and maintenance of cilia length, but not for maintenance of existing basal bodies. Additionally, we show that endogenous levels of SAS6b are not able to rescue loss of SAS6a in basal body assembly or cilia length maintenance.
SAS6a Is Required for Assembly of the Cartwheel
Tetrahymena basal bodies are different from mammalian basal bodies in that every mature basal body possesses a cartwheel and electron-dense lumen (Allen, 1969). The basal body is physically separated from the cytoplasm by its microtubule walls, the cartwheel at the proximal end, and the transition zone at the distal end. We were interested whether the instances of an Atu1 signal lacking a Sas6a signal (Supplemental Figure S3a, −SAS6a) represented abnormal basal bodies. Therefore, we examined +SAS6a and −SAS6a cells by electron microscopy to determine if there were specific basal body structural defects associated with loss of SAS6a. Cells expressing SAS6a (+SAS6a) displayed all of the normal features associated with Tetrahymena basal bodies: the cartwheel (Figure 4b, unfilled arrows), electron-dense luminal material (Figure 4b, arrowheads), and ribosomes excluded from lumen of the basal body (Figure 4b, white arrows), illustrating the physical separation of the basal body interior from the cytoplasm. In basal bodies of −SAS6a cells, the cartwheel was absent in 67% of basal bodies examined (Figure 4b, unfilled arrow, n = 36) and the lumen of the basal body where the electron-dense material would normally reside was instead replaced by what are presumably ribosomes in 40% of basal bodies examined (Figure 4b, white arrow, n = 173), showing that the cytoplasm had infiltrated the lumen of the basal body. Examination of serial sections through adjacent basal bodies in the −SAS6a cells (Figure 4b and Supplemental Figure S4) shows that loss of the cartwheel (unfilled arrows) is correlated with appearance of ribosomes in the lumen of the basal body (white arrows). Both of the basal bodies in cross section display the ninefold symmetry typical of normal basal bodies and are also associated with markers of mature Tetrahymena basal bodies, postciliary microtubules (Figure 4b, PC), and kinetodesmal fibers (Figure 4b, KF). We therefore conclude that SAS6a is required for the assembly of the cartwheel and that without the cartwheel the physical separation of the basal body from the cytoplasm is compromised, which correlates with the loss of normal luminal structures.
SAS6b Influences the Timing and Location of New Basal Body Assembly
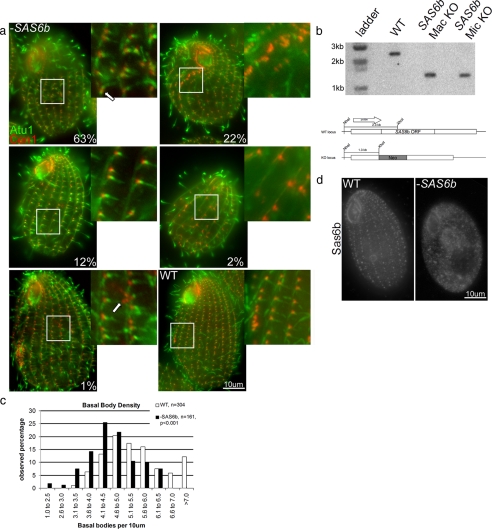
Having shown that SAS6a is an essential gene in Tetrahymena, we were curious as to whether or not SAS6b was also essential. Surprisingly, we discovered that SAS6b was in fact not an essential gene, as mating two SAS6b knockout heterokaryon strains to each other produced viable progeny. We were also able to generate SAS6b macronuclear null cells using paromomycin resistance as a selectable marker for deletion of SAS6b. Southern blot analysis (Figure 5b), immunofluorescence microscopy using Sas6b antibodies (Figure 5d), and PCR (data not shown) confirmed that wild-type SAS6b was completely absent from both SAS6b null strains. Furthermore, SAS6b null cells are capable of multiple rounds of cell division with little or no reduction in growth rate (data not shown). SAS6b is therefore not essential for cell division and the presence of SAS6a may be sufficient to sustain viability.
Figure 5.
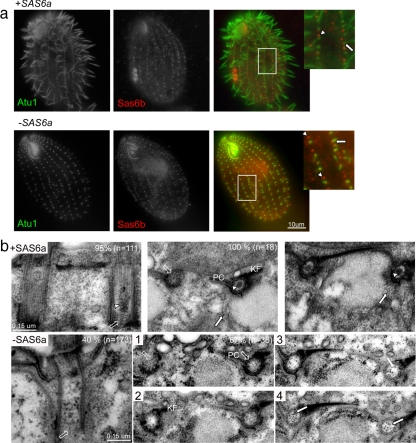
SAS6b is important for but not essential to vegetative cell growth and new basal body assembly. (a) Loss of SAS6b is correlated with appearance of cells with fewer basal bodies and/or lacking complete oral apparatus. Fixed wild-type (WT) and SAS6b KO cells (−SAS6b) were labeled with antibodies to Cen1 (left panels, red merge) and Atu1 (middle panels, green merge). Cells (n = 300) that resembled the cells in the images shown in this figure were counted. The percentage of cells that matched these representative images is shown in the bottom right-hand corner. The arrow in the first cell (63%) indicates a branching of a cortical row. The arrow in the fifth panel (1%) indicates a cluster of basal bodies present along a cortical row. (b) SAS6b is not required for vegetative cell growth. Southern blot showing genomic DNA from indicated strains digested with XhoI and NheI. The blot was probed with DNA 1 kb 5′ of the SAS6b start site. Diagram below blot shows wild-type (WT) SAS6b locus and null (KO) locus. The wild-type fragment recognized by the probe is 2.3 kb and the null fragment is 1.3 kb. (c) SAS6b null cells have fewer basal bodies per 10 μm than wild type. Basal bodies were counted based on instances of Cen1 and Atu1 colocalization. (d) Sas6b basal body signal is absent in SAS6b KO cells. Fixed wild-type (WT) or SAS6b KO cells (−SAS6b) were labeled with the anti-Sas6b antibody.
Although it appeared that SAS6b is not essential, we were curious if there was a phenotype associated with these knockout cells. Therefore, we examined SAS6b null cells by immunofluorescence microscopy using antibodies to Cen1 and Atu1 to mark basal bodies. Cells were categorized into five different phenotypic classes, examples of which are shown in Figure 5a. Sixty-three percent of cells had obvious defects in cortical organization. These defects ranged from mild bends in the cortical rows (data not shown) to the spurring of an additional cortical row off of an exiting one (Figure 5a, top left, arrow). Twenty-two percent of SAS6b null cells possessed an abnormal existing oral apparatus (data not shown) or a prematurely assembled new oral apparatus (Figure 5a, top right). 12% of cells had visible gaps between exiting basal bodies with otherwise normal cortical organization (Figure 5a, middle left). A small fraction of cells (2%) were categorized as severely abnormal. These cells had many fewer basal bodies and lacked an oral apparatus altogether (Figure 5a, middle right). Even fewer cells (1%) were categorized as normal, despite the presence of basal body clusters along a cortical row (Figure 5a, bottom left, arrow). Cortical row basal bodies are aligned along the anterior posterior axis, and the presence of a basal body cluster such as the one shown in Figure 5a represents an abnormal situation. As was done with −SAS6a cells, the number of basal bodies per 10 μm were counted in −SAS6b and wild-type cells based on the number of Atu1 foci colocalizing with Cen1 foci. −SAS6b cells had an average of 4.6 ± 0.9 (n = 161) basal bodies per 10 μm compared with 5.5 ± 1.1 basal bodies per 10 μm for wild-type cells (n = 304, p < 0.001). The distribution of basal body densities observed in wild-type and −SAS6b cells is shown graphically in Figure 5c. The mild decrease in average basal body density in SAS6b null cells indicates that although the spatial arrangement of basal bodies is altered, new basal bodies are nonetheless being assembled, sometimes earlier than expected.
Loss of SAS6b Does Not Significantly Affect Cartwheel Formation
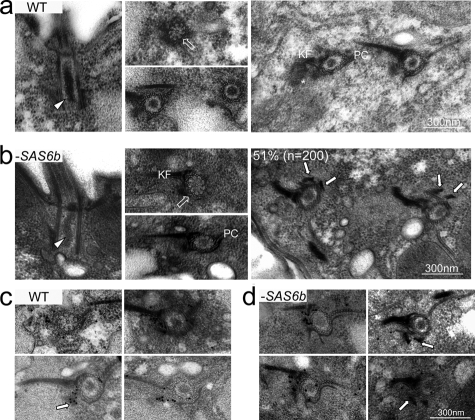
Having shown that loss of SAS6a results in the absence of the cartwheel from basal bodies, we were curious if loss of SAS6b produced a similar result. If Sas6a and Sas6b both functioned as structural components of the cartwheel, then we might expect that the loss of SAS6b would affect the formation of the cartwheel in the same manner as loss of SAS6a. However, based on all of the data presented thus far, it seemed more likely that loss of SAS6b would not affect the formation or maintenance of the cartwheel. To test this prediction, we analyzed SAS6b null cells by thin-section transmission electron microscopy. SAS6b null cells (−SAS6b) possessed all of the structural features found in wild-type basal bodies: the cartwheel at the proximal end (Figure 6b, unfilled arrow), the electron-dense luminal material (arrowhead), postciliary microtubules (PC), and kinetodesmal fibers (KF). Although all of the normal structural elements were present in SAS6b null basal bodies, 51% (n = 200) of basal bodies examined had electron-dense regions at or near the site of new assembly (Figure 6b, arrows), which was significantly higher than the 15% (n = 189) observed for wild-type basal bodies. New basal bodies form just adjacent to the kinetodesmal fiber and opposite of the postciliary microtubules (Figure 6a, asterisk). It is therefore possible that the electron-dense regions seen in the SAS6b null cells represent new basal body assembly intermediates. Alternatively, the electron densities seen more commonly in the SAS6b null cells could be distorted basal body collar regions. To help exclude this second possibility, we examined the localization of Cen1 in SAS6b null cells. Cen1 localizes to the site of new basal body assembly in Tetrahymena (Stemm-Wolf et al., 2005). If the aggregates seen in the SAS6b null are associated with new basal body assembly events, then the localization of Cen1 to this density could reflect this possibility. As in wild-type cells, Cen1 localized to the site of new assembly in SAS6b null cells (−SAS6b). More interestingly, Cen1 was also found on the electron densities or in close proximity to them (Figure 6d, arrows). The localization of Cen1 to the electron densities may reflect a role for the protein in determining the site of new basal body assembly. It also supports the possibility that the electron densities are somehow related to new basal body assembly.
Figure 6.
Loss of SAS6b does not affect cartwheel assembly, but does result in accumulation of electron densities near the site of new basal body assembly. (a) Electron micrographs showing wild-type (WT) basal bodies in longitudinal or cross section. Arrowhead, the electron-dense lumen; unfilled arrow, a cartwheel in cross section. A new basal body (asterisk) is seen in the panel on the right just below the kinetodesmal fiber (KF) and opposite of the postciliary microtubules (PC). (b) Basal bodies from SAS6b null cells (−SAS6b) possess all of the structures associated with wild-type basal bodies. Relevant structures are indicated as in panel a. Fifty-one percent of basal bodies examined (n = 200) showed electron densities (arrows) at or near the site of new assembly. (c) Cen1 localizes to the site of new basal body assembly in wild-type (WT) cells. Immunoelectron micrographs using a 25-nm gold–conjugated secondary antibody showing the localization of Cen1. The arrow indicates Cen1 localization at the site of new assembly. (d) Cen1 localizes to the site of new assembly in SAS6b null cells (−SAS6b) and is associated with the electron densities. Immunoelectron micrographs labeled as in panel c. The arrows point to Cen1 association with the electron densities.
Overexpression of SAS6b Results in Inappropriate New Basal Body Assembly
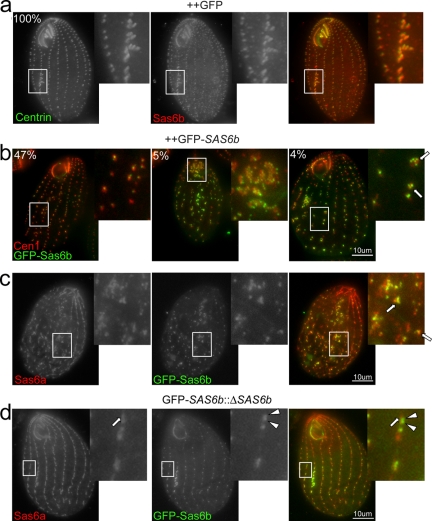
Overexpression of SAS-6 proteins has been shown to cause the formation of excess centrioles in human cells as well as in Drosophila (Leidel et al., 2005; Peel et al., 2007; Strnad et al., 2007). Overexpression of SAS6a in Tetrahymena results in aggregations of the protein along the cortex of the cell, but not in ectopic basal bodies (data not shown). However, it is possible that over the course of evolution the basal body seeding property observed for other Sas-6 proteins has been assumed by the SAS6b gene. In controls, all cells overexpressing GFP (n = 200) displayed normal localization patterns for both Sas6b and centrin: Sas6b was enriched at the developing oral apparatus, which is always found along the equator of the cell below the existing oral apparatus (Figure 7a, ++GFP). In contrast, after approximately two cell divisions cells overexpressing GFP-SAS6b displayed three classes of cortical organization defects when labeled with antibodies to GFP and Cen1. Forty-seven percent of cells examined (n = 203) had disorganized cortical row basal bodies (Figure 7b, left panel). 5% of cells had an abnormal existing oral apparatus (Figure 7b, middle panel), and 4% of cells displayed clusters of basal bodies in abnormal locations. The right panel in Figure 7b shows an example of one such cell; the arrows point to clusters of basal bodies present along a cortical row where such accumulations of basal bodies is not observed in wild-type cells. Additionally, cells with the most severe cortical architecture abnormalities had more intense GFP-Sas6b signal. As further evidence that the clusters of GFP-Sas6b represent bona fide basal bodies, these clusters were also labeled with Sas6a antibody (Figure 7c).
Figure 7.
SAS6b helps dictate sites of new basal body assembly and acts upstream of SAS6a. (a) Overexpression of GFP does not affect cellular morphology. Fixed cells overexpressing GFP (++GFP) were labeled with antibodies to centrin (left panel, green in merge) and Sas6b (middle panel, red in merge). Sas6b is enriched at the developing oral apparatus (inset), which is always found immediately below the existing oral apparatus along the equator of the cell (100%, n = 200). (b) Overexpression of GFP-SAS6b causes the formation of clusters of basal bodies in abnormal locations. Fixed cells overexpressing GFP-SAS6b were labeled with antibodies to Cen1 (red) and GFP (green). Forty-seven percent of cells in the population had disorganized cortical rows and large gaps between existing basal bodies (left panel). Five percent of cells had an abnormal oral apparatus (middle panel), and 4% of cells had clusters of basal bodies in abnormal locations (right panel, n = 203). Arrows point to clusters of basal bodies in abnormal locations. (c) Sas6a colocalizes with GFP-Sas6b clusters. Fixed cells overexpressing GFP-SAS6b were labeled with antibodies to Sas6a and GFP. Arrows indicate Sas6a and GFP colocalization. (d) GFP-Sas6b localizes to presumptive sites of new basal body assembly before Sas6a. Fixed SAS6b macronuclear null cells carrying an inducible GFP-SAS6b allele at a different locus were induced to express GFP-SAS6b and labeled with antibodies to Sas6a (left panel, red in merge) and GFP (middle panel, green in merge). The arrow shows Sas6a kinetodesmal fiber localization. The arrowheads show GFP-Sas6b doublets representing a new basal body assembly event.
To investigate the temporal localization pattern of Sas6a and Sas6b in the same cell, we induced GFP-SAS6b expression in a SAS6b macronuclear knockout. As stated previously, new basal bodies assemble anterior to existing basal bodies adjacent to the kinetodesmal fiber. If Sas6a and Sas6b are recruited to basal bodies at the same time then new basal body assembly events would be observed as a colocalization of GFP-Sas6b and Sas6a at the new anterior basal body. If either protein is recruited to sites of new assembly in advance of the other, then this would be observed as the presence of either Sas6a or GFP-Sas6b in the absence of the other. GFP-Sas6b appears at sites of new basal body assembly in the absence of Sas6a. At higher magnification two GFP-Sas6b foci (Figure 7d, arrowheads) are seen just to the cells left of the Sas6a kinetodesmal fiber signal (Figure 7d, arrow). This data shows that GFP-Sas6b localizes to sites of new basal body assembly before Sas6a basal body incorporation.
DISCUSSION
The defining feature of basal bodies and centrioles is their ninefold symmetrical arrangement of microtubules. However, the molecular mechanism(s) by which this symmetry is faithfully established have only recently been addressed. In C. elegans, SAS-6 is required for the formation of the central tube, the scaffold around which the microtubules of the centriole assemble (Pelletier et al., 2006). In many other eukaryotes, a central hub with nine spokes (the cartwheel) is present at the base of the basal body or procentriole. Each spoke makes contact with the “A” tubule of a microtubule triplet (Lange and Gull, 1996). These observations imply that the cartwheel is important for establishment of ninefold symmetry. Chlamydomonas and possibly Drosophila SAS-6 mutants assemble centrioles lacking the cartwheel, and these centrioles often had greater than or less than nine microtubule triplets (Nakazawa et al., 2007; Rodrigues-Martins et al., 2007a). Furthermore, SAS-6 has been shown to localize to the cartwheel hub in Tetrahymena and Chlamydomonas (Kilburn et al., 2007; Nakazawa et al., 2007). In human cells SAS-6 is only found at the immature centriole (Strnad et al., 2007), the only centriole of the pair with a cartwheel (Vorobjev and Chentsov Yu, 1982). Based on the cumulative observations from all these systems, it seems likely that the central tube of C. elegans is analogous to the cartwheel and that SAS-6 is required for the formation of both, either as a structural component (Strnad and Gonczy, 2008) and/or as a recruitment factor for other components.
It is now clear that SAS-6 is component of the hub of the cartwheel in Tetrahymena and most other organisms (Kilburn et al., 2007; Nakazawa et al., 2007). However, endogenous Sas6a and Sas6b not only localize to the cartwheel hub, but also to the spokes and electron-dense lumen and at the site of new basal body assembly. These localizations were not observed for CrSas-6 (Nakazawa et al., 2007). This discrepancy may be due to the specificity of our antibodies or it may be due to CrSas-6 being localized in biochemically isolated centrioles, whereas our study used whole cell preparations. It may also be that the noncartwheel hub localization patterns observed herein are Tetrahymena specific.
On repression of SAS6a, the cartwheel hub and basal body luminal structures were absent from mature basal bodies. This result provides confirmation in another model system that Sas6 proteins function at centrioles and basal bodies as components of the cartwheel. The presence of mature basal body structures associated with basal bodies lacking the cartwheel combined with the overall decrease in basal body number observed in −SAS6a cells leaves open the possibility that after SAS6a depletion the protein was turned over at mature basal bodies and subsequently the cartwheel was disassembled. Recently, it has been shown that GFP-Sas6a can be recruited to the cartwheel in the absence of new basal body assembly (Pearson et al., 2008), showing that there is a dynamic population of Sas6a at the cartwheel. Furthermore, in humans, the cartwheel is lost from procentrioles at the onset of mitosis concomitant with the disappearance of HsSas6 (Lange and Gull, 1996; Strnad et al., 2007). These observations support the possibility that loss of Sas6 from centrioles results in disassembly of the cartwheel. Our observation that Cen1 localizes to foci absent of all or significant Atu1 signal suggests that Cen1 acts upstream of Sas6a, contrary to what has been reported in human cells (Strnad et al., 2007). There are a few possible alternative explanations for this observation: Tetrahymena may employ different assembly mechanisms than human cells, these basal bodies may have progressed beyond early assembly intermediates and then fallen apart, or basal body assembly may not proceed in a linear pathway. The last possibility seems particularly attractive as both Drosophila and Chlamydomonas SAS-6 mutants assemble triplet microtubules in the absence of the cartwheel.
Apart from its role in central tube or cartwheel assembly, overexpression of SAS-6 has been shown in a few systems to lead to the formation of excess centrioles (Leidel et al., 2005; Peel et al., 2007; Strnad et al., 2007), suggesting a role for the protein in the initiation of new centriole assembly. However, it is unclear as to whether or not this potential role for the protein is specifically related to or separate from its cartwheel localization. If the Sas6 proteins not only function as structural components of the cartwheel, but also as an initiation factor for new basal body or centriole assembly, then it is possible that the two SAS-6 genes present in Tetrahymena reflect these potential functions. On the basis of multiple lines of evidence, we conclude that the Tetrahymena SAS-6 genes have in fact evolved such that SAS6b is predominantly responsible for the timing and spatial determination of new basal body assembly, whereas SAS6a has more of a structural role in the basal body. Although Sas6a and Sas6b localize to all of the same structural domains within the basal body, Sas6a is a component of the kinetodesmal fiber, whereas Sas6b is not. Sas6b is specifically enriched at immature basal bodies, which implies an increased requirement for the protein at immature basal bodies, whereas the levels of Sas6a are nearly constant at all basal bodies. Loss of SAS6b does not seem to affect cartwheel morphology, but does affect the distribution of basal bodies within the cell. The significant increase in basal bodies with Cen1-labeled electron densities near the site of new assembly in SAS6b null cells provides an attractive explanation for the cortical disorganization seen in SAS6b null cells. It is possible that these densities represent sites of new basal body assembly from a single mother. The assembly of a new basal body at one of these sites may result in the branching off of an additional cortical row as basal body assembly proceeds, thus resulting in cortical disorganization. Additionally, GFP-SAS6b was found to localize to presumptive sites of new assembly in the absence of Sas6a. Furthermore, Cen1 and Sas6b do not depend on the presence of Sas6a for their localization. Overexpression of GFP-SAS6b caused the formation of clusters of new basal bodies in abnormal locations within the cell; no such activity for Sas6a was observed. This finding would not be unique to Tetrahymena as it has been observed in Drosophila as well as mammalian cell culture systems (Leidel et al., 2005; Peel et al., 2007; Strnad et al., 2007). Cumulatively our observations suggest a model in which SAS6b normally acts to spatially and temporally dictate where new basal body assembly will occur, whereas SAS6a normally functions as a structural protein required for microtubule incorporation into basal bodies.
The presence of two SAS-6 homologues in Tetrahymena leaves open the possibility of functional redundancy between SAS6a and SAS6b. However, this functional redundancy is likely not mutual, as SAS6a is an essential gene, whereas SAS6b is not. This situation is not without precedent; for example, only one of the two α-tubulin genes, TUB1, is essential for viability in yeast (Schatz et al., 1986). The pleiotropy of SAS6b null phenotypes likely results from the same cause; in the absence of SAS6b the spatial and temporal cues provided by Sas6b are absent and so basal bodies assemble at inappropriate locations. This could manifest in a disorganized cortex, a misplaced oral apparatus, or a complete lack of assembly. Cells derived from a division of a cell with a misplaced oral apparatus may therefore not inherit an oral apparatus. It is likely that given enough cell divisions, SAS6b null cells will all eventually appear as the most severe phenotype shown in Figure 5a.
Loss of SAS-6 has been shown to inhibit cilia assembly (Rodrigues-Martins et al., 2007a; Vladar and Stearns, 2007). However, it was unclear whether this requirement was in the establishment of cilia or the maintenance of their length. We conclude that SAS6a is not only required for the generation of cilia, but also for the maintenance of their length based on the following logic. Cells expressing SAS6a have wild-type cilia lengths and no defects in new basal body assembly. However, upon repression of SAS6a expression the cells become unable to assemble new basal bodies and there is a concomitant shortening of the cilia they do possess. If SAS6a was not required for the maintenance of cilia length, then cilia present before repression of SAS6a should not be affected by the absence of SAS6a. Consistent with a ciliary role for SAS6a, the protein was seen in cilia by immunofluorescence, immunoelectron microscopy, and Western blot.
We have presented evidence here that the Tetrahymena SAS-6 genes have evolved separate functions in both basal body assembly and maintenance of cilia length. SAS6a is required for maintenance of cilia length and cartwheel assembly, whereas SAS6b is important for spatially determining the site of new basal body assembly. In the future we look forward to pursuing the mechanism by which each protein performs the potentially separate functions of initiation and cartwheel assembly. Specifically, we would like to pursue the identification of complexes containing each or both proteins with the goal of linking or separating the cartwheel assembly from new basal body assembly initiation.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank all members of the Winey lab for their helpful and advice and discussion during the preparation of this manuscript, particularly Michele H. Jones, Alexander Stemm-Wolf, and Chad Pearson. B.P.C. also specifically thanks M.B.M. and O.B. for their support during this period. This work was funded by National Institutes of Health Grant GM074746 to M.W.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0838) on January 21, 2009.
REFERENCES
- Allen R. D. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 1969;40:716–733. doi: 10.1083/jcb.40.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Brown T. Southern blotting. In: Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. Current Protocols in Molecular Biology. Vol. 1. Hoboken, NJ: John Wiley and Sons; 1999. pp. 2.9.1–2.9.15. [Google Scholar]
- Bruns P. J., Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 2000;62:501–512. doi: 10.1016/s0091-679x(08)61553-8. [DOI] [PubMed] [Google Scholar]
- Callaini G., Whitfield W. G., Riparbelli M. G. Centriole and centrosome dynamics during the embryonic cell cycles that follow the formation of the cellular blastoderm in Drosophila. Exp. Cell Res. 1997;234:183–190. doi: 10.1006/excr.1997.3618. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardtii. J. Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Muller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Delattre M., Canard C., Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Dippell R. V. The development of basal bodies in paramecium. Proc. Natl. Acad. Sci. USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen E. R. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J. Cell Biol. 1971;51:286–302. doi: 10.1083/jcb.51.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 1997;46:101–111. doi: 10.1093/sysbio/46.1.101. [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y. D., Wilkinson C. J., Nigg E. A. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hyams J. S., King C. A. Identification of proteins of the striated rootlet of Tetrahymena by immunofluorescence microscopy and immunoblotting with an anti-rootlet serum. Eur. J. Cell Biol. 1985;38:102–105. [Google Scholar]
- Kilburn C. L., Pearson C. G., Romijn E. P., Meehl J. B., Giddings T. H., Jr, Culver B. P., Yates J. R., 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., Nigg E. A. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Lange B. M., Gull K. Structure and function of the centriole in animal cells: progress and questions. Trends Cell Biol. 1996;6:348–352. doi: 10.1016/0962-8924(96)10033-7. [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Nanney D. L. Patterns of basal body addition in ciliary rows in Tetrahymena. J. Cell Biol. 1975;65:503–512. doi: 10.1083/jcb.65.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Giddings T. H., Jr, Winey M. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol. Biol. Cell. 19:904–914. doi: 10.1091/mbc.E08-08-0835. Epub December 3, 2008. DOI: 10.1091/mbc.E08-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N., Stevens N. R., Basto R., Raff J. W. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D. M. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 2007a;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D. M., Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007b;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Satir P., Guerra C., Bell A. J. Evolution and persistence of the cilium. Cell Motil. Cytoskelet. 2007;64:906–913. doi: 10.1002/cm.20238. [DOI] [PubMed] [Google Scholar]
- Schatz P. J., Solomon F., Botstein D. Genetically essential and nonessential alpha-tubulin genes specify functionally interchangeable proteins. Mol. Cell. Biol. 1986;6:3722–3733. doi: 10.1128/mcb.6.11.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Li B., Gorovsky M. A. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J. Cell Biol. 2002a;158:1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Song X., Bowen J., Corstanje R., Gao Y., Gaertig J., Gorovsky M. A. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA. 2002b;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A. J., Morgan G., Giddings T. H., Jr, White E. A., Marchione R., McDonald H. B., Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Gonczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K. R., Cole E. S. Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 2000;62:291–311. doi: 10.1016/s0091-679x(08)61538-1. [DOI] [PubMed] [Google Scholar]
- Thompson G. A., Jr, Baugh L. C., Walker L. F. Nonlethal deciliation of Tetrahymena by a local anesthetic and its utility as a tool for studying cilia regeneration. J. Cell Biol. 1974;61:253–257. doi: 10.1083/jcb.61.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E. K., Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I. A., Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.