Abstract
Dbl-related oncoproteins are guanine nucleotide exchange factors (GEFs) specific for Rho-family GTPases and typically possess tandem Dbl (DH) and pleckstrin homology (PH) domains that act in concert to catalyze exchange. Although the exchange potential of many Dbl-family proteins is constitutively activated by truncation, the precise mechanisms of regulation for many Dbl-family proteins are unknown. Tim and Vav are distantly related Dbl-family proteins that are similarly regulated; their Dbl homology (DH) domains interact with N-terminal helices to exclude and prevent activation of Rho GTPases. Phosphorylation, substitution, or deletion of the blocking helices relieves this autoinhibition. Here we show that two other Dbl-family proteins, Ngef and Wgef, which like Tim contain a C-terminal SH3 domain, are also activated by tyrosine phosphorylation of a blocking helix. Consequently, basal autoinhibition of DH domains by direct steric exclusion using short N-terminal helices likely represents a conserved mechanism of regulation for the large family of Dbl-related proteins. N-Terminal truncation or phosphorylation of many other Dbl-family GEFs leads to their activation; similar autoinhibition mechanisms could explain some of these events. In addition, we show that the C-terminal SH3 domain binding to a polyproline region N-terminal to the DH domain of the Tim subgroup of Dbl-family proteins provides a unique mechanism of regulated autoinhibition of exchange activity that is functionally linked to the interactions between the autoinhibitory helix and the DH domain.
The activation cycle of Rho GTPases1 is tightly controlled by three families of proteins: GTPase-activating proteins (GAPs), guanine nucleotide dissociation inhibitors (GDIs), and guanine nucleotide exchange factors (GEFs). The Dbl family of proteins constitutes the largest group of GEFs specific for Rho GTPases. Dbl-family proteins are characterized by a Dbl homology (DH) domain, which contacts the Rho GTPase to catalyze nucleotide exchange by promoting and stabilizing an intermediate, nucleotide-free GTPase state, and an associated pleckstrin homology (PH) domain, which fine-tunes the exchange process. The 69 human Dbl-family proteins are divergent in regions outside the DH/PH module and contain additional protein domains that dictate unique cellular functions and regulate exchange activity (1).
Many Dbl-family proteins play a role in neuronal development. For example, Kalirin has been shown to affect neurite outgrowth and both neuronal and dendritic spine morphogenesis (2). In addition, P-rex was shown to be involved in neurotrophin-derived signaling and neuronal migration (3). FRG (also known as FARP2 or FIR) is expressed in the ventricular zone during mouse brain development and may play a role in neurogenesis, asymmetric cell division, neuronal migration, and neurite remodeling (4). Plexin A1 binds to the N-terminal FERM domain of FRG, and activation of plexin A1 by its ligand, semaphorin 3A, leads to an increase in the exchange activity of FRG and subsequent growth cone collapse (5). Finally, Tiam1 binding to the polarity protein, Par3, spatially restricts this Dbl-family GEF to dendritic spines. This restricted localization is required for dendritic spine morphogenesis (6).
Of the 69 human Dbl-family proteins, approximately one-third contain an SH3 domain in addition to the DH/PH module, and many of these SH3 domain-containing Dbl-family proteins also play a role in neuronal development. Several of these SH3 domain-containing Dbl-family proteins also contain a proline rich region, indicating that intramolecular interactions between an SH3 domain and a polyproline region may be one mechanism by which Dbl-family proteins are autoinhibited. In fact, the Rac-specific exchange activity of Kalirin, which contains an SH3 domain between its two DH/PH cassettes, is negatively regulated by an intramolecular interaction between that SH3 domain and three different polyproline-containing regions (7). However, the Dbl-family proteins Asef and Intersectin-L, which contain SH3 domains N-terminal to their DH/PH cassettes, are negatively regulated by SH3 domains directly binding to the DH domain in a manner independent of the polyproline binding site of the SH3 domain (8–10). Dbl-family GEFs, then, are able to be regulated by intramolecular interactions involving SH3 domains in both polyproline-dependent and -independent manners.
Although many Dbl-family proteins are involved in diverse facets of neuronal development, the mechanisms by which these proteins are regulated in this context are largely unknown. We have recently shown that Tim, a small Dbl-family protein, is autoinhibited by a putative helix N-terminal to the DH domain, which directly binds the DH domain to sterically exclude Rho GTPases and prevent their activation. This autoinhibition is relieved by truncation or mutation of the autoinhibitory helix or by a mutation on the conserved surface of the DH domain that disrupts interactions with the autoinhibitory helix. Src and EphA4 phosphorylate Tim on two tyrosines in the autoinhibitory helix, Tyr 19 and Tyr 22, which activates the exchange potential of Tim. Finally, a peptide comprising the autoinhibitory helix is able to inhibit a truncated version of Tim in trans (11).
Tim consists of an SH3 domain C-terminal to the DH and PH domains, and this domain architecture is shared with a small group of Dbl-family members, including Wgef, Sgef, Vsm-RhoGEF, neuroblastoma, and Ngef. In contrast to Vav and other Dbl-family members, relatively little is known about the physiological importance and molecular regulation of this subgroup. For example, Ngef is expressed primarily in the brain, is localized to chromosomal region 2q37, and is transforming in cell culture (12, 13). The mouse ortholog of Ngef, ephexin, is expressed in the central nervous system during development and was originally cloned on the basis of its ability to interact with the EphA4 receptor tyrosine kinase. Ephexin was shown to activate RhoA, Rac1, and Cdc42 in cell-based assays and mediate ephrinA-induced growth cone collapse (14). In addition, ephexin was shown to be phosphorylated by Src-family kinases downstream of EphA4 receptor activation (13). Tyrosine phosphorylation enhances the ability of ephexin to activate RhoA but does not affect its ability to activate Rac1 and Cdc42 (15). Most recently, ephexin was shown to be phosphorylated in its N-terminus by Cdk5 downstream of EphA4 receptor activation (16). The precise mechanism by which ephexin undergoes its specificity switch and the mechanisms by which Cdk5 and Src coordinate to modulate ephexin activity are poorly understood.
Here we show that Ngef, like Tim, is regulated by phosphorylation of the autoinhibitory helix. Unlike the case for ephexin, truncation, mutation, or phosphorylation of the autoinhibitory helix activates the exchange potential of Ngef toward RhoA, Rac1, and Cdc42 in vitro and in cell-based assays. In contrast to Tim, Ngef is phosphorylated by Src on only one of the tyrosines in the autoinhibitory helix, and Ngef is not phosphorylated by EphA4. In addition, we show that Ngef, like ephexin, interacts with and is phosphorylated downstream of signaling from ligated EphA4. This phosphorylation leads to an increase in the exchange activity of Ngef toward Rac1. Finally, we show that the related protein Wgef is similarly autoinhibited and activated by phosphorylation. These results highlight a common mode of regulation shared by Tim, Ngef, Wgef, and, likely, all members of the clade of Tim-related Dbl-family members. In addition, we show that intramolecular interactions between the C-terminal SH3 domain of Tim and a polyproline region immediately N-terminal to the DH domain are similarly autoinhibitory. The SH3 domain of the Tim paralog, Ngef, functions in a manner similar to that of Tim. These data demonstrate that the exchange potentials of Tim and its paralogs are negatively regulated by two distinct intramolecular interactions, and each of these interactions must be disrupted to fully activate these Dbl-family proteins.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Full-length and truncated versions of human Ngef and Wgef were PCR-amplified and ligated into pET-21a (Novagen) between NdeI and XhoI in frame with a C-terminal, noncleavable hexazhistidine tag. The cDNAs for these exchange factors were obtained as IMAGE clones (ATCC) (accession numbers BC031573 and BC040640).
Both the Ngef and Wgef constructs were expressed in Escherichia coli strain BL21(DE3) (Novagen). Cell cultures were grown at 37 °C in LB/ampicillin (100 µg/mL) and induced with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) for 5 h at 27 °C. Cell pellets were resuspended in 20 mM Tris (pH 8), 300 mM NaCl, and 10% glycerol (buffer A) with 10 mM imidazole, lysed using an Emulsiflex C5 cell homogenizer (Avestin), and clarified by centrifugation at 40000g for 45 min at 4 °C. Clarified supernatants were loaded on a nickel-charged metal chelating column (GE Healthcare) equilibrated with buffer A containing 10 mM imidazole, washed with buffer A containing 50 mM imidazole, and eluted with buffer B containing 400 mM imidazole. Eluted proteins were subsequently loaded onto an S-200 size exclusion column equilibrated with 20 mM Tris (pH 7.5), 150 mM NaCl, 2 mM DTT, 1 mM EDTA, and 5% glycerol. Fractions containing monomeric Ngef or Wgef were pooled, concentrated, and stored at −80 °C. Mutations were introduced into wild-type Ngef or Wgef using the Quikchange site-directed mutagenesis kit (Stratagene) as per the manufacturer’s instructions, and these mutant proteins were expressed and purified as described above. Mutations were also introduced into a pET-21a (Novagen) construct encoding full-length Tim or Tim (Δ22) using the Quikchange site-directed mutatgenesis kit (Stratagene) by following the manufacturer’s instructions. The PCR-amplified product for Tim (Δ57) was ligated into pET-21a between NdeI and XhoI. These mutant Tim proteins were expressed and purified as described previously (11). DNA sequences of all expression constructs were verified by automated sequencing. RhoA, Cdc42, and Rac1 were purified as described previously (17–19). The kinase domains of Src and EphA4 were also purified as described previously (11).
Guanine Nucleotide Exchange Assays
Fluorescence spectroscopic analysis of incorporation of N-methylanthraniloyl (mant)-GTP into RhoA was carried out as described previously (20). In brief, assay mixtures containing 20 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 µM mant-GTP (Molecular Probes), and 2 µM RhoA were allowed to equilibrate with continuous stirring. After equilibration, 400 nM Ngef or 50 nM Wgef was added and nucleotide loading was monitored as the decrease in the tryptophan fluorescence (λex = 295 nm; λem = 335 nm) of RhoA or an increase in mant-GTP fluorescence (λex = 360 nm; λem = 440 nm) as a function of time using a Perkin-Elmer LS 55 spectrophotometer. Rates of guanine nucleotide exchange (kobs) were determined from intrinsic tryptophan fluorescence data sets by fitting the data to a single-exponential decay model with GraphPad Prism. Fold activation represents the ratio of kobs in the presence of an exchange factor to the spontaneous nucleotide exchange rate for RhoA. Data were normalized to yield percent GDP released, and assays were performed in duplicate.
Fluorescence spectroscopic analyses of release of mant-GTP from Cdc42 and Rac1 were also carried out as described previously (20). In brief, assay mixtures containing 20 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 20 µM GTP, and 250 nM Rac1 or Cdc42 preloaded with mant-GTP were allowed to equilibrate with continuous stirring. After equilibration, 400 nM Ngef was added and mant-GDP release was monitored as the decrease in mant-GDP fluorescence (λex = 360 nm; λem = 440 nm) as a function of time. Rates of guanine nucleotide exchange (kobs) were determined from the single-exponential decay best fits as described above.
Cell Culture and Mammalian Expression Constructs
COS-7 cells were maintained in DMEM supplemented with penicillin/streptomycin and 10% fetal bovine serum (Sigma). PCR-amplified Ngef or Wgef was ligated into pcDNA 3.1 Hygro (Invitrogen) between EcoRI and XhoI such that an expressed HA tag was encoded at the N-terminus of each construct. In addition, PCR-amplified HA-tagged Ngef was also ligated into the pCIG2 vector (21), which contains a (cDNA)–IRES–eGFP complex under the control of a CMV promoter and a chicken β-actin promoter, between EcoRI and XhoI. Mutations were introduced into wild-type Ngef and Wgef as described above.
GTPase Activation Assays
Affinity purifications of RhoA, Rac1, and Cdc42 were carried out as described previously (22). In brief, the Rho binding domains (RBDs) of Pak (amino acids 70–132) and Rhotekin (amino acids 7–89) were expressed as a GST fusion proteins in BL21(DE3) cells and immobilized on glutathione-coupled Sepharose 4B beads (GE Healthcare). COS-7 cells were transiently transfected in 100 mm dishes with 12 µg of various pCIG2-Ngef constructs using LipofectAMINE 2000 (Invitrogen) according to the manufacturer’s protocol. Immediately post-transfection, these cells were serum starved in DMEM supplemented with 0.1% FBS and incubated for 16 h. Cells were washed in ice-cold PBS and lysed in lysis buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 30 mM MgCl2, 1.0% Triton X-100, and protease inhibitors]. Lysates were clarified by centrifugation at 16000 rpm for 10 min. The total protein concentration of the lysates was determined by a colorimetric assay (Bio-Rad). One milligram of clarified COS-7 lysate was incubated with 120 µg of GST-Pak-RBD beads for 1 h at 4 °C. The beads were washed three times in lysis buffer. Total and affinity-purified lysates were subjected to SDS-PAGE and Western blot analysis using anti-Cdc42 (Transduction Laboratories) monoclonal antibodies. Each experiment was performed two or three times.
Activated GEF Assays
The activated GEF assays were performed essentially as described previously (23). In brief, nucleotide-free mutants of RhoA (G17A), Rac1 (G15A), and Cdc42 (G15A) were expressed as GST fusion proteins in BL21 (DE3) cells and immobilized on glutathione-coupled Sepharose 4B beads (GE Healthcare). COS-7 cells were transiently transfected in 100 mm dishes with 12 µg of various pCIG2-Ngef constructs using LipofectAMINE 2000 (Invitrogen) according to the manufacturer’s protocol. Immediately post-transfection, these cells were serum starved in DMEM supplemented with 0.1% FBS and incubated for 16 h. Cells were washed in ice-cold PBS and lysed in lysis buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 2 mM MgCl2, 1.0% Triton X-100, and protease inhibitors]. Lysates were clarified by centrifugation at 16000 rpm for 10 min. The total protein concentration of the lysates was determined by a colorimetric assay (Bio-Rad); 0.5 mg of clarified COS-7 lysate was incubated with 120 µg of GST–GTPase beads for 1 h at 4 °C. The beads were washed three times in lysis buffer. Total and affinity-purified lysates were subjected to SDS–PAGE and Western blot analysis using anti-HA (Covance) monoclonal antibodies. Each experiment was performed two or three times.
In Vitro Kinase Assay
Thirty micrograms of purified Ngef protein was incubated with 250 ng of recombinant Src or EphA4 kinase domain in kinase buffer [100 mM Tris (pH 7.5) and 125 mM MgCl2] supplemented with 100 µM ATP 30 min at 37 °C. Samples were subjected to SDS–PAGE and immunoblot analysis using an anti-phosphotyrosine monoclonal antibody (BD Transduction Laboratories).
Phosphorylation in COS-7 Cells
COS-7 cells were trans-fected with various combinations of pCIG2-HA-Ngef and murine EphA4 (generous gift of K. Kullander, Uppsala Universitet, Uppsala, Sweden) using LipofectAMINE 2000 (Invitrogen) according to the manufacturer’s protocol. Eighteen hours post-transfection, the cells were serum starved and treated for 30 min with either 2 µg/mL ephrin-A3-Fc (R&D Systems) that had been preclustered with human anti-Fc antibody or the anti-Fc antibody itself. The cells were then lysed in 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, and 1% Triton X-100 with protease inhibitor tablets (Roche). The lysates were subjected to immunoprecipitation analysis using anti-HA-conjugated affinity matrix (Roche) or GTPase activation assays.
Analytical Gel Filtration Chromatography
One milligram of purified full-length Tim was loaded onto a calibrated Superdex S-75 column (GE Healthcare) in 100 µL of 20 mM Tris (pH 7.5), 300 mM NaCl, 2 mM DTT, 1 mM EDTA, and 5% glycerol and was eluted at a flow rate of 0.5 mL/ min.
RESULTS
The Exchange Activity of Ngef toward Its Cognate GTPases Is Regulated by Autoinhibition
We have previously shown that the Dbl-family protein, Tim, is autoinhibited by a short, putative helix N-terminal to its DH domain. Tim shares domain architecture and a high level of sequence identity with five other human Dbl-family proteins: neuroblastoma, Sgef, Vsm-RhoGEF, Wgef, and Ngef. All members of this subgroup possess a putative autoinhibitory region N-terminal to the DH domain with a high degree of sequence identity to the short autoinhibitory region mapped in Tim (Figure 1A). Consequently, it is likely that other members of this subgroup are regulated in a manner similar to that of Tim, and we tested this hypothesis with Ngef.
FIGURE 1. Ngef is regulated by autoinhibition.
(A) The N-terminal portions of Ngef and its closest homologues are highlighted in the multiple-sequence alignment. Colored bars bordering the alignment demark regions in Ngef corresponding to the autoinhibitory helix (red), an intervening region (green), and the start of the DH domain (yellow). Tyrosines 179 and 182 of Ngef are numbered. The region in Ngef and its close homologues that is strongly predicted to be helical using standard algorithms is indicated (α). The peptide inhibitor used in Figure 2C and Figure 5C was derived from the autoinhibitory helix of Tim (SQLLYQEYSDV). The position of the DH domain mutation described in Figure 5A is also indicated (asterisk). The domain architecture of Ngef and its closest homologues is shown above the alignment. (B) Full-length (wt) and truncated (Δ166) forms of Ngef that retain the conserved autoinhibitory helix modestly activate RhoA, Rac1, and Cdc42. In contrast, Ngef forms in which the autoinhibitory helix is truncated (Δ185) or mutated (Y179E) have dramatically enhanced capacities to catalyze nucleotide exchange on all three GTPases. The RhoA assays monitor the quenching of tryptophan fluorescence due to mant-GTP binding and use 400 nM GEF and 2 µM GTPase, while the Rac1 and Cdc42 assays monitor the quenching of mant-GTP fluorescence as it is released into solution and use 400 nM GEF and 250 nM GTPase. Equal amounts (5 µg) of purified proteins used in the exchange assays are shown at the bottom right following SDS–PAGE and staining with Coomassie brilliant blue.
The mouse ortholog of Ngef, ephexin, has been shown to activate RhoA, Rac1, and Cdc42 in cell-based activity assays (14), and similarly, Ngef activated these GTPases in vitro (Figure 1B and Table 1). However, this activation required deletion of the putative autoinhibitory helix, since a construct lacking this helix exhibited robust activity (Δ185) while the longer but still N-terminally truncated form of Ngef (Δ166) and the full-length version (wt) were essentially inert in RhoA, Rac1, and Cdc42 exchange assays. Furthermore, substitution of Tyr 179 with Glu (Y179E) within the putative autoinhibitory helix activated Ngef to the same extent as removing this region by truncation. The RhoA assays were performed by monitoring the quenching of fluorescence of tryptophan residues intrinsic to RhoA upon binding of the fluorescently labeled GTP, while the Rac and Cdc42 assays were performed by monitoring the quenching of fluorescence of the prebound GTP analogue as it is replaced by unlabeled GTP. These two assays involve differing concentrations of both GTPase and GEF; therefore, we cannot use this experiment to determine the GTPase on which Ngef is most efficient at exchange of guanine nucleotide. However, we do show that like Tim, Ngef is activated toward its full repertoire of Rho GTPase substrates by removal or substitution of a small, conserved segment with high helical propensity located N-terminal to its DH domain. Similar to the correlation seen with Tim, the in vitro guanine nucleotide exchange activity of Ngef also tracked with its capacity to transform NIH 3T3 cells (data not shown).
Table 1.
Activation of Ngef by Mutation of the Autoinhibitory Helixa
| RhoA |
Rac1 |
Cdc42 |
||||
|---|---|---|---|---|---|---|
| Ngef | kobs (×10−3 s−1) | fold stimulation | kobs (×10−3 s−1) | fold stimulation | kobs (×10−3 s−1) | fold stimulation |
| - | 0.51 | 1.00 | 0.01 | 1.00 | 0.13 | 1.00 |
| wt | 1.47 | 2.85 | 0.23 | 17.5 | 0.22 | 1.70 |
| Δ166 | 1.62 | 3.15 | 0.68 | 51.7 | 0.32 | 2.47 |
| Δ185 | 18.2 | 35.3 | 1.40 | 106 | 3.27 | 25.2 |
| Y179E | 16.7 | 32.5 | 1.20 | 90.9 | 4.53 | 35.0 |
An analysis of the representative exchange data presented in Figure 1 is shown. The rates of nucleotide loading (kobs) were estimated from the single-exponential decay best fits using GraphPad Prism. Stimulation is relative to the intrinsic exchange rate of each GTPase (i.e., RhoA, Rac1, or Cdc42 without added Ngef).
To confirm that the N-terminal region is necessary for Ngef inhibition in the context of an intact cell, we performed affinity purifications of active Ngef using GST-tagged versions of nucleotide-free RhoA, Rac1, and Cdc42 [RhoAG17A, Rac1 G15A, and Cdc42 G15A (Figure 2A)]. Since Dbl-family proteins bind preferentially to nucleotide-free Rho-family GTPases, these constructs are expected to affinity purify any active GEF specific for these GTPases present in a cell lysate. HA-tagged Ngef (Δ166), when expressed in serum-starved COS-7 cells, did not appreciably interact with RhoA G17A, Rac1 G15A, or Cdc42 G15A. In contrast, Ngef (Δ166+Y179E), in which a key tyrosine in the autoinhibitory helix is mutated, was affinity purified by all three of the nucleotide-free GTPases, suggesting that the region of Ngef immediately N-terminal to its DH domain, and Y179 in particular, leads to Ngef autoinhibition by preventing its interaction with its cognate GTPases. These experiments were performed in the context of Δ166 Ngef because full-length Ngef harboring the Y179E mutation expresses inconsistently in COS-7 cells.
FIGURE 2. Form of Ngef in which the Autoinhibitory Helix is Mutated is Activated in Cell-Based Assays.
(A) The activity of Ngef constructs transiently expressed in quiescent COS-7 cells was assessed by a pull-down assay using GST-tagged nucleotide-free RhoA, Rac1, or Cdc42 as the affinity purification matrix. Levels of active Ngef were determined by immunoblotting the pull-down samples. Immunoblots of the total cell lysate (2.5% input shown) show approximately equal expression of the Ngef constructs (n.s., nonspecific). The experiments were performed three times, and representative examples are shown. (B) Expression constructs for the phosphomimetic form of Ngef or vector alone were transfected into COS-7 cells, which were subsequently serum starved in DMEM supplemented with 0.1% FBS for 16 h prior to affinity purification of active Rho GTPases with the effector domain of either Rhotekin (RTK) or PAK. The levels of active and total GTPases were determined by immunoblotting. In the same experiment, expression of the HA-tagged Ngef variant was verified by immunoblotting (n.s., nonspecific). The experiments were performed three times, and representative examples are shown. (C) Ngef (Δ185) was incubated with a peptide corresponding to the N-terminal autoinhibitory region of Tim (WT, biotin-SQLLYQEYSDV-amide) or a mutant peptide (MT, biotin-SQLLEQEYSDV-amide) at room temperature for 20 min. The resulting complexes were assayed for their ability to stimulate loading of mant-GTP onto RhoA. The peptide concentrations were 100 µM.
Affinity purification of active RhoA and Rac1 from the lysates of COS-7 cells (Figure 2B) confirmed the direct correlation between the capacity of Ngef to activate RhoA and Rac1 in vitro and in cells. Transfection of Ngef (Δ166+Y179E) into COS-7 cells significantly stimulated loading of GTP onto endogenously expressed RhoA and Rac1 relative to the empty vector. We were unable to detect anincrease in levels ofCdc42 · GTP due to Ngef (Δ166+Y179E) expression (Figure 2B), indicating that either Ngef expression does not lead to Cdc42 activation in COS-7 cells or Ngef expression is associated with a transient activation of Cdc42 that we are unable to detect by affinity precipitation of steady-state levels of activated Cdc42. Another Dbl-family protein, Dbs, has been shown to be an exchange factor specific for RhoA and Cdc42 in vitro, but its expression in NIH 3T3 cells does not lead to an increase in the level of Cdc42 · GTP as determined by affinity purification assays (24). Interestingly, a peptide corresponding to the autoinhibitory helix of Tim (11) effectively inhibits both the RhoA and Cdc42-specific exchange activity of Ngef in vitro (Figure 2C and data not shown), confirming that Tim and Ngef are regulated similarly.
The Exchange Potential of Ngef Is Directly Activated by Src
Also like Tim, Ngef containing the autoinhibitory region (Δ166) was robustly phosphorylated by Src in vitro, while further truncation (Δ185) to remove the autoinhibitory region led to loss of phosphorylation by Src (data not shown). Since Tyr 179 and 182 are the only tyrosines removed by the larger truncation, these data indicate that Ngef (Δ166) is phosphorylated by Src exclusively within the autoinhibitory region at either of these tyrosines or both. Interestingly, mutation of Y179 led to a decrease in the level of Ngef phosphorylation by Src (Figure 3A), while mutation of Y182 led to a substantial increase in the level of phosphorylation of Ngef by Src. These data indicate that both Y179 and Y182 are important for the maintenance of Ngef in the autoinhibited state, but in contrast to the situation described for Tim, only Y179 is a substrate for direct phosphorylation by Src.
FIGURE 3. Phosphorylation by Src directly activates Ngef.
(A) Src specifically phosphorylates tyrosine 179 within the autoinhibitory helix of Ngef. Purified forms of Ngef were incubated with recombinant Src and ATP for 30 min prior to SDS-PAGE (bottom panel), and immunoblotting was used to assess levels of phosphorylation (top panel). Substitution of Tyr 179 abrogates phosphorylation. This image represents a composite of several gels. (B) Phosphorylation of Ngef by Src promotes the capacity of Ngef to catalyze nucleotide exchange on RhoA. Ngef (Δ166) was incubated with combinations of kinase and ATP as indicated for 30 min prior to the addition of the mixtures to exchange reaction mixtures.
Relative to its unphosphorylated form, phosphorylation of Ngef (Δ166) by Src led to an 8-fold increase in the rate of catalyzed guanine nucleotide exchange using RhoA as a substrate in vitro (Figure 3B and Table 2). This increase in the guanine nucleotide exchange rate due to Src phosphorylation is relatively modest due to the fact that only approximately 10% of the Ngef input is estimated to be phosphorylated in the in vitro kinase assay (data not shown). Using Cdc42 as the substrate, the equivalent enhancement was 3-fold (Table 2). Reminiscent of the situation with Tim, Src most likely also forms a metastable complex with Ngef, since Src in the absence of ATP was able to stimulate guanine nucleotide exchange catalyzed by Ngef (Δ166) approximately 3- and 1.5-fold for RhoA and Cdc42 substrates, respectively. These rate enhancements required both Ngef and Src since the kinase alone is incapable of stimulating exchange with either RhoA or Cdc42 (Figure 3B and Table 2). Interestingly, and in contrast to Tim, Ngef is neither directly phosphorylated nor activated by the kinase domain of EphA4 in vitro (Table 2 and data not shown).
Table 2.
Phosphorylation by Src but Not EphA4 Activates the Exchange Activity of Ngefa
| RhoA |
Cdc42 |
|||||
|---|---|---|---|---|---|---|
| Ngef | Src | ATP | kobs (×10−3s−1) | fold stimulation | kobs (×10−3s−1) | fold stimulation |
| − | − | − | 0.120 | 1.0 | 0.0377 | 1.0 |
| + | − | − | 0.164 | 1.3 | 0.114 | 3.0 |
| + | − | + | 0.198 | 1.7 | 0.147 | 3.4 |
| + | + | − | 0.466 | 3.9 | 0.184 | 4.9 |
| + | + | + | 1.21 | 10.2 | 0.298 | 8.4 |
| − | + | + | 0.112 | 0.9 | 0.0682 | 1.8 |
| RhoA |
Cdc42 |
|||||
| Ngef | EphA4 | ATP | kobs (×10−3s−1) | fold stimulation | kobs (×10−3s−1) | fold stimulation |
| + | + | − | 0.0704 | 0.6 | ND | |
| + | + | + | 0.0587 | 0.5 | ND | |
| − | + | + | 0.201 | 1.7 | ND | |
Ngef Interacts with and Is Phosphorylated Downstream of EphA4
We have established that Ngef is directly tyrosine phosphorylated by Src in the autoinhibitory helix. However, ephexin, the mouse ortholog of Ngef, is known to be tyrosine phosphorylated in a Src-dependent manner as a consequence of EphA4 stimulation. To determine if Ngef also interacts with EphA4 and is phosphorylated as a result of EphA4 stimulation, we performed co-immunoprecipitation experiments from COS-7 cells overexpressing both Ngef and EphA4 (Figure 4). Overexpression of EphA4 in COS-7 cells leads to significant autophosphorylation and activation of this receptor tyrosine kinase in the absence of exogenous ephrin A3 ligand (data not shown). This situation presumably arises due to spontaneous clustering and transactivation of over-expressed EphA4 receptors. In the same vein, Ngef is significantly tyrosine phosphorylated when coexpressed with EphA4 in either the presence or absence of exogenous ephrin A. Furthermore, Ngef and EphA4 were shown to interact without the application of ephrin A (Figure 4A).
FIGURE 4. Ngef is phosphorylated and activated upon stimulation of EphA4.
(A) Ngef (Δ166) is phosphorylated when coexpressed with EphA4 in COS-7 cells, either in the presence or in the absence of ephrin A3-Fc ligand preclustered with anti-Fc antibody (ephrinA3+Fc) as determined by immunoprecipitation of HA-Ngef and performance of immunoblot experiments using an anti-phosphotyrosine antibody (top panel). The bands resulting from phosphorylated Ngef (p-Ngef) and the IgG heavy chain (h.c.) are indicated. In addition, EphA4 interacts with Ngef as determined by immunoblotting these same immunoprecipitation reaction mixtures with an anti-EphA4 antibody (BD Transduction Laboratories; bottom panel). In the same experiment, expression of HA-Ngef and EphA4 was verified (middle panels; 2.5% input shown). (B) COS-7 cells were cotransfected with various combinations of HA-Ngef (Δ166) and EphA4 and serum starved prior to affinity purification of active Rac1. The levels of active and total GTPases were determined by immunoblotting. In the same experiment, expression of the HA-tagged Ngef variant and EphA4 was verified.
Despite the pluripotent capacity of ephexin to activate Cdc42, Rac1, and RhoA, it has been reported that EphA4-and Src-dependent tyrosine phosphorylation of ephexin results primarily in RhoA activation in vivo (15). Therefore, the capacity of EphA4 to restrict the exchange profile of Ngef by phosphorylation was also tested. Various combinations of Ngef and EphA4 were transfected into COS-7 cells prior to affinity purification of active Rac (Figure 4B). These cells were not treated with ephrin A3 since it was shown earlier that overexpressed EphA4 constitutively phosphorylates Ngef under these conditions. Transfection of EphA4 and Ngef significantly stimulated loading of GTP onto Rac1 compared to transfection of Ngef alone (Figure 4B), indicating that Ngef is able to activate Rac1 when stimulated by EphA4 in the context of an intact cell.
Wgef Is Similarly Regulated by Autoinhibition and Phos-phorylation
The studies described above establish that Ngef is regulated like Tim in that Ngef possesses a putative helical motif that is required for autoinhibition and that this autoinhibition is relieved by phosphorylation of the helical motif. To extend this mode of regulation to other Tim-related members, Wgef was studied. Wgef is expressed primarily in liver, heart, and kidney and activates RhoA, Rac1, and Cdc42 in cell-based activity assays (25) but is specific for RhoA and Cdc42 in vitro (26). Truncation of the autoin-hibitory helix of Wgef (Δ302) is activating in vitro and in affinity purification assays using nucleotide-free RhoA as the affinity matrix (Figure 5A,B). In addition, Wgef behaved like Tim and Ngef in that mutation of its autoinhibitory helix (Y295E, analogous to Y19E in Tim; Y179E in Ngef) activated a truncated form of Wgef (Δ285) containing the autoinhibitory region (Figure 5A and Table 3). Also, it was previously shown that mutation of the DH domain of Tim (S114A) designed to disrupt specifically interactions with its autoinhibitory helix was activating and similar mutation of Wgef (Δ285+S394A) was also activating. Interestingly, Wgef (Δ302+S239A) has a slower rate of exchange than Wgef (Δ302), indicating that this residue may be important for the DH domain of Wgef engaging RhoA as well as for engaging the autoinhibitory helix. The analogous mutation in Ngef (Δ166+S291A) was insoluble in E. coli, and thus, the activity of this mutant protein could not be assessed. Moreover, Wgef, like Tim and Ngef, is inhibited by a peptide derived from the autoinhibitory helix of Tim (Figure 5C). Wgef is also similar to Tim in its activation by both Src and EphA4 phosphorylation in vitro (Figure 5D and Table 3), while Ngef is only directly activated by Src (Figure 2B and Table 2). These data indicate that autoinhibition mediated by short helical motifs and autoinhibition relieved by phosphorylation are general mechanisms of regulation for Tim and related Dbl-family members.
FIGURE 5. Wgef is regulated by autoinhibition and phosphorylation.
(A) The exchange of GDP bound to RhoA and catalyzed by 50 nM Wgef was assessed using a standard fluorescence-based assay (17, 20). Equal amounts (2 µg) of purified proteins used in the exchange assays are shown at the right following SDS–PAGE and staining with Coomassie brilliant blue. (B) The activity of Wgef constructs transiently expressed in quiescent COS-7 cells was assessed by a pull-down assay using GST-tagged nucleotide-free RhoA as the affinity purification matrix. Levels of active Wgef were determined by immunoblotting the pull-down samples. Immunoblots of the total cell lysate (2.5% input shown) show approximately equal expression of the Wgef constructs. (C) Wgef (Δ302) was incubated with either a peptide corresponding to the N-terminal autoinhibitory region of Tim (WT, biotin-SQLLYQEYSDV-amide) or a mutant peptide (MT, biotin-SQLLEQEYSDV-amide) at room temperature for 20 min. The resulting complexes were assayed for their ability to stimulate loading of mant-GTP onto RhoA. The peptide concentrations were 100 µM, and the Wgef concentrations were 50 nM. (D) Phosphorylation of Wgef by Src promotes the capacity of Wgef to catalyze nucleotide exchange on RhoA. Autoinhibited Wgef (Δ285) was incubated with combinations of kinase and ATP as indicated for 30 min prior to addition of the mixtures to exchange reaction mixtures.
Table 3.
Wgef Is Regulated Like Ngef and Tima
| RhoA |
||||
|---|---|---|---|---|
| Wgef | kobs (×10−3s−1) | fold stimulation | ||
| - | 0.07 | 1.00 | ||
| Δ285 | 0.11 | 1.53 | ||
| Δ302 | 4.59 | 65.7 | ||
| Δ285+S394A | 0.43 | 6.19 | ||
| Δ302+S394A | 2.72 | 39.0 | ||
| Δ285+Y295E | 3.54 | 50.7 | ||
| RhoA |
||||
| Wgef | Src | ATP | kobs (×10−3 s−1) | fold stimulation |
| − | − | − | 0.04 | 1.00 |
| + | − | − | 0.09 | 2.06 |
| + | − | + | 0.11 | 2.52 |
| + | + | − | 0.13 | 3.12 |
| + | + | + | 0.31 | 7.36 |
| − | + | + | 0.05 | 1.14 |
| RhoA |
||||
| Wgef | EphA4 | ATP | kobs (×10−3 s−1) | fold stimulation |
| + | + | − | 0.05 | 1.11 |
| + | + | + | 0.19 | 4.47 |
| − | + | + | 0.08 | 1.86 |
Tim Is Autoinhibited by a Proline-Containing Region in Its N-Terminus
The N-terminus of Tim consists of an extended region of low complexity and no known domain structure. Truncation of the 22 N-terminal amino acids of Tim leads to a robust increase in its exchange activity in vitro (11). To determine if additional regions of the Tim N-terminus play a role in its regulation, we tested the exchange activity of Tim mutants in which the N-terminus had been further truncated. A shorter version of Tim [Δ57 (Figure 6 and Table 4)] was able to catalyze nucleotide exchange on RhoA 45% more efficiently than the previously characterized Tim (Δ22). Further N-terminal truncations destabilized Tim upon overexpression in E. coli (data not shown), most likely reflecting the loss of important secondary structural elements necessary for the integrity of the extended DH domain, which begins at residue 70 in full-length Tim. This DH domain extension is also observed in two RhoA-specificDbl-familyproteins, LARGandPDZ-RhoGEF(27,28).
FIGURE 6. Intramolecular interaction between the SH3 domain and the N-terminal polyproline region negatively regulates the exchange potential of Tim and Ngef.
(A) Multiple-sequence alignment of the polyproline regions found in Tim and related GEFs. Positions 22, 49, and 57 of human Tim are indicated above the alignment. (B) In vitro exchange assays indicate that removal (Δ57) or substitution (Δ22+P49A) of the polyproline region of Tim is activating. Purified proteins (5 µg) used in all exchange assays are shown at the right after SDS–PAGE and staining with Coomassie brilliant blue. (C) Purified Tim at an initial concentration of 166 µM elutes from an analytical gel filtration column as a single peak at a volume corresponding to a molecular mass of 60 kDa. Both the chromatogram (top panel) and SDS–PAGE analysis of the resulting fractions (bottom panel) are shown. (D) In vitro exchange assays indicate that Ngef (µ185) possesses an activated exchange potential relative to the spontaneous loading (black trace) of guanine nucleotides onto Cdc42; W649R further activates Ngef (Δ185). Equal amounts (5 µg) of purified proteins used in the exchange assays are shown in the bottom panel following SDS-PAGE and staining with Coomassie brilliant blue.
Table 4.
Activation of Tim-Related GEFs by Mutation of the N-Terminal Polyproline Region or the C-Terminal SH3 Domaina
| RhoA |
|||||
|---|---|---|---|---|---|
|
kobs (×10−3 s−1) |
fold stimulation |
||||
| Tim | mean | deviation | mean | deviation | p value |
| Δ22 | 6.97 | 0.46 | 51.5 | 3.41 | |
| Δ57 | 9.56 | 0.26 | 70.6 | 1.93 | 0.0005 |
| Δ22+P46A | 7.77 | 2.35 | 57.4 | 17.37 | 0.3000 |
| Δ22+P49A | 10.1 | 0.37 | 74.8 | 2.73 | 0.0004 |
| Cdc42 |
|||||
|
kobs (×10−3 s−1) |
fold stimulation |
||||
| Ngef | mean | deviation | mean | deviation | p value |
| Δ185 | 2.01 | 0.29 | 6.67 | 0.96 | |
| Δ185+W649R | 3.38 | 0.51 | 11.24 | 1.70 | 0.0078 |
An analysis of the exchange data presented in Figure 6 is shown. The mean and standard deviation for both kobs and fold stimulation for three independent experiments are given. p values were calculated using the pairwise t test assuming equal variances and are relative to Tim (Δ22) and Ngef (Δ185), respectively.
Primary sequence analysis (Figure 6A) indicated that the region of Tim between residues 23 and 57 contains a polyproline sequence that fits the consensus for type 1A ligands of SH3 domains, namely PXФXXPXXP, where P represents proline, X represents any amino acid, and Ф represents a hydrophobic amino acid (29). To test if the integrity of this region is important for maintenance of Tim autoinhibition, prolines within this sequence were individually mutated to alanine within the context of both full-length Tim and Tim (Δ22), and the mutated proteins were tested for an elevated capacity to activate RhoA in vitro. Tim (Δ22+P49A), similar to Tim (Δ57), was approximately 45% more efficient at catalyzing nucleotide exchange than Tim (Δ22) (Figure 6B and Table 4). P49A Tim was itself 3-fold more efficient than wild-type Tim at catalyzing nucleotide exchange (data not shown). Mutation of either proline 46 or proline 52 to alanine had no effect on the exchange potential of full-length Tim or Tim (Δ22) (Table 4 and data not shown). These data indicate that the polyproline region of Tim, proline 49 in particular, is involved in the regulation of Tim by autoinhibition.
The SH3 Domain of the Tim-Related Proteins Is Involved in Their Autoinhibition
Tim clusters with five other human Dbl-family proteins, neuroblastoma, Sgef, Wgef, Ngef, and Vsm-RhoGEF, on the basis of the high level of sequence identity among DH domains and overall domain architecture that includes an SH3 domain carboxyl-terminal to the canonical DH/PH module. While not readily evident using standard sequence alignment algorithms such as PSI-BLAST, three other members of this subgroup, namely, Ngef, Wgef, and Vsm-RhoGEF, possess a proline rich region with sequence homology to the polyproline region of Tim shown above to mediate Tim autoinhibition and engagement of its SH3 domain (Figure 6A). Because the polyproline region in the N-terminus was determined to be required for Tim autoinhibition, we wanted to test the hypothesis that the C-terminal SH3 domain and the N-terminal polyproline region are involved in an intramolecular SH3 domain–ligand interaction. Our preliminary data indicate that the isolated GST-tagged SH3 domain of Tim interacts with full-length Tim in affinity purification assays and that an untagged version of the SH3 domain is able to interact with a peptide comprised of the polyproline region of Tim (data not shown). We therefore wanted to ask if the SH3 domain of Tim was important for its autoinhibition. We were unable to assess the consequences of removal of the SH3 domain on the in vitro exchange activity of Tim, since both truncation of the SH3 domain and mutation of a conserved tryptophan (W470R) known to be critical for polyproline ligand binding destabilized Tim upon overexpression in E. coli (data not shown). In addition, expression of Tim (Δ22+W470R) in NIH 3T3 cells did not induce formation of foci relative to background levels, despite the fact that Tim (Δ22) did induce formation of foci in the same experiment (data not shown). Given these results, it seems likely that forms of Tim mutated within its SH3 domain are unstable in vivo as well as in vitro.
To assess whether the SH3 domain of Tim mediated intramolecular interactions or possibly homo-oligomerization, we performed an analytical gel filtration chromatography experiment (Figure 6C). Tim eluted as a single peak from this column at a volume corresponding to 60 kDa. These data indicate that Tim is 100% monomeric in solution. Because this experiment was performed at a Tim concentration higher than the binding affinity of a physiologically relevant intermolecular interaction (166 µM), it is likely that the SH3 domain of Tim contributes to its autoinhibition through intramolecular, rather than intermolecular, interactions.
Similar to the case of Tim, removal of the SH3 domain of Ngef renders the protein inactive in vitro (data not shown). Therefore, to test if this domain plays a role in the autoinhibition of Ngef, we introduced a point mutation (W649R) in the context of truncated, active Ngef (Δ185) that is expected to abrogate the ability of this domain to bind polyproline-containing ligands (30). The W649R mutation led to an approximately 2-fold increase in the exchange activity of Ngef (Δ185) in vitro (Figure 6D and Table 4), suggesting that an interaction between the C-terminal SH3 domain and the N-terminal polyproline region of Ngef plays a role in regulating the exchange activity of Ngef.
DISCUSSION
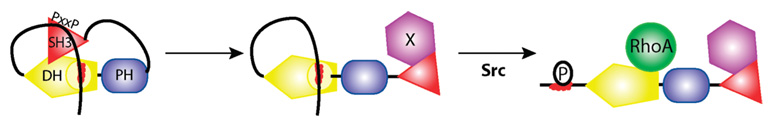
A model for the regulation of Tim and related GEFs by intramolecular and intermolecular interactions is depicted in Figure 7. In the basal state (left), the exchange potential of the GEF is autoinhibited by two sets of intramolecular interactions: (i) the autoinhibitory helix packing into a conserved pocket on the DH domain and (ii) the SH3 domain binding to the N-terminal polyproline region. These two sets of interactions are mutually cooperative and serve to tightly suppress access of Rho GTPases to the surface of the DH domain needed to catalyze guanine nucleotide exchange. Binding of another protein to the SH3 domain, with an affinity higher than that of the intramolecular interaction, would remove the SH3 domain from that polyproline region (center). This binding interaction could also localize the GEF to the appropriate subcellular compartment for nucleotide exchange. Interactions between another as yet unidentified protein and the polyproline region of the GEF would similarly serve to modulate GEF activity and localization.
FIGURE 7.
Model for the regulation of Tim and its paralogs by intramolecular and intermolecular interactions. In the basal state (left), the exchange potential of Tim is autoinhibited by two sets of intramolecular interactions. First, the autoinhibitory helix (red) packs against a conserved pocket on the DH domain. Second, the SH3 domain binds to the N-terminal polyproline region. This second set of interactions both stabilizes the autoinhibitory helix on the DH domain and potentially occludes the DH domain. Intermolecular competition by an unknown protein (purple) disrupts the intramolecular association of the SH3 domain for the polyproline region (center) to increase the rate of dissociation of the autoinhibitory helix from the DH domain. The increased solvent exposure of the autoinhibitory helix favors phosphorylation by Src, thereby preventing reassociation of the autoinhibitory helix with the DH domain and fully activating the exchange potential of Tim (right).
With interactions between the SH3 domain and the polyproline region disrupted, interaction of the autoinhibitory helix with the DH domain would be destabilized, increasing the frequency that the autoinhibitory helix is solvent-exposed. Src could then phosphorylate the tyrosine residues in the autoinhibitory helix as previously described (11), both preventing a rebinding event and fully activating the exchange potential of this Dbl-family GEF (Figure 7, right panel).
Activation of Rho GTPases downstream of transmembrane receptors such as Ephs occurs through either inactivation of GAPs or activation of GEFs. While EphB receptors are able to interact with Intersectin (31), Kalirin (2), and Vav2 (32), the best-characterized interaction between an EphA receptor and a Dbl protein is that of EphA4 and ephexin. Ephexin is tyrosine phosphorylated in an N-terminal motif with a significant degree of sequence identity to the autoinhibitory helix described here for Ngef and related Dbl-family members. Phosphorylation of ephexin in this motif is reported to change the specificity of this GEF for cognate GTPases. Specifically, when ephexin was not tyrosine phosphorylated, it activated RhoA, Rac1, and Cdc42, but when ephexin was tyrosine phosphorylated, ephexin activated RhoA exclusively (15). These results, based largely upon examination of the cellular morphology of fibroblasts transiently transfected with mutants of ephexin and EphA4, are in contrast to those shown here.
We have used in vitro guanine nucleotide exchange assays and cell-based GTPase activity assays to show mechanistically that phosphorylation or phosphomimetic mutation of Ngef leads to an activation of the exchange potential of this GEF toward all three GTPases that were studied: RhoA, Rac1, and Cdc42. This discrepancy between human Ngef and it mouse ortholog, ephexin, could be explained in several different ways. First, phosphorylated Ngef may be differentially localized in the growth cone to activate Rac1 and Cdc42 in restricted microdomains. High-resolution microscopy studies examining the intracellular localization of phosphorylated Ngef and the activated cognate GTPases need to be carried out to examine this idea. Second, ephrin A binding to EphA4 could independently activate an as yet unidentified GAP for Rac1 and Cdc42 such that the net result of this signaling pathway is a preferential activation of RhoA. Finally, activation of Rac1 and Cdc42 by Ngef could play a role in growth cone collapse. Rac1 activity has been shown to be required for growth cone collapse since it promotes internalization of the Eph-ephrin complex from the plasma membrane (32, 33). Additionally, active Rac1 and Cdc42 are critical for axon retraction, branching, and defasiculation following growth cone collapse (34). The role of Ngef in these processes has yet to be determined.
Further work is necessary to identify potential intermolecular binding partners for the SH3 domains and polyproline regions of the members of this subfamily. One potential binding partner for the polyproline region of Tim is the SH3 domain of Src itself. In this scenario, the interaction between Src and the polyproline region of Tim would relieve the autoinhibitory interactions between the SH3 domain of Src and its intramolecular ligand in the SH2 kinase linker, thus allowing the kinase domain to phosphorylate and activate Tim. This positive feedback loop would allow for rapid, specific activation of Tim.
Phenotypic diversity and complexity in biological systems often arise from new combinations of proteins and independently folding protein domains working together in networks, and not from de novo generation of new protein functions (35). In fact, multiple-sequence analysis of Dbl-family proteins from diverse animal species revealed that GEFs are composed of an N-terminal SH3 domain followed by a DH/ PH cassette, such as Asef, and GEFs are composed of a DH/PH cassette followed by an SH3 domain, such as Tim, evolved from a common ancestor composed of only a DH/PH cassette through independent insertion of the SH3 domain (36). Since the SH3 domains of both Tim and Asef function to regulate the exchange potential of these GEFs by autoinhibition, it is likely that the evolutionary pressure for the insertion of the SH3 domain was to achieve finer control of the exchange activity of these Dbl-family proteins.
The stabilization of the autoinhibitory motif on the DH domain of Tim by intramolecular interactions between the SH3 domain and polyproline regions, like the autoinhibitory motif itself, is reminiscent of the mechanism of autoinhibition of the Vav isozymes. For example, recent structural characterization of full-length Vav3 by electron microscopy showed that the CH domain of this protein stabilizes the acidic region, including the autoinhibitory helix, through intramolecular interactions with the zinc finger domain (37). For both the Tim and Vav subfamilies, then, multiple domains are functioning together to stabilize the inactive state of the catalytic DH domain. While the first set of interactions involving the autoinhibitory helix is conserved between the Tim and Vav subfamilies (11), the second set of interactions is divergent in both primary and tertiary structure. If autoinhibition by interaction of the autoinhibitory helix with the DH domain is a conserved mechanism of regulation among Dbl-family GEFs, it seems likely that the second, cooperative set of stabilizing interactions dictates the activating inputs and fine-tunes the dynamics of the guanine nucleotide exchange process. In this sense, the two sets of autoinhibitory interactions with Tim and Vav approximate a logical AND gate (38). Ultimately, full activation of these Dbl-family proteins cannot occur until both sets of autoinhibitory interactions are removed, thus allowing a large degree of spatiotemporal control over the activation of GTPases by Dbl-family proteins. We anticipate similar coordinated regulation to be found in other members of the Dbl family.
In conclusion, we have demonstrated that Ngef is activated toward its full repertoire of cognate GTPases, namely RhoA, Rac1, and Cdc42, by removal, substitution, or Src-dependent tyrosine phosphorylation of a small, conserved sequence N-terminal to its DH domain. Similarly, EphA4-dependent phosphorylation of Ngef promotes the activation of multiple Rho GTPases. Ngef exchange activity is inhibited by a peptide derived from the autoinhibitory sequence of the related protein, Tim, indicating that these two proteins are regulated in a nearly identical manner. A third member of this subfamily, Wgef, is regulated similarly. Tim, Ngef, and Wgef cluster with Sgef, Vsm-RhoGEF, and neuroblastoma on the basis of common domain architecture and sequence similarity. Given that each of these Dbl-family members shares highly conserved autoinhibitory motifs, it is highly likely that all members of this clade are regulated by steric occlusion of their DH domains by short helical motifs and that this autoinhibition is relieved physiologically by phosphorylation of these motifs. Finally, we have identified a second mechanism of autoinhibition that is likely to be conserved among the members of this clade, which is mediated by interactions between the C-terminal SH3 domain and N-terminal polyproline region and may function to increase the degree of spatiotemporal control over the activation of the Tim-related proteins.
Footnotes
J. S. is supported by the National Institutes of Health (Grants GM65533 and GM62299).
Abbreviations: GTPase, guanosine triphosphatase; Tim, transforming immortalized mammary; GEF, guanine nucleotide exchange factor; DH, Dbl homology; PH, pleckstrin homology; SH3, src homology 3; GST, glutathione S-transferase; mant-GTP, methylanthraniloyl-GTP; HEK, human embryonic kidney; DMEM, Dulbecco’s modified Eagle’s medium; FBS, fetal bovine serum; HA, hemagglutinin; Ngef, neuronalspecific GEF; Wgef, widely expressed GEF.
REFERENCES
- 1.Rossman KL, Der CJ, Sondek J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 2.Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- 3.Yoshizawa M, Kawauchi T, Sone M, Nishimura YV, Terao M, Chihama K, Nabeshima Y, Hoshino M. Involvement of a Rac activator,P-Rex1, in neurotrophin-derived signaling and neuronal migration. J. Neurosci. 2005;25:4406–4419. doi: 10.1523/JNEUROSCI.4955-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubo T, Yamashita T, Yamaguchi A, Sumimoto H, Hosokawa K, Tohyama M. A novel FERM domain including guanine nucleotide exchange factor is involved in Rac signaling and regulates neurite remodeling. J. Neurosci. 2002;22:8504–8513. doi: 10.1523/JNEUROSCI.22-19-08504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyofuku T, Yoshida J, Sugimoto T, Zhang H, Kumanogoh A, Hori M, Kikutani H. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat. Neurosci. 2005;8:1712–1719. doi: 10.1038/nn1596. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat. Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- 7.Schiller MR, Chakrabarti K, King GF, Schiller NI, Eipper BA, Maciejewski MW. Regulation of RhoGEF activity by intramolecular and intermolecular SH3 domain interactions. J. Biol. Chem. 2006;281:18774–18786. doi: 10.1074/jbc.M512482200. [DOI] [PubMed] [Google Scholar]
- 8.Zamanian JL, Kelly RB. Intersectin 1L guanine nucleotide exchange activity is regulated by adjacent src homology 3 domains that are also involved in endocytosis. Mol. Biol. Cell. 2003;14:1624–1637. doi: 10.1091/mbc.E02-08-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitin N, Betts L, Yohe ME, Der CJ, Sondek J, Rossman KL. Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression. Nat. Struct. Mol. Biol. 2007;14:814–823. doi: 10.1038/nsmb1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murayama K, Shirouzu M, Kawasaki Y, Kato-Murayama M, Hanawa-Suetsugu K, Sakamoto A, Katsura Y, Suenaga A, Toyama M, Terada T, Taiji M, Akiyama T, Yokoyama S. Crystal structure of the rac activator, asef, reveals its autoinhibitory mechanism. J. Biol. Chem. 2007;282:4238–4242. doi: 10.1074/jbc.C600234200. [DOI] [PubMed] [Google Scholar]
- 11.Yohe ME, Rossman KL, Gardner OS, Karnoub AE, Snyder JT, Gershburg S, Graves LM, Der CJ, Sondek J. Auto-inhibition of the Dbl family protein Tim by an N-terminal helical motif. J. Biol. Chem. 2007;282:13813–13823. doi: 10.1074/jbc.M700185200. [DOI] [PubMed] [Google Scholar]
- 12.Rodrigues NR, Theodosiou AM, Nesbit MA, Campbell L, Tandle AT, Saranath D, Davies KE. Characterization of Ngef, a novel member of the Dbl family of genes expressed predominantly in the caudate nucleus. Genomics. 2000;65:53–61. doi: 10.1006/geno.2000.6138. [DOI] [PubMed] [Google Scholar]
- 13.Knoll B, Drescher U. Src family kinases are involved in EphA receptor-mediated retinal axon guidance. J. Neurosci. 2004;24:6248–6257. doi: 10.1523/JNEUROSCI.0985-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shamah SM, Lin MZ, Goldberg JL, Estrach S, Sahin M, Hu L, Bazalakova M, Neve RL, Corfas G, Debant A, Greenberg ME. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell. 2001;105:233–244. doi: 10.1016/s0092-8674(01)00314-2. [DOI] [PubMed] [Google Scholar]
- 15.Sahin M, Greer PL, Lin MZ, Poucher H, Eberhart J, Schmidt S, Wright TM, Shamah SM, O’Connell S, Cowan CW, Hu L, Goldberg JL, Debant A, Corfas G, Krull CE, Greenberg ME. Eph-Dependent Tyrosine Phosphorylation of Ephexin1 Modulates Growth Cone Collapse. Neuron. 2005;46:191–204. doi: 10.1016/j.neuron.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 16.Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, Ip NY. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat. Neurosci. 2007;10:67–76. doi: 10.1038/nn1811. [DOI] [PubMed] [Google Scholar]
- 17.Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2000;408:682–688. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- 18.Rossman KL, Worthylake DK, Snyder JT, Siderovski DP, Campbell SL, Sondek J. A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 2002;21:1315–1326. doi: 10.1093/emboj/21.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, Der CJ, Sondek J. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat. Struct. Biol. 2002;9:468–475. doi: 10.1038/nsb796. [DOI] [PubMed] [Google Scholar]
- 20.Rojas RJ, Kimple RJ, Rossman KL, Siderovski DP, Sondek J. Established and emerging fluorescence-based assays for G-protein function: Ras-superfamily GTPases. Comb. Chem. High Throughput Screening. 2003;6:409–418. doi: 10.2174/138620703106298509. [DOI] [PubMed] [Google Scholar]
- 21.Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, Schuurmans C, Guillemot F, Polleux F. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 22.Solski PA, Wilder RS, Rossman KL, Sondek J, Cox AD, Campbell SL, Der CJ. Requirement for C-terminal sequences in regulation of Ect2 guanine nucleotide exchange specificity and transformation. J. Biol. Chem. 2004;279:25226–25233. doi: 10.1074/jbc.M313792200. [DOI] [PubMed] [Google Scholar]
- 23.Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J. Biol. Chem. 2002;277:42964–42972. doi: 10.1074/jbc.M207401200. [DOI] [PubMed] [Google Scholar]
- 24.Cheng L, Rossman KL, Mahon GM, Worthylake DK, Korus M, Sondek J, Whitehead IP. RhoGEF specificity mutants implicate RhoA as a target for Dbs transforming activity. Mol. Cell. Biol. 2002;22:6895–6905. doi: 10.1128/MCB.22.19.6895-6905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Suzuki H, Yokoo T, Tada-Iida K, Kihara R, Miura M, Watanabe K, Sone H, Shimano H, Toyoshima H, Yamada N. WGEF is a novel RhoGEF expressed in intestine, liver, heart, and kidney. Biochem. Biophys. Res. Commun. 2004;324:1053–1058. doi: 10.1016/j.bbrc.2004.09.153. [DOI] [PubMed] [Google Scholar]
- 26.Smith WJ, Hamel B, Yohe ME, Sondek J, Cerione RA, Snyder JT. A Cdc42 mutant specifically activated by intersectin. Biochemistry. 2005;44:13282–13290. doi: 10.1021/bi050591b. [DOI] [PubMed] [Google Scholar]
- 27.Kristelly R, Gao G, Tesmer JJ. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated RhoGEF. J. Biol. Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 28.Derewenda U, Oleksy A, Stevenson AS, Korczynska J, Dauter Z, Somlyo AP, Otlewski J, Somlyo AV, Derewenda ZS. The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca2+ sensitization pathway in smooth muscle. Structure. 2004;12:1955–1965. doi: 10.1016/j.str.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Cesareni G, Panni S, Nardelli G, Castagnoli L. Can we infer peptide recognition specificity mediated by SH3 domains? FEBS Lett. 2002;513:38–44. doi: 10.1016/s0014-5793(01)03307-5. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez-Ballester G, Blanes-Mira C, Serrano L. The tryptophan switch: Changing ligand-binding specificity from type I to type II in SH3 domains. J. Mol. Biol. 2004;335:619–629. doi: 10.1016/j.jmb.2003.10.060. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura T, Yamaguchi T, Tokunaga A, Hara A, Hamaguchi T, Kato K, Iwamatsu A, Okano H, Kaibuchi K. Role of numb in dendritic spine development with a Cdc42 GEF intersectin and EphB2. Mol. Biol. Cell. 2006;17:1273–1285. doi: 10.1091/mbc.E05-07-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowan CW, Shao YR, Sahin M, Shamah SM, Lin MZ, Greer PL, Gao S, Griffith EC, Brugge JS, Greenberg ME. Vav Family GEFs Link Activated Ephs to Endocytosis and Axon Guidance. Neuron. 2005;46:205–217. doi: 10.1016/j.neuron.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Jurney WM, Gallo G, Letourneau PC, McLoon SC. Rac1-mediated endocytosis during ephrin-A2- and semaphorin 3A-induced growth cone collapse. J. Neurosci. 2002;22:6019–6028. doi: 10.1523/JNEUROSCI.22-14-06019.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thies E, Davenport RW. Independent roles of Rho-GTPases in growth cone and axonal behavior. J. Neurobiol. 2003;54:358–369. doi: 10.1002/neu.10135. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, Motifs, and Scaffolds: The Role of Modular Interactions in the Evolution and Wiring of Cell Signaling Circuits. Annu. Rev. Biochem. 2006 doi: 10.1146/annurev.biochem.75.103004.142710. (in press) [DOI] [PubMed] [Google Scholar]
- 36.Sun QL, Zhou HJ, Lin K. The evolutionary relationship of the domain architectures in the RhoGEF-containing proteins. Genomics, Proteomics Bioinf. 2005;3:94–106. doi: 10.1016/S1672-0229(05)03014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Llorca O, Arias-Palomo E, Zugaza JL, Bustelo XR. Global conformational rearrangements during the activation of the GDP/GTP exchange factor Vav3. EMBO J. 2005;24:1330–1340. doi: 10.1038/sj.emboj.7600617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim WA. The modular logic of signaling proteins: Building allosteric switches from simple binding domains. Curr. Opin. Struct. Biol. 2002;12:61–68. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]