Abstract
Bacteriophage P2 encodes a scaffolding protein, gpO, which is required for correct assembly of P2 procapsids from the gpN major capsid protein. The 284 residue gpO protein also acts as a protease, cleaving itself into an N-terminal fragment, O*, that remains in the capsid following maturation. In addition, gpO is presumed to act as the maturation protease for gpN, which is N-terminally processed to N*, accompanied by DNA packaging and capsid expansion. The protease activity of gpO resides in the N-terminal half of the protein. We show that gpO is a classical serine protease, with a catalytic triad comprised of Asp 19, His 48 and Ser 107. The C-terminal 90 amino acids of gpO are required and sufficient for capsid assembly. This fragment contains a long α-helical segment between residues 197 and 257 and exists as a multimer in solution, suggesting that oligomerization is required for scaffolding activity. Correct assembly requires the C-terminal cysteine residue, which is most likely involved in transient gpN interactions. Our results suggest a model for gpO scaffolding action in which the N-terminal half of gpO binds strongly to gpN, while oligomerization of the C-terminal α-helical domain of gpO and transient interactions between Cys 284 and gpN lead to capsid assembly.
Keywords: virus, structure, morphogenesis, capsid, maturation, chaperone
Introduction
Most of the known double-stranded (ds) DNA bacteriophages use scaffolding proteins for the assembly of their capsids. These proteins act as chaperones for the assembly process, but are degraded or removed from the head during capsid maturation and DNA packaging. Some scaffolding proteins act as direct catalysts of the assembly process, while others serve more to prevent incorrect interactions, or affect the assembly kinetics (Dokland, 1999; Fane and Prevelige, 2003). The scaffolding protein gene typically resides in the same operon as the major capsid protein gene, immediately preceding it in the phage genome. Many bacteriophages also encode a protease that is involved in processing the structural proteins during capsid maturation. The protease gene usually precedes the scaffolding protein gene in the genome (Hendrix, 2003).
Bacteriophage P2 forms a 60 nm diameter, T=7 icosahedral capsid from 415 copies of a 36.7 kDa capsid protein (N*) derived from gene product N (gpN) (Dokland et al., 1992; Lindqvist et al., 1993). Assembly of the 55 nm P2 procapsid is dependent on the 31.7 kDa scaffolding protein gpO, which also acts as a protease during capsid maturation (Chang et al., 2008; Lengyel et al., 1973; Wang et al., 2006). Formation of viable procapsids in addition requires the assembly of 12 copies of the portal protein gpQ into a ring-like connector at a unique vertex of the capsid, through which the DNA is packaged and to which the tail is subsequently attached (Doan and Dokland, 2007; Rishovd et al., 1994). Following assembly, gpO is cleaved autoproteolytically between residues 141 and 142 into a 15.5 kDa N-terminal fragment, O*, that remains in the mature capsid (Chang et al., 2008; Lengyel et al., 1973; Rishovd and Lindqvist, 1992). Capsid maturation also involves processing of gpN into N* through the removal of the N-terminal 31 residues (Rishovd and Lindqvist, 1992). In the presence of the genetically unrelated bacteriophage P4, 235 copies of gpN-derived protein are assembled into a smaller, 45 nm capsid with T=4 symmetry (Dokland et al., 1992; Lindqvist et al., 1993). The size of the P4 capsid is determined by the P4-encoded Sid protein, which forms an external scaffold around the P4 procapsids (Barrett et al., 1976; Marvik et al., 1995). The volume of the P4 capsid only permits encapsidation of the smaller P4 genome.
Full-length gpO undergoes autoproteolytic degradation upon expression in E. coli; however, truncation of the first 25 resides from the N-terminus of the protein abolishes the proteolytic activity (Wang et al., 2006), suggesting that one of the active site residues for the protease activity resides within these 25 residues. The truncated protein is capable of catalyzing the formation of P2 procapsid-like particles upon co-expression with gpN. Additionally, a C-terminal fragment that includes only residues 195–284 is able to promote the formation of correctly formed P2 procapsids (Chang et al., 2008). This fragment includes a long predicted αhelical sequence between residues 197 and 257 (Fig. 1). Such an α-helical nature is similar to the scaffolding proteins of other viruses such as P22 and φ29 (Morais et al., 2003; Sun et al., 2000).
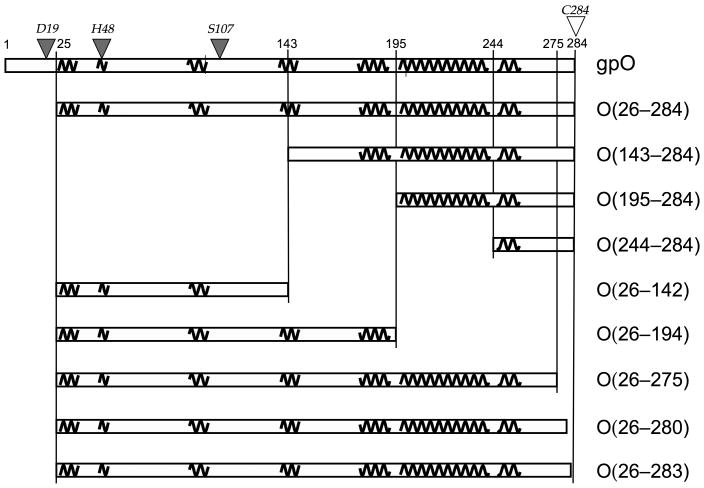
Figure 1.
Design of gpO truncation constructs. The sequence of gpO from 1–284 is represented by the rectangular box at the top. Pertinent residue numbers are shown. The predicted α-helical structures are shown as squiggly lines. Selected gpO truncation constructs that were made and the segments of α-helical structure included in each are shown in the boxes below. The filled triangles on top of the sequence indicate the residues that were shown by point mutations to correspond to the protease active site, while the C-terminal Cys residue that was shown by mutation to be essential for assembly is shown as an open triangle.
In this paper, the functional domain organization of gpO is further characterized for its protease activity and its ability to bind gpN and promote capsid formation. We have identified the residues that comprise the catalytic triad of the gpO proteolytic activity, consistent with a classic serine protease residing in the N-terminal half of the protein. Although the protease domain itself binds strongly to the gpN capsid protein, correct assembly depends on the α-helical domain within the C-terminal 90 amino acids of gpO. The C-terminal Cys residue is essential for scaffolding activity and is most likely involved in transient gpO–gpN interactions. Our results suggest a model for gpO action in which the N-terminal protease domain binds strongly to gpN, but that assembly depends on oligomerization of the C-terminal α-helical domain and transient interaction with gpN. Autoproteolytic cleavage succeeds assembly and is probably required to inactivate the C-terminal scaffolding domain.
Results
Domain organization of gpO
The 284 residue gpO protein serves two functions in the in the P2 assembly process: scaffolding and proteolytic cleavage. The scaffolding activity resides in the C-terminal part of the protein, while the protease activity is found in the N-terminal half (Chang et al., 2008; Wang et al., 2006). We previously found that residues 195–284 of gpO were sufficient for capsid formation (Chang et al., 2008). This sequence includes a region of predicted α-helical structure between residues 197 and 257, which is divided into a long predicted α-helix from 197–240 and a shorter segment from 247–257, separated by a six residue loop (Fig. 1). Such an α-helical nature is common to most scaffolding proteins (Dokland, 1999; Fane and Prevelige, 2003). The scaffolding proteins of both bacteriophages P22 and φ29 contain helix-loop-helix (HLH) motifs, which were found to be essential for assembly (Fu et al., 2007; Morais et al., 2003; Parker et al., 1998; Sun et al., 2000). By analogy, the C-terminal α-helical region of gpO might contain a similar HLH motif.
To further dissect the scaffolding activity of gpO, a set of clones expressing various truncated forms of gpO alone or together with gpN was generated, as shown in Fig. 1. Constructs O(26–284), O(143–284) and O(195–284) were previously described: O(143–284) represents the entire C-terminal sequence that is proteolytically removed from gpO during capsid maturation, while O(195–284) includes the predicted α-helical region from amino acid 197 to 257 that appears to be crucial for capsid formation (Fig. 1) (Chang et al., 2008). The clone O(244–284) includes only the C-terminal 41 residues of gpO (residues 244 to 284), thereby removing the long α-helix between residues 197 and 240 (Fig. 1). The respective co-expression clones that express the truncated gpO protein together with gpN are denoted as O(26–284)+N, O(143–284)+N, O(195–284)+N and O(244–284)+N.
In addition, a series of gpO constructs with truncations from the C-terminus were made, including O(26–142), O(26–194), O(26–275), O(26–280) and O(26–283) (Fig. 1). The clones start at residue 26, as we had previously determined that removal of the first 25 amino acids abolished the proteolytic activity of gpO without an adverse effect on assembly. All gpO truncations were subsequently cloned into co-expression constructs with gpN and denoted as O(26–142)+N etc. (Fig. 1).
Other constructs were made with point mutations in residues found to be relevant for either scaffolding or protease activity, as described in further detail below. These point mutations are indicated by triangles over the sequence in Fig. 1.
The C-terminal 90 residues of gpO are required and sufficient for assembly
All gpN-expressing clones produce capsid-related particles. These particles were harvested by centrifugation at 100,000×g, followed by separation on 10–40% sucrose gradients and analysis by SDS-PAGE to assay for capsid formation and gpO incorporation. Capsid-related structures sediment near the middle of the gradient under these conditions, at about 150S or more, while unbound gpO protein remains in the 100,000×g supernatant or at the top of the gradient. The capsid protein-containing fractions were pooled in order to capture all capsid-related structures in the sample, concentrated by centrifugation and analyzed by negative stain electron microscopy (EM) and cryo-EM as previously described (Dokland and Ng, 2006). The particles in the micrographs were counted and classified according to whether they were aberrant in size and shape, or closed and isometric. Isometric shells generally fall into discrete size classes according to the rules of triangulation (Caspar and Klug, 1962). Most of the isometric shells produced by the gpO + gpN co-expressions fell into two size classes, namely T=7 or “P2-like” shells of 55 nm diameter and T=4 or “P4-like” shells of around 40 nm diameter. For the purpose of classification, shells of ≥46 nm in diameter were considered “P2-like”, while those ≤45 nm in diameter were classified as “P4-like”. These data are tabulated in Table 1.
Table 1.
Percentages of different types of shells found in micrographs of gpO truncations and mutation clones.
| Clone | n | Aberrant (%) | P2-like (%) | P4-like (%) |
|---|---|---|---|---|
| N | 237 | 71.7 | 23.6 | 4.6 |
| O(26–284)+N | 787 | 6.7 | 92.5 | 0.8 |
| O(143–284)+N | 807 | 8.3 | 91.7 | 0.0 |
| O(195–284)+N | 762 | 13.6 | 82.3 | 4.1 |
| O(244–284)+N | 874 | 90.6 | 5.5 | 3.9 |
| O(26–142)+N | 672 | 83.4 | 8.8 | 7.8 |
| O(26–194)+N | 1551 | 95.6 | 3.6 | 0.8 |
| O(26–275)+N | 1073 | 82.0 | 5.3 | 12.7 |
| O(26–280)+N | 306 | 81.0 | 10.8 | 8.2 |
| O(26–283)+N | 143 | 45.5 | 21.7 | 32.9 |
| O(26–C284A)+N | 633 | 86.9 | 6.6 | 6.5 |
| O(26–C284S)+N | 866 | 88.8 | 7.4 | 3.8 |
n = number of particles counted. Aberrant includes both unclosed shells and closed shells of aberrant shape. For the purpose of classification, P2-like shells are defined as isometric shells with a size ≥46 nm, while P4-like shells are isometric with size ≤ 45 nm.
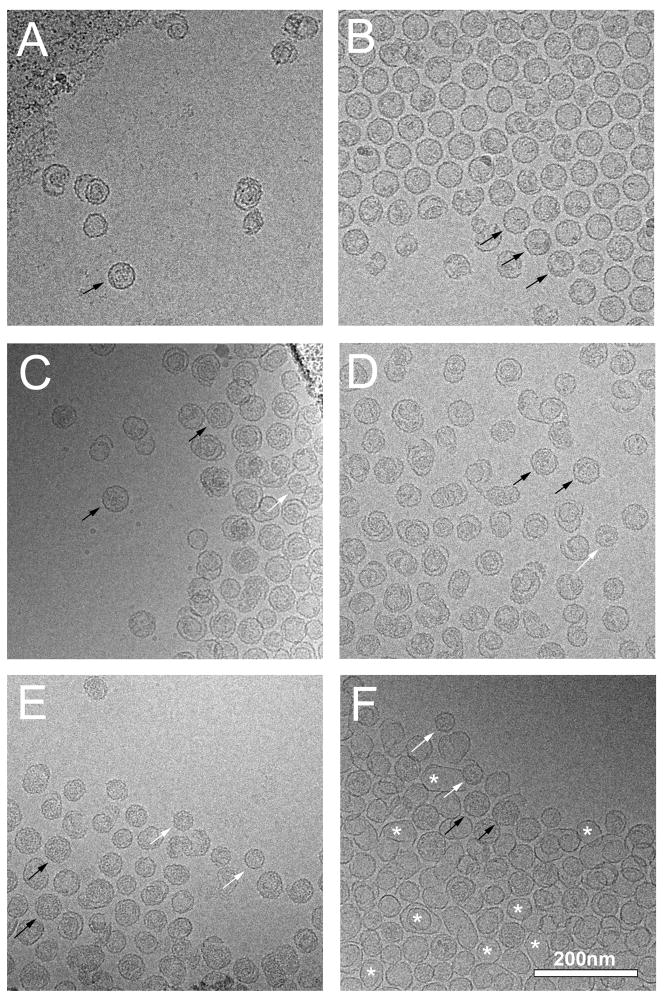
Expression of gpN alone yielded mostly aberrant shells (71.7 %), with the remainder being closed, isometric shells predominantly of P2 size (Fig. 2A). As previously described, both O(26–284) and O(142–284) promote the formation of predominantly (>90 %) morphologically correct P2 procapsids upon co-expression with gpN from clones O(26–284)+N and O(142–284)+N, respectively (Chang et al., 2008). Expression of O(195–284)+N also resulted in the formation of P2 procapsids, albeit at slightly lower efficiency (82.3 %), showing that the C-terminal α-helical domain is sufficient for assembly (Fig. 2B). Cryo-EM of the particles formed by O(195–284)+N revealed the typical rounded, thick-walled and serrated appearance of P2 procapsids, as well as a few broken, incomplete or otherwise aberrant shells (Fig. 2B). Most of these aberrant shells had a serrated outline similar to the complete procapsids. Surprisingly, the O(195–284) protein remained in the supernatant after the 100,000×g spin and therefore did not enter the gradient with gpN, suggesting that the interaction between O(195–284) and gpN is only transient (Chang et al., 2008).
Figure 2.
Cryo-EM of structures produced by expression of gpN alone (A), expression of O(195–284)+N (B), O(244–284)+N (C), O(26–194)+N (D), O(26–283)+N (E), and O(26–C284S)+N (F). Examples of well-formed particles of P2 and P4 size are indicated in each panel (where available) by black (P2) and white (P4) arrows. Some of the many thin-walled shells in (F) are indicated with asterisks. Scale bar, 200 nm.
Expression of O(244–284)+N, on the other hand, yielded predominantly (90.6%) aberrant, unclosed shells (Fig. 2C). The few closed, isometric capsids were about evenly distributed between P2- and P4-like shells (Table 1). Thus, O(244–284) produced even fewer closed, isometric shells than gpN alone, suggesting that the gpO fragment interferes with the assembly process.
These results show that the C-terminal 90 residues are sufficient for scaffolding activity and that the long α-helical segment between residues 194 and 243 is essential for assembly. Although it is still possible that the entire predicted α-helical segment is not required, truncations within this region would likely disrupt the integrity of the helix; hence, no constructs with truncations within these residues were generated.
Cys 284 is essential for correct assembly
To further test the requirements for the scaffolding activity of the C-terminal domain, a series of clones co-expressing full-length gpN and gpO truncated from the C-terminus were generated (Fig. 1). As previously shown, constructs that include the N-terminal half of gpO (residues 26–142) bind strongly to gpN and co-sediment with gpN on sucrose gradients (Chang et al., 2008). As expected, C-terminal truncations that did not include the α-helical segment between residues 197 and 257, such as O(26–194)+N (Fig. 1), did not promote the formation of procapsids upon co-expression with gpN (Fig. 2D), even though they bound strongly to gpN and co-sedimented on sucrose gradients (data not shown). This construct produced >95% aberrant shells, worse than with gpN alone. However, even constructs with short C-terminal truncations that left the entire α-helical segment intact, such as O(26–275)+N and O(26–280)+N yielded mostly aberrant shells (>80%; Table 1). In contrast, O(26–283), in which only the C-terminal Cys residue was removed, yielded far more well-formed procapsid shells (54.5%) than the shorter truncations. Assembly into procapsids was thus better from this construct than with gpN alone, though not as good as from constructs with an intact C-terminus. Furthermore, the majority of these shells were small, P4-size shells (Fig. 2E; Table 1). These results show that an intact C-terminus is essential for scaffolding activity, but that the residues immediately preceding Cys 284 also play an important role in capsid size determination.
The C-terminal cysteine residue of gpO, Cys 284, was mutated to either Ala or Ser in the O(26–284)+N construct, generating co-expression constructs O(26–C284A)+N and O(26–C284S)+N, respectively (Fig. 1). The mutant constructs were expressed and analyzed as described above. Both mutants exhibited aberrant phenotypes comparable to O(26–280)+N, producing close to 90% aberrant shells (Table 1). However, in the mutant expressions, 25–30% of the shells had a thin-walled appearance similar to that of expanded (mature) capsid shells (Fig. 2F). By comparison, O(26–283)+N and O(26–284)+N yielded only 1–3% thin-walled shells. Thus, the nature of this residue is crucial for the correct assembly of procapsid shells.
The assembly domain of gpO forms oligomers in solution
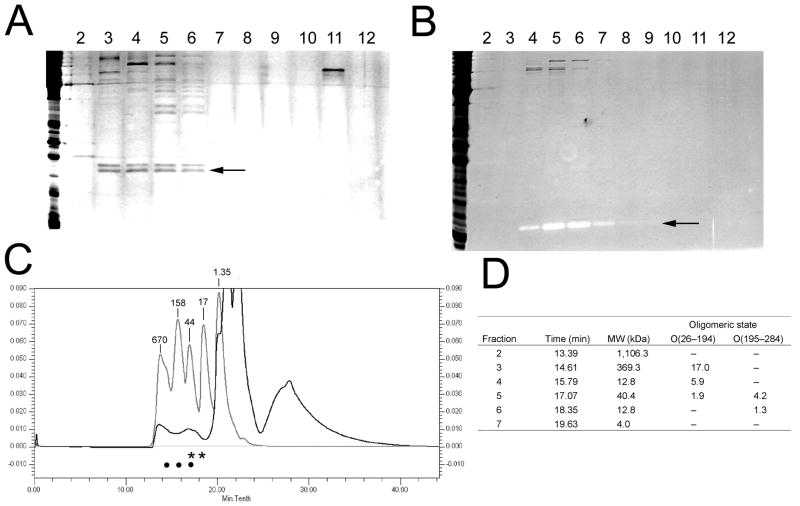
Residues 200 to 240 within the predicted α-helical region of gpO have a strong predicted propensity for coiled-coil formation, suggesting an involvement in gpO oligomerization. Such oligomerization might be conceivably involved in the capsid assembly process. To assess the role of oligomerization in gpO function, O(26–194) and O(195–284) proteins expressed in E. coli were analyzed by size exclusion chromatography (SEC). SEC analysis of O(26–194) yielded a peak broad peak of protein eluting at between 14 and 17 min (Fig. 3A, C), consistent with a mass in the range of 40 to 400 kDa. This indicates that the 21.6 kDa O(26–194) protein exists in a mixture of dimers and higher oligomers in solution (Fig. 3D).
Figure 3.
Size exclusion chromatography of gpO truncations. Silver stained SDS-PAGE of fractions from the SEC separation of O(26–194) (A) and O(195–284) (B). The fraction numbers are shown above each lane. The bands corresponding to the truncated proteins are indicated (arrows). Note the contrast reversal for O(195–284) in (B). (C) Chromatogram of O(26–194) separation (black plot) superimposed on the chromatogram of a MW standard (light gray). UV absorption at 280 nm is plotted against the time of elution. The corresponding MW for each standard peak is indicated (in kDa). The large peak beyond 20 min elution time results from detergent in the buffer. The positions of the protein peaks determined from the gels of O(26–194) and O(195–284) in (A) and (B) are shown as bullets and asterisks, respectively. (D) Table showing the oligomeric state of O(26–194) and O(195–284) corresponding to the SEC fractions in which they were contained.
The SEC analysis of O(195–284) was complicated by the lack of any aromatic residues that absorb UV light. However, silver stained SDS-PAGE gels of the fractions collected from the SEC separation showed the presence of O(195–284) in a peak with a maximum at about 17 to 18 min of elution (Fig. 3B). Curiously, O(195–284) shows up in negative contrast by silver stain (light bands). Comparison with Coomassie-stained gels confirmed that the reverse contrast bands corresponded to O(195–284) (not shown). An elution time of 17.5 min would be consistent with a mass of 27.4 kDa, corresponding to a dimer or trimer of the 9.7 kDa O(195–284) protein (Fig. 3D).
These results indicate that both the N- and C-terminal halves of gpO form oligomers in solution, consistent with previous observations that O(26–284) aggregates. Considering that O(195–284) promotes procapsid assembly, oligomerization is likely to be involved in capsid assembly.
Protease activity of gpO
We previously showed that the protease activity of gpO resides in the N-terminal half of the protein (residues 1–141, O*) and that removing the first 25 N-terminal residues of the protein abolished the protease activity (Wang et al., 2006). We originally hypothesized that a cysteine residue at position 18 (Cys 18) might constitute part of a catalytic triad (or dyad) of a cysteine protease activity. This hypothesis was tested by mutating Cys 18 in the full-length protein to either Ser or Ala, generating clones O(C18S) and O(C18A), respectively. However, neither mutation abolished the autoproteolytic activity of gpO and the expression pattern of both mutant proteins was similar to that of the wild-type protein described previously (Wang et al., 2006), i.e. there was no visible expression the mutant protein (data not shown). This demonstrated that Cys 18 was not involved in the proteolytic activity of gpO.
Cheng et al. (2004) aligned multiple sequences of the bacteriophage procapsid protease superfamily, including that of the P2 gpO protein, against the herpesvirus protease family. In the human cytomegalovirus (HCMV) protease, His 63, Ser 132, and His 157 constitutes the catalytic triad of a serine peptidase (Chen et al., 1996). Two of these residues, His 63 and Ser 132, were found to be conserved throughout the procapsid protease superfamily, strongly suggesting structural and functional homology between the proteases of herpesviruses and bacteriophages (Cheng et al., 2004). The third residue, His 157, is distinctive to the catalytic triad of HCMV, as either Asp or Glu normally occupy the third position in classical serine proteases (Chen et al., 1996). The corresponding conserved catalytic residues in gpO, as determined by the sequence alignment, are His 48 and Ser 107, both of which reside in the N-terminal domain of the protein.
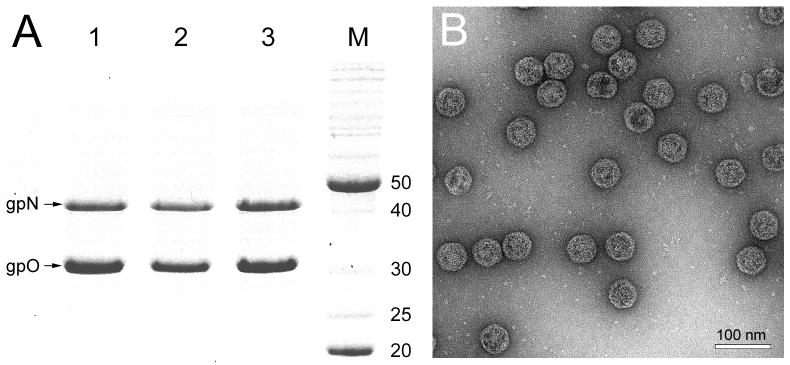
To test this assertion, His 48 and Ser 107 of gpO were individually changed to Ala in a clone expressing full-length gpO and gpN proteins in tandem. These clones were denoted as O(H48A)+N and O(S107A)+N, respectively. Co-expression of both O(H48A)+N and O(S107A)+N resulted in bands on SDS-PAGE corresponding to the expected size for full-length gpO and gpN proteins (Fig. 4A). Autoproteolytic activity was not observed in either case, demonstrating that His 48 and Ser 107 are two critical residues for protease activity. Both co-expressions led to the production of a fast-sedimenting fraction containing the mutant gpO protein and gpN, which was further purified on 10–40% sucrose gradients. The resulting particles were analyzed by electron microscopy, revealing a homogeneous population of P2 procapsid-like particles (Fig. 4B).
Figure 4.
Expression of mutant gpO. (A) Coomassie-stained SDS-PAGE showing expression of gpN and gpO from the clones O(D19A)+N (lane 1), O(H48A)+N (lane 2) and O(S107A)+N (lane 3). Lane M, marker; MW as indicated (kDa). (B) Electron micrograph of negatively stained procapsid particles produced in the O(D19A)+N co-expression. Scale bar, 100 nm.
Based on the results from expression of truncated gpO proteins described earlier (Wang et al., 2006), we suspected that the third residue of the catalytic triad resided in the N-terminal 25 residues. In a classical serine protease, the third residue is either a Glu or Asp. gpO contains three candidate residues in this region, namely Glu 14, Asp 16 and Asp 19. Each of these residues were mutated in turn to Ala in the full-length gpO protein and co-expressed with gpN. Mutations of either Glu 14 or Asp 16 failed to abolish the autoproteolytic activity of gpO, producing no visible expression. When Asp 19 was changed to Ala, however, a band corresponding to the size of the expected full-length gpO was seen on SDS-PAGE (Fig. 4A). Co-expression of O(D19A)+N resulted in the production of a homogenous population of P2 procapsid-like particles (Fig. 4B). It is worth noting that this Asp residue is completely conserved within the group of proteases that includes gpO (Cheng et al., 2004). Thus, Asp 19 most likely comprises the third member of the catalytic triad of a gpO serine protease activity.
Discussion
While many bacteriophages encode separate scaffolding and protease proteins, variations on this theme exist. In bacteriophage HK97, for example, the scaffolding function resides in an N-terminal “delta” domain of the capsid protein (Hendrix, 2005), while in phage λ, the scaffolding protein gene is embedded in-frame into the 3’ end of the C protease gene (Hendrix and Casjens, 2006). Similarly, in bacteriophage P2, the protease and scaffolding activities are fused together in the gpO protein. Thus, gpO can be divided into two functional domains: an N-terminal protease domain that comprises residues 1–141, and a C-terminal scaffolding domain (residues 195–284) that is required for P2 procapsid assembly and which is removed from the capsid after assembly has completed (Chang et al., 2008).
The N-terminal protease domain of gpO is a classical serine protease with a catalytic triad comprised of Asp 19, His 48 and Ser 107. This protease activity acts on itself to remove the C-terminal half of the protein, residues 142–284, and presumably cleaves gpN between residues 31 and 32 (Chang et al., 2008). The N-terminal portion of gpO, O*, retains its protease activity after cleavage. It binds strongly to gpN, but does not promote the formation of P2 procapsids. Such targeting of gpO may be important to sequester the strong and potentially toxic protease activity to the capsid shells.
Conversely, the C-terminal domain that includes residues 195–284 is sufficient to promote capsid assembly, but appears to bind only transiently to gpN during assembly. Its proteolytic removal during capsid maturation indicates that it is not integral to capsid formation once its obligations as a chaperone to gpN have been fulfilled. This interaction most likely involves Cys 284, which appears to be the critical residue for gpO activity. The effect of this interaction on gpN is unclear, but it is likely to involve gpO–induced conformational changes in gpN. Very small alterations in the nature of this interaction, such as those caused by changing Cys 284 to Ser, led to large perturbations in the resulting shell, including a predominance of small P4-like shells, as in O(26–283)+N, or the formation of large, thin-walled shells of abnormal shape, as in O(26–C284A)+N and O(26–C284S)+N.
Both the N- and C-terminal domains of gpO form oligomers in solution. The dependence of capsid formation on the α-helical, predicted coiled-coil region of gpO between residues 197 and 257 suggests that oligomerization is a requirement for scaffolding action. This α-helical nature and propensity for coiled-coil formation is shared with the scaffolding proteins of other phages (Dokland, 1999; Fane and Prevelige, 2003), suggesting that the mechanism of scaffolding activity may be similar within this group of viruses.
Together, these data suggest a model in which the gpO scaffolding protein is targeted to gpN by strong interactions in the N-terminal domain. Assembly then depends on oligomerization via the α-helical region (residues 197–257) that might bring the gpN subunits together (Fig. 5A,C). This oligomerization can also occur in the absence of the N-terminal domain, without which assembly is still mostly correct (Figs. 2B, 5B). The required specificity may be provided by a transient interaction between Cys 284 and gpN. In the absence of this residue, the fidelity of assembly is reduced and the size of the capsids is affected (Figs. 2E, 5D). Once assembly has completed, autoproteolytic cleavage causes removal of the C-terminal scaffolding domain, while in situ cleavage of gpN by gpO completes the maturation process, priming the procapsid for DNA packaging and expansion.
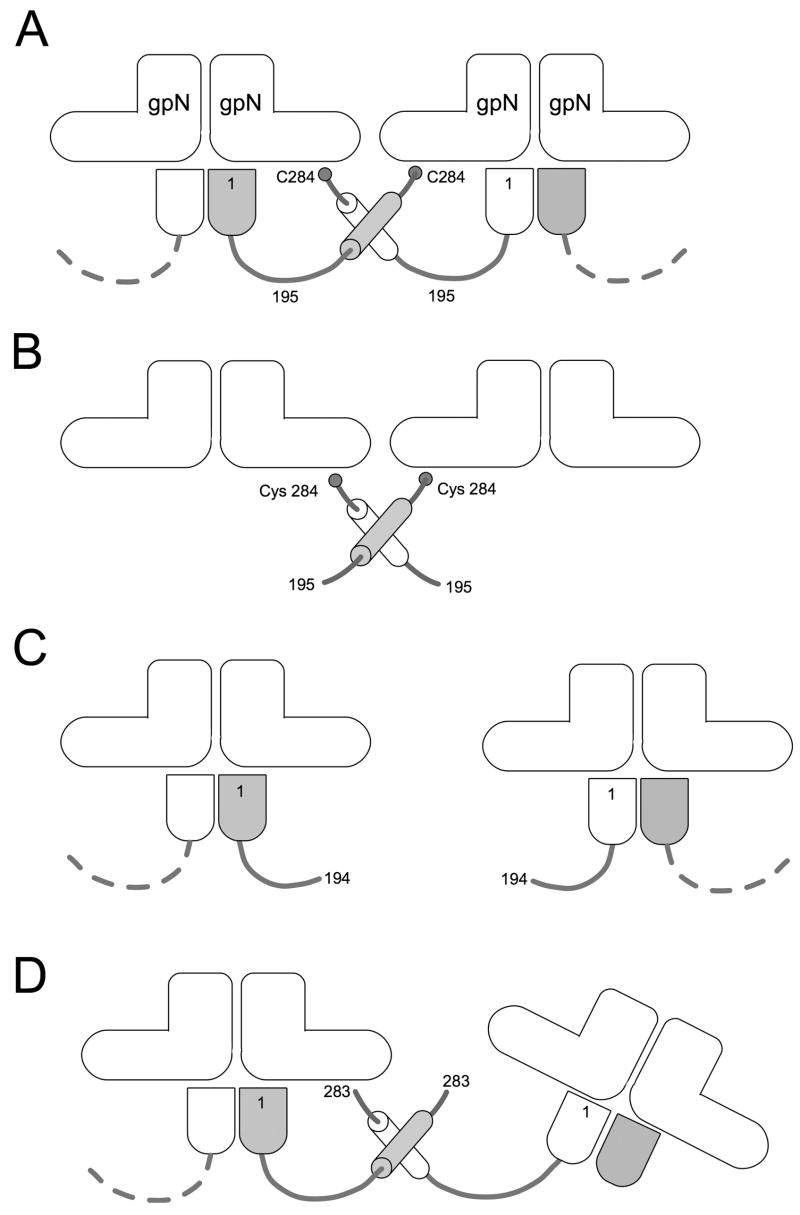
Figure 5.
Model for gpO action. The “L” shapes represent the gpN monomers. In the full-length gpO protein (A), the N-terminal protease domain (bullet shape) binds (as an oligomer) to the underside of gpN, while interactions between the α-helical domain (cylinders) and between Cys 284 and gpN lead to oligomerization and capsid assembly. In the absence of the N-terminal domain (B), O(195–284) is sufficient to promote correct assembly. O(26–194), however, lacks the required C-terminal domain, and assembly fails (C). The removal of only the C-terminal Cys 284 affects the fidelity of assembly and the size distribution of the procapsids (D). In this case, however, interactions between the α-helical domains can still occur, and about 55% of the shells formed are isometric.
One open question is whether gpO–gpN binding causes conformational changes in the gpN protein or whether its effect is mainly a matter of bringing together gpN through binding and oligomerization. It will also be interesting to identify the target residue(s) for Cys 284 on gpN and determine in which way this interaction affects gpN conformation. Indeed, it is the conformational flexibility of gpN and its ability to take multiple conformations that allows P4 and its external scaffolding protein Sid to usurp the assembly machinery of P2 and assemble small capsids with very high efficiency (Dokland et al., 2002; Lindqvist et al., 1993). One benefit of this study is that the availability of full-length protease-deficient gpO protein will now allow the investigation of other P2 assembly processes that may depend on the full-length protein, such as incorporation of the connector.
Materials and Methods
Cloning and mutagenesis
Prediction of secondary structure was done with the PROF (Ouali and King, 2000) and GOR (Garnier et al., 1996) servers, while coiled coil prediction was done with the COILS server (Lupas et al., 1991).
The truncated O, full-length O and N genes were generated by PCR from genomic P2 DNA. The O-derived sequences were inserted between the NcoI and NdeI sites of the vector pET16b (Novagen) under the control of the T7 promoter, as previously described (Chang et al., 2008; Wang et al., 2006). Tandem clones expressing truncated gpO with full-length gpN were produced by inserting the N gene, preceded by an artificial ribosome binding site, between the NdeI and XhoI sites in the appropriate O deletion construct. Cloning was performed in the E. coli recA− strain DH5α (Invitrogen). Site-directed mutagenesis of full-length O was carried out using the QuikChange Lightning Site-Directed Mutagenesis kit (Stratagene).
Expression and purification
For expression, the pET16b-derived clones were transformed into E. coli strain BL21(DE3) (Novagen). The cells were grown at 37 °C in LB with 100 μg/ml ampicillin until D600 reached 0.4–0.6, induced by the addition of 0.5 mM IPTG, and harvested after 2 hr. The harvested cells were resuspended in lysis buffer (10 mM Tris-HCl pH 7.4, 200 mM NaCl, 1 mM PMSF, 1% Triton X-100, and 0.5% deoxycholate) and frozen overnight at −20 °C. The thawed, resuspended cells were lysed by sequential passages at 1,000; 5,000; 10,000 and 15,000 psi through an Emulsiflex EF-C3 high-pressure cell disruptor (Avestin Inc., Ottawa). The lysates were clarified by centrifugation at 17,000×g for 30 min in a Beckman JA-25.50 rotor.
Procapsid purification
To collect the formed particles, the clarified supernatants were centrifuged at 40,000 rpm (114,000×g) for 1 hr in a Beckman Type 60Ti rotor. The pellets were resuspended in procapsid buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM MgCl2), loaded onto 10–40% sucrose gradients prepared in the same buffer and centrifuged for 2 h at 30,000 rpm (110,000×g) in a Beckman SW 41 rotor. 1 ml gradient fractions were collected manually. Total lysates, pellets and sucrose gradient fractions were analyzed by SDS-PAGE. Fractions were pooled and then pelleted by centrifugation for 40 min at 50,000 rpm (178,000×g) in a Beckman Type 60Ti rotor. The resulting pellet was resuspended in procapsid buffer and prepared for EM.
Size exclusion chromatography
Lysates of O(26–194) and O(195–284) were clarified by centrifugation at 40,000 rpm (114,000×g) for 1 hr in a Beckman Type 60Ti rotor and concentrated, as needed, by centrifugation at 14,000×g in a Microcon YM-2 concentrator (Millipore) to a final concentration of 1–2 mg/ml. The samples (50 or 100 μl) were loaded on a Bio-Silect SEC 250–5 column (Bio-Rad) that had been equilibrated in 0.1 M Na phosphate, pH 6.8, 150 mM NaCl, 0.01 M Na azide, 5% DMSO and run at 0.5 ml/min in the same buffer. The fractions were analyzed by SDS-PAGE and stained using the Silver Stain Plus kit (Bio-Rad).
Electron microscopy
Virus samples from the sucrose gradients were dialyzed against EM buffer (10 mM Tris-HCl, pH 8.0, 20 mM NaCl, 2 mM MgCl2) on a 0.025 μm filter for 30 min. A 3 μl sample was applied to carbon-coated grids (Electron Microscopy Sciences), rinsed 3μwith EM buffer and stained with 1 % uranyl acetate (Dokland and Ng, 2006). Cryo-EM was done by standard methods: 3μl of sample was applied to C-flat holey film (Electron Microscopy Sciences), blotted briefly before plunging into liquid ethane and transferred to a Gatan cryo-sample holder (Dokland and Ng, 2006). All samples were observed in an FEI Tecnai F20 electron microscope operated at 200 kV, and images were captured on a 4k × 4k Gatan Ultrascan CCD camera at magnifications of 38,000× or 65,500×.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrett KJ, Marsh ML, Calendar R. Interactions between a satellite bacteriophage and its helper. J Mol Biol. 1976;106:683–707. doi: 10.1016/0022-2836(76)90259-x. [DOI] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Chang JR, Poliakov A, Prevelige PE, Mobley JA, Dokland T. Incorporation of scaffolding protein gpO in bacteriophages P2 and P4. Virology. 2008;370:352–361. doi: 10.1016/j.virol.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Tsuge H, Almassy RJ, Gribskov CL, Katoh S, Vanderpool DL, Margosiak SA, Pinko C, Matthews DA, Kan CC. Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell. 1996;86:835–843. doi: 10.1016/s0092-8674(00)80157-9. [DOI] [PubMed] [Google Scholar]
- Cheng H, Shen N, Pei J, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004;13:2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan DNP, Dokland T. The gpQ portal protein of bacteriophage P2 forms dodecameric connctors in crystals. J Struct Biol. 2007;157:432–436. doi: 10.1016/j.jsb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Dokland T. Scaffolding proteins and their role in viral assembly. Cell Mol Life Sci. 1999;56:580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T, Lindqvist BH, Fuller SD. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2–P4 bacteriophage system. EMBO J. 1992;11:839–846. doi: 10.1002/j.1460-2075.1992.tb05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T, Ng ML. Electron microscopy of biological samples. In: Dokland T, Hutmacher DW, Ng ML, Schantz JT, editors. Techniques in microscopy for biomedical applications. World Scientific Press; Singapore: 2006. [Google Scholar]
- Dokland T, Wang S, Lindqvist BH. The structure of P4 procapsids produced by coexpression of capsid and external scaffolding proteins. Virology. 2002;298:224–231. doi: 10.1006/viro.2002.1485. [DOI] [PubMed] [Google Scholar]
- Fane BA, Prevelige PE. Mechanism of scaffolding-assisted viral assembly. Adv Prot Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- Fu CY, Morais MC, Battisti AJ, Rossmann MG, Prevelige PE. Molecular dissection of phi29 scaffolding protein in an in vitro assembly system. J Mol Biol. 2007;366:1161–1173. doi: 10.1016/j.jmb.2006.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J, Gibrat JF, Robson B. GOR secondary structure prediction method version IV. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage HK97: assembly of the capsid and evolutionary connections. Adv Virus Res. 2005;64:1–14. doi: 10.1016/S0065-3527(05)64001-8. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Casjens S. Bacteriophage lambda and its genetic neighborhood. In: Calendar R, editor. The bacteriophages. 2. Oxford University Press; New York: 2006. [Google Scholar]
- Lengyel JA, Goldstein RN, Marsh M, Sunshine MG, Calendar R. Bacteriophage P2 head morphogenesis: cleavage of the major capsid protein. Virology. 1973;53:1–23. doi: 10.1016/0042-6822(73)90461-3. [DOI] [PubMed] [Google Scholar]
- Lindqvist BH, Deho G, Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev. 1993;57:683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Marvik OJ, Dokland TE, Nøkling RH, Jacobsen E, Larsen T, Lindqvist BH. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J Mol Biol. 1995;251:59–75. doi: 10.1006/jmbi.1995.0416. [DOI] [PubMed] [Google Scholar]
- Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BAL, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- Ouali M, King RD. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 2000;9:1162–1176. doi: 10.1110/ps.9.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MH, Casjens S, Prevelige PE. Functional domains of bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- Rishovd S, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: protein processing and capsid size determination. Virology. 1992;187:548–554. doi: 10.1016/0042-6822(92)90457-z. [DOI] [PubMed] [Google Scholar]
- Rishovd S, Marvik OJ, Jacobsen E, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: identification and characterization of the portal protein. Virology. 1994;200:744–751. doi: 10.1006/viro.1994.1238. [DOI] [PubMed] [Google Scholar]
- Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PE, Krishna NR. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J Mol Biol. 2000;297:1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- Wang S, Chang JR, Dokland T. Assembly of bacteriophage P2 and P4 procapsids with internal scaffolding protein. Virology. 2006;348:133–140. doi: 10.1016/j.virol.2005.12.021. [DOI] [PubMed] [Google Scholar]