Abstract
The commonly used, monomeric EYFP enabled imaging of intracellular protein structures beyond the optical resolution limit (‘super-resolution’ imaging) in living cells. By combining photoinduced activation of single EYFP fusions and time-lapse imaging, we obtained sub–40 nm resolution images of the filamentous superstructure of the bacterial actin protein MreB in live Caulobacter crescentus cells. These studies demonstrated that EYFP is a useful emitter for in vivo super-resolution imaging.
As is well known, optical fluorescence microscopy is an important tool for cell biology because light can be used to noninvasively probe a sample with relatively small perturbation of the specimen, allowing dynamical observation of the motions of internal structures in living cells but with resolution usually limited to ~250 nm by optical diffraction. Single-molecule widefield fluorescence microscopy achieves nanometer-scale localization accuracy (super-localization) by taking advantage of the fact that the point-spread function (PSF) of an isolated nanoscale emitter can be fit to a precision far greater than the standard diffraction limit1. To apply this idea to experiments with high concentrations of label in biologically relevant, room-temperature studies, a control variable is needed2, and photoactivation or photoswitching have been used to maintain the concentration of emitters at the ‘single-molecule level’, where the PSFs of the individual molecules do not overlap3–5. For example, in photoactivated localization microscopy (PALM)3, structures labeled by an ensemble of photoactivatable fluorescent proteins too dense to be imaged simultaneously are resolved by repeated cycles in each of which only a sparse subset of the fluorophores is activated. The final, super-resolution image is reconstituted from a superposition of the single-molecule positions.
In previous PALM-type imaging, the photoactivatable fluorescent protein has been selected from various sophisticated constructs such as PA-GFP, Dronpa, Kaede, tdEosFP, Dendra2 and rsFastLime3,6,7. However, immobilized and apparently bleached single yellow fluorescent proteins (containing mutations S65G,S72A,T203Y or S65G,S72A,T203F) have been shown to reactivate with violet light more than 10 years ago8, and the possibility of controllable reactivation suggested that PALM-type imaging should be feasible with the closely related EYFP (which contains mutations S65G,V68L,S72A,T203Y). EYFP-MreB fusions have been previously shown to be functional in C. crescentus and in other bacteria, making this a physiologically relevant system9–11. Furthermore, single-molecule imaging of EYFP-MreB with 514-nm excitation has previously shown that this fluorescent protein is a very good, bright single-molecule emitter for live-cell imaging12. Also, EYFP fusions are widely used in cell biology owing to the absence of physiological perturbations with such fusions; thus PALM with EYFP fusions would vastly expand the number of biological specimens immediately available for super-resolution imaging.
The micrometer-scale size of bacterial cells combined with growing interest in the complex protein localization patterns that control their biology make bacteria important targets for super-resolution imaging. Caulobacter crescentus is a particularly interesting prokaryote because each division is asymmetric, and the progression of the cell cycle requires the dynamic localization of both structural and regulatory proteins13. The actin homolog MreB is a bacterial structural protein critical for cell shape, polarity and chromosome segregation in C. crescentus. Low-resolution imaging shows that this protein forms a superstructure that is dynamic over the course of the cell cycle, suggesting filamentous helices in stalked cells and midplane rings in pre-divisional cells, but super-resolution imaging of this polymerized protein chain has thus far been limited to the ‘single-molecule concentration’ observation of tracks of individual molecules treadmilling through filaments12. A recent study applied photoactivation tracking to single molecules of FtsZ in Escherichia coli, but resolution beyond the diffraction limit had not been obtained7. Because the cell cycle in C. crescentus lasts only ~200 min in minimal medium at room temperature, capturing the MreB protein superstructure at different stages of the cell cycle, with its slow but finite dynamics, requires the implementation of live-cell, super-resolution imaging with acquisition times on the order of minutes.
Notably, the resolution criterion for super-resolution imaging of structures is more rigorous than the super-localization that arises from single-molecule imaging. In the latter case, localization beyond the diffraction limit is determined by the goodness of fit of the single-molecule PSF, which is ultimately limited by the number of photons collected from that emitter14. However, to obtain super-resolution images, not only must each single molecule be localized with the desired accuracy, but the Nyquist criterion, which requires that the sampling interval along the structure be at least half the desired resolution, must also be satisfied15. In this work, we used time-lapse imaging to allow single-molecule motions to fill in filamentous structures, increasing the effective labeling concentration without physiologically perturbing the cell. As the MreB proteins treadmill along polymeric chains at a rate of 6.0 nm/s, by introducing a dark period between each imaging frame, these natural dynamics can be exploited to detect more molecular localization events.
We incubated cultures of a C. crescentus merodiploid containing the wild-type mreB gene and a single chromosomal copy of a eyfp-mreB fusion under the control of a xylose-inducible promoter in the presence of a xylose concentration selected to produce a high concentration of fluorescently labeled MreB fusion proteins (~100 times greater concentration than needed for isolated single-molecule imaging). We used 514-nm irradiation to illuminate the sample until the density of emissive fluorophores was reduced to the single-molecule level. We imaged this low-concentration sub-ensemble until all remaining fluorophores bleached, and we localized and recorded single EYFP-MreB positions in each frame (Supplementary Methods online). We imaged at 10 frames/s, a rate at which freely diffusing cytoplasmic EYFP-MreB molecules smear out and become unobservable12. In this fashion, we observed only those EYFP-MreB molecules that were incorporated into a polymerized MreB chain (filament) or other quasi-static structure. We verified this assumption in control experiments with the depolymerizing agent A22 (ref. 11); after the addition of this reagent we observed only a negligible number of localized EYFP-MreB (Supplementary Fig. 1 online).
The previously reported photoinduced EYFP reactivation in vitro8 can be achieved in live cells (Fig. 1). After the initial bleaching step, a cell showed two single EYFP-MreB molecules (Fig. 1a). Additional imaging with 514-nm light bleached all fluorophores (Fig. 1b). A 2-s dose of 407-nm laser illumination reactivated EYFP (Fig. 1c,e,g,i,k) after this and all subsequent bleaching steps (Fig. 1d,f,h,j,l). We chose this pulse length and a reactivation intensity of 103–104 W/cm2, such that at most one EYFP molecule was reactivated in each diffraction-limited region. Additional imaging (5 s) and reactivation turned on different single molecules. We determined the positions and amplitudes of localized proteins in each imaging frame (Fig. 1a,c,e,g,i,k) and at the end of the 4-min acquisition period, summed these localization events. To form the image, we depicted each localized molecule as a unit-area Gaussian profile with fixed width between 30 and 40 nm.
Figure 1.
Reactivation of EYFP-MreB fusions in the same live C. crescentus cell. (a–l) Fluorescence images of single EYFP-MreB molecules overlaid on a reversed-contrast white-light image of the cell being examined. Initial ‘single-molecule concentration’ image showing two isolated single molecules (a). A nonemissive cell after exposure to 514-nm excitation for a few seconds (b). A short 407-nm reactivation pulse was administered after frames b, d, f, h and j in all of which no molecules are in the emissive state. Reactivated single molecules are observed in subsequent frames (c,e,g,i,k). Scale bar, 1 µm.
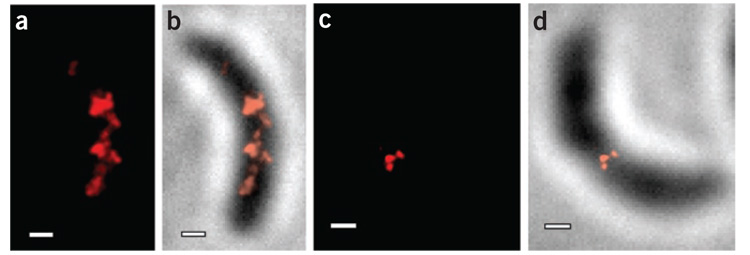
Live-cell PALM allowed us to image single molecules along MreB filamentous structures with sub-40-nm localization accuracy (Fig. 2). We calculated this accuracy as the statistical 96% confidence interval of the center of each single-molecule PSF fit, a more conservative criterion than a theoretical evaluation of the resolution based on number of collected photons14. The system stability did not contribute to the localization error as confirmed by 2,610 successive localizations of a stationary fiduciary marker (Supplementary Fig. 2 online). Our PALM images of EYFP-MreB in C. crescentus showed the presence of two different structures at different stages in the cell cycle: a regularly spaced band-like arrangement of MreB molecules that suggested the presence of a helix in the stalked cell (Fig. 2a,b) and a ring of MreB molecules at the cell midplane in the predivisional cell (Fig. 2c,d). The C. crescentus cells we imaged were ~2 µm long and ~0.5 µm in diameter. As the depth of field in our system was similar to the thickness of the cells, the PALM images represent a two-dimensional projection of the MreB structures. Notably, these images have a much smaller field of view than most images of mammalian cells. Bulk epifluorescence images of EYFP-MreB in stalked cells or in cells treated with A22 show no EYFP-MreB structure whatsoever (Supplementary Fig. 3a online).
Figure 2.
PALM images of EYFP-MreB in C. crescentus cells. (a,b) Banded structure in a stalked cell. (c,d) Midplane ring in a predivisional cell. Fluorescence PALM images are shown in a and c. The PALM images in b and d are the same cells as in a and c, respectively, overlaid on a reversed-contrast white-light transmission image of the cell. Scale bars, 300 nm.
Understanding composite images derived from many acquisitions requires careful consideration of the emitter photophysics and the dynamics, if any, of the underlying structure. We obtained the continuous-acquisition PALM images with 10–20 cycles of a 2-s 407-nm activation pulse followed by 5 s of fluorescence image acquisition (fifty 100-ms frames; Fig. 2). Previous single-molecule studies have found that MreB filaments have an average length of 390 nm, and single MreB units may be observed treadmilling along such polymeric MreB filaments at a rate of 6.0 nm (1.2 monomer additions) per second12. As the MreB molecules travel slowly along polymer chains, the treadmilling motion of each activated molecule during a 5-s acquisition is only 30 nm, which is on the order of the localization precision. As every frame in which single molecules can be localized yields Gaussian profiles for the final image, then for the imaging times chosen, the same single molecule can be included in the image multiple times: that is, it may be oversampled. In this situation, multiple determinations of the same activated molecule would appear as relatively bright punctate spots along with other dimmer molecules that do not emit over multiple frames, and the full structure of the filament would be distorted. To prevent these problems and maximize the information content of the data, we removed this oversampling in a post-processing step (Supplementary Fig. 3).
However, analytically removing the oversampling as described above leads to a smaller number of localization events. As a result, although each molecule contributing to the images in Figure 2 was localized to within 40 nm, the helix in the image in Figure 2a and the ring in the image in Figure 2c were made up of only 145 and 72 molecules, respectively. According to the Nyquist criterion, to resolve a superstructure with 40-nm accuracy, one molecule must be localized every 20 nm along the length of the superstructures. A helix with 0.5-µm diameter that makes four turns along a 2-µm cylinder is 6.6 µm in contour length and thus requires 330 localization events for 40-nm super-resolution. A ring 0.5 µm in diameter is 1.57 µm in contour length and thus requires 79 localization events for 40-nm super-resolution. Our two-dimensional projections were shorter than the full contour length and would require fewer points in principle, but we used the conservative metric. Thus, the continuous-acquisition PALM images in Figure 2 did not provide 40-nm super-resolution. In nonbiological samples this concern could be addressed by increasing the number of fluorophores, but the maximum labeling concentration in live-cell experiments was limited because of changes in the cell morphology at high concentrations of fusion protein; this is an important limit to the resolution of live-cell PALM experiments15. However, as the cells were live, the dynamics of MreB monomers treadmilling along filaments in the present experiment can be exploited to provide more positional information for the same number of fluorophores, thus addressing the Nyquist criterion requirements while maintaining a biologically relevant system.
For this purpose, we implemented time-lapse PALM (TL-PALM; Fig. 3). In TL-PALM, we introduced a 0.9-s delay between each imaging frame and illuminated the sample with 514-nm light only during the short acquisition period such that the time to acquire fifty 100-ms frames was lengthened to 50 s. Over this longer time, each activated molecule traced out up to ~300 nm of path along the filaments so that the distinct localization events elucidated more of the underlying superstructure. The effective labeling concentration was thus increased tenfold. With TL-PALM, as with continuous-acquisition PALM, we identified two distinct MreB superstructures in the cells: a quasi-helical arrangement in a stalked cell (Fig. 3a,b) and a midplane ring in the predivisional cell (Fig. 3c,d). Clearly, the images obtained in this manner were more continuous than those shown in Figure 2 and provided more information on the protein localization patterns. In particular, in the case of the stalked cell (Fig. 3a,b), the improved resolution of TL-PALM made visible strands that join the bands observed in the image in Figure 2a,b, showing that such bands were likely part of a continuous structure. Additionally, use of time-lapse allowed us to localize 487 and 285 molecular positions in the images in Figure 3a,c, respectively, meeting the Nyquist criterion for sub-40-nm super-resolution imaging.
Figure 3.
TL-PALM images of EYFP-MreB in C. crescentus cells showing fewer punctuate spots than continuous-acquisition PALM. (a,b) Quasi-helical structure in a stalked cell. (c,d) Midplane ring in a predivisional cell. Fluorescence PALM images are shown in a and c. The PALM images in b and d are the same cells as in a and c, respectively, overlaid on a reversed-contrast white-light transmission image of the cell. Scale bars, 300 nm.
Here we showed that EYFP provides a useful photoactivatable emitter for super-resolution imaging by active control of single molecules. The fact that EYFP has not yet been fully optimized for photoswitching leaves open the possibility for future genetic engineering of a new class of photoswitchable fluorophores based on this bright, monomeric, single-molecule emitter. Attaining a sufficiently high labeling concentration for super-resolution imaging in live cells is a challenge. By taking advantage of the potential for EYFP molecules to be reactivated multiple times and of the natural dynamics MreB treadmilling in C. crescentus with time-lapse imaging, we obtained super-resolution (≤40 nm) PALM images of the MreB filaments in C. crescentus at different stages of the cell cycle. We also identified issues that must be considered in a general imaging application. First, can the concentration of fluorescent protein fusions be made high enough to fully report the structure of interest? This must be evaluated in each case, and smaller, less-perturbative small-molecule labels may be necessary. Second, in live cells, one should always carefully tune the time scales of imaging, time-lapse and reactivation to sample appropriate time slices of a possibly dynamic cellular structure.
Supplementary Material
Note: Supplementary information is available on the Nature Methods website.
ACKNOWLEDGMENTS
This work was supported in part by the US National Institutes of Health grants 1P20HG003638 (Roadmap for Medical Research), 1R01GM085437 (W.E.M.), R01GM051426 (L.S.) and F32GM080008 (postdoctoral fellowship, G.R.B), and a National Science Foundation Graduate Research Fellowship (M.A.T.).
Footnotes
Published online at http://www.nature.com/naturemethods/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Moerner WE. Nat. Methods. 2006;3:781–782. doi: 10.1038/nmeth1006-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betzig E. Opt. Lett. 1995;20:237–239. doi: 10.1364/ol.20.000237. [DOI] [PubMed] [Google Scholar]
- 3.Betzig E, et al. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 4.Rust MJ, Bates M, Zhuang X. Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess ST, Girirajan TPK, Mason MD. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler C, et al. Appl. Phys. A. 2007;88:223–226. [Google Scholar]
- 7.Niu L, Yu P. Biophys. J. 2008;95:2009–2016. doi: 10.1529/biophysj.108.128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 9.Figge RM, Divakaruni AV, Gober JW. Mol. Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 10.Carballido-López R, Errington J. Dev. Cell. 2003;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- 11.Gitai Z, Dye N, Shapiro L. Proc. Natl. Acad. Sci. USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Proc. Natl. Acad. Sci. USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thanbichler M, Shapiro L. Nat. Rev. Microbiol. 2008;6:28–40. doi: 10.1038/nrmicro1795. [DOI] [PubMed] [Google Scholar]
- 14.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Nat. Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Methods website.