Abstract
Intestinal epithelial cells undergo progressive cell maturation as they migrate along the crypt-villus axis. To determine molecular signatures that define this process, proteins differentially expressed between the crypt and villus were identified by 2D-DIGE and MALDI-MS. Forty-six differentially expressed proteins were identified, several of which were validated by immunohistochemistry. Proteins upregulated in the villus were enriched for those involved in brush border assembly and lipid uptake, established features of differentiated intestinal epithelial cells. Multiple proteins involved in glycolysis were also upregulated in the villus, suggesting increased glycolysis is a feature of intestinal cell differentiation. Conversely, proteins involved in nucleotide metabolism, and protein processing and folding were increased in the crypt, consistent with functions associated with cell proliferation. Three novel paneth cell markers, AGR2, HSPA5 and RRBP1 were also identified. Notably, significant correlation was observed between overall proteomic changes and corresponding gene expression changes along the crypt-villus axis, indicating intestinal cell maturation is primarily regulated at the transcriptional level. This proteomic profiling analysis identified several novel proteins and functional processes differentially induced during intestinal cell maturation in vivo. Integration of proteomic, immunohistochemical, and parallel gene expression datasets demonstrate the coordinated manner in which intestinal cell maturation is regulated.
Keywords: Intestine, proteomics, crypt, villus
Introduction
Epithelial cells in the small intestine undergo a spatially and temporally regulated maturation program as they migrate along the crypt-villus axis1. Stem cells at the base of the crypt give rise to transient amplifying cells which then differentiate into either cells of the secretory (goblet, enteroendocrine or paneth cells) or enterocytic cell lineage. Enterocytes, goblet and enteroendocrine cells differentiate as they migrate upwards along the crypt-villus axis, while paneth cell precursors migrate downwards towards the crypt-base1. A similar renewal process occurs in the colonic epithelium with the exception that the colon is devoid of villi and paneth cells.
The signaling pathways and transcription factors that regulate small intestinal and colon cell maturation are becoming increasingly well understood. Signaling pathways implicated in the regulation of cell fate determination and lineage specific differentiation in the intestine include Wnt -β-catenin-TCF2–4, Notch and its downstream effectors HES1 and Math15, BMP-TGF-β-SMAD 6, 7 and hedgehog signaling8. A number of transcription factors, many of which are downstream targets of these signaling pathways, including cdx-1 and cdx-2, kruppel-like factor 4, GATA4, 5 and 69, and several forkhead family members2, 5, 10–12, have also been implicated in regulating components of the maturation program. Finally, E-cadherin-mediated cell-cell and integrin-mediated cell-substratum adhesion, chemotactic gradients, extracellular matrix and mesenchymal components, and a range of cytokines, hormones and growth factors, have each been implicated in the regulation of intestinal cell maturation11, 13–15.
Importantly, the signaling pathways which regulate intestinal cell maturation are the same pathways perturbed during colorectal tumorigenesis. Foremost among these is the Apc-β-catenin-TCF pathway, which is perturbed either by inactivating mutations in the Apc tumor suppressor gene or activating mutations in β-catenin in over 90% of colorectal cancers 16. Likewise, a high frequency of inactivating mutations in the TGF-βRII-SMAD4 signaling pathway, have been described 17 . Understanding the molecular events which regulate normal intestinal cell maturation may therefore provide important insights into intestinal tumorigenesis.
To dissect the complexity of the transcriptional reprogramming that regulates intestinal cell maturation, we have previously used cDNA microarray analysis to profile gene expression changes in several in vitro models of intestinal cell differentiation18–20, and more recently, by characterizing the transcriptional changes that define cell maturation along the crypt-villus axis21. These analyses demonstrated that genes with a role in cell cycle progression, RNA processing and protein synthesis were enriched among those preferentially expressed in the crypt, while genes with a role in cytoskeleton assembly, and lipid uptake and transport, were enriched among those preferentially expressed in the villus21. This analysis implicated a prominent role for c-myc in the regulation of intestinal cell differentiation as evidenced by the high percentage of myc-regulated genes among those altered in expression along the cryptvillus axis21. In the present study we extend this transcriptional profiling analysis by determining the parallel changes in the cell proteome during intestinal cell maturation in vivo.
Experimental Procedures
Fractionation of cells along the crypt-villus axis
The sequential isolation of mouse small intestinal epithelial cells along the crypt-villus axis was based on the method of Ferraris et al 22, which was a modification of the original method described for rats by Weiser et al 23, 24. Briefly, mice were sacrificed by CO2 asphyxiation and cervical dislocation. The entire small intestine (duodenum to terminal ileum) was removed and flushed once with phosphate buffered saline (PBS). The small intestine was then everted, tied off at one end, and filled to distension with PBS prior to closing the open end. The small intestine was then incubated at 37°C for 15 minutes in 15 ml of buffer B (96 mM NaCl, 1.5 mM KCl, 27 mM Na-citrate, 8 mM KH2PO4, and 5.6 mM Na2HPO4, pH 7.3), and for 10 minutes in 15 ml of buffer C (PBS plus 1.5 mM EDTA, 0.5 mM dithiothreitol, and 1 mg/ml bovine serum albumin), in a shaking 37°C incubator. At the completion of the 15-minute incubation, detached enterocytes were collected (Fraction 1) and 15 ml of fresh buffer C added to the tissue. This procedure was repeated four more times, the steps lasting 25, 25, 25 and 30 minutes, respectively (Fractions 2, 3, 4 and 5), for a total of 120 minutes of incubation time 22. Approximate cell numbers isolated at each fraction were as follows: F1 (13,480,000), F2 (33,290,000), F3 (34,980,000), F4 (35,240,000) and F5 (43,290,000). Light microscopy demonstrated that the majority of cells isolated in each fraction were intact, with a smooth rounded morphology. At the completion of the final incubation period, cells collected from each of the 5 fractions were harvested by centrifugation at 1500 rpm at 4°C for 5 minutes. Cell pellets were washed twice by resuspension and centrifugation in 15 ml of ice cold PBS, snap frozen in liquid nitrogen, and stored at −80°.
2D DIGE (2-dimensional fluorescence difference gel electrophoresis)
2D DIGE experiments were performed in duplicate with a reverse labeling approach, using protein extracts from fraction 1 (villus) and fraction 5 (crypt), from 4 different male C57Bl6 mice. All animals were 12 weeks old and had been maintained on a standard laboratory chow diet. To maximize solubility of hydrophobic proteins cells were solubilized in a lysis buffer containing 7M Urea, 2M thiourea, 4% CHAPS, and 25mM Tris, pH 8.5, and lysates centrifuged at 12,000 rpm for 4 min 25. The resulting supernatant was utilized for determination of protein concentration and labeling for 2D gel analysis. Protein concentration was determined by amino acid analysis using a Beckman Model 7300 ion-exchange instrument. For each mouse, protein samples (50 ug) from the villus (F1) and crypt (F5) were labeled with 400 pmol of Cy3 or Cy5 N-hydroxysuccinimidyl ester dyes respectively, for 30 min (GE Healthcare). For each mouse, a second gel was run in which villus and crypt fractions were reverse labeled (8 gels in total). The labeling reaction was quenched with 0.2 mM lysine and an equal volume of sample buffer containing 7M Urea, 2M thiourea, 4% CHAPS, 2% DTT and 2.0 % Pharmalyte (GE Healthcare). An internal standard was generated by pooling an aliquot of all samples which was then labeled with cy2 in a similar fashion. The three differently labeled samples (crypt, villus and internal standard) were mixed together and an additional 50µg of unlabeled protein from each sample added to increase the amount of protein available for identification.
Rehydration of the IPG strip (non-linear immobiline DryStrip pH 3–10, 24cm) with protein samples was performed in rehydration buffer containing 8M Urea, 4% CHAPS, bromophenol blue, 1% Phamalyte, and 2 mg/ml DTT using the IPGphor IEF system (GE Healthcare). Rehydration was carried out at 20°C for 12 hours followed by isoelectric focusing for a total of 120 kVh at 20 °C.
The gel strips were then equilibrated for 2 × 15 minutes with gentle shaking in equilibration solution containing 50mM Tris-Cl buffer, 6M urea, 30% glycerol, 2% SDS, and bromophenol blue. DTT (1% w/v) was added to the first, and iodoacetamide (2.5 % w/v) to the second equilibration step. SDS polyacrylamide gel electrophoresis in the second dimension was carried out using homogenous 12.5% polyacrylamide gels and the Ettan Dalt II large vertical system (GE Healthcare).
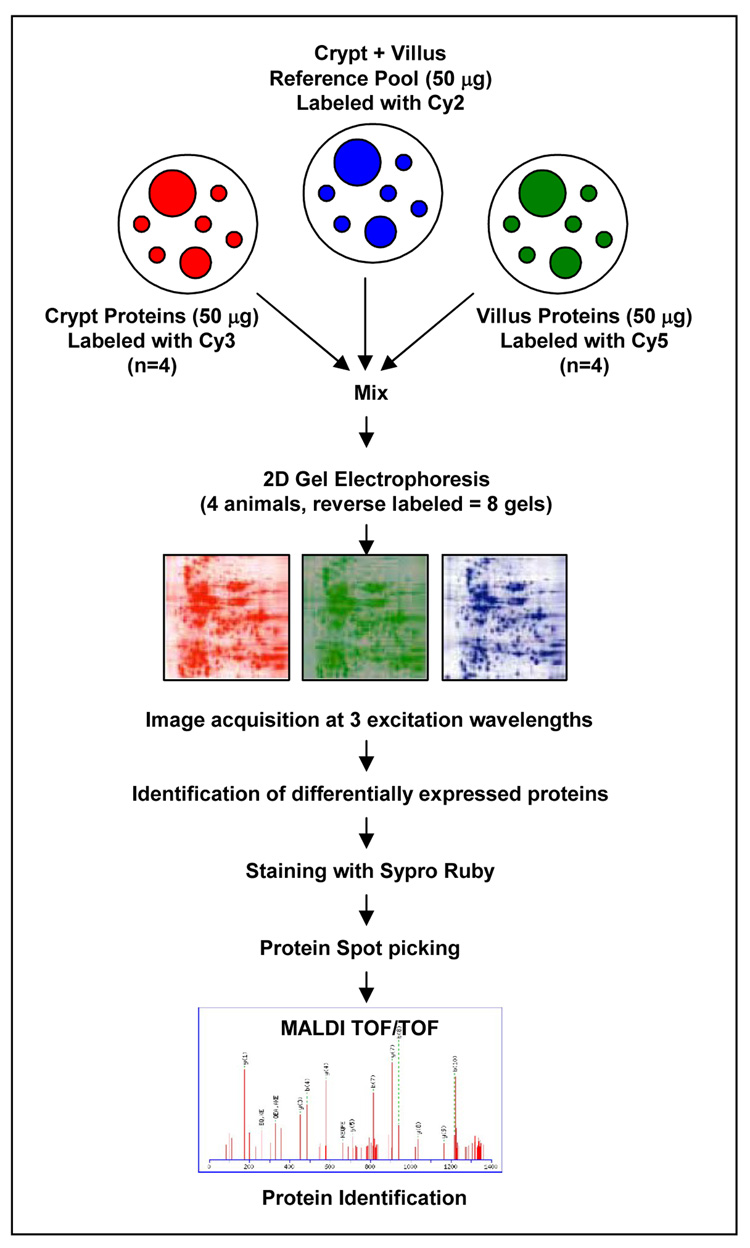
Labeled proteins were visualized using the Typhoon 9410 imager (GE Healthcare). The fluorescence images were individually imaged using excitation/emission wavelengths of 488/520 nm, 532/580 nm, and 633/670 nm for cy2, cy3, and cy5 respectively. DeCyder software v.5 (GE Healthcare) was used for image analysis including spot detection, background subtraction, matching, and normalization. For each spot, the average value from the 2 reverse labeled gels run were computed, log transformed, and used for subsequent statistical analyses. Differentially expressed proteins were identified as those with a statistically significant P value based upon a paired t test, and which had a mean volume ratio cutoff of >1.5. Prior to excision of the proteins of interest, the gel was fixed in 10% methanol and 7% acetic acid for 10 minutes, followed by SYPRO Ruby staining overnight. An overview of the procedure is provided in Figure 1.
Figure 1.
Schematic representation of proteomic analysis procedure as described in the materials and methods. A detailed description is provided in the materials and methods section.
Proteins of interest were excised using an Ettan Spot Picker instrument (GE Healthcare) and gel pieces were digested using sequencing grade trypsin (Promega Corp., Madison, WI) on the Ettan TA Digeste (GE Healthcare). Digested peptides with matrix (50% acetonitrile, 0.1% TFA containing 3 mg/ml alpha-cyano-4-hydroxy cinnamic acid matrix) were then spotted onto a MALDI-MS target on a Micromass MassPrep Sample Handler. Samples were analyzed with a Model 4700 Proteomics Analyzer (Applied Biosystems) using software 4000 series explorer v.3.0, based on a combination of Peptide Mass Fingerprinting and MALDI-TOF/TOF MS/MS spectra. MS spectra were acquired in reflectron positive ion mode. 1 fmol bradykinin, which has a protonated, monoisotopic mass of 1060.5692, and 2 fmol ACTH clip 18–39, which has a protonated, monoisotopic mass of 2465.1989, were used as internal calibrants. MS/MS spectra were acquired for the 10 most abundant ions present in each MS spectrum excluding known contaminant peaks such as keratin and trypsin autolysis peaks by averaging 10,000 laser shots per MS/MS spectrum. Protein identification was determined by searching NCBInr 20051029 database containing 2972605 sequences and 1023918613 residues using MASCOT v.2.0.05 26 via the search program GPS EXPLORER v.3.0 (Applied Biosystems). Searches were performed with carbamidomethylation of cysteine, with partial oxidation of methionine, with one missed cleavage allowed, and with mass tolerance of 50 ppm and 0.2 Da for MS and MS/MS, respectively. A probability-based protein score of >77 (p<0.05), based on combined mass and mass/mass spectra, was considered significant. Two proteins with a probability score >60 but <77 were identified by manual inspection of MS/MS spectra as previously described 27. Redundant proteins which appeared in the databases under different names and or accession numbers were eliminated following manual inspection of the sequences.
Enrichment and Pathway analysis
To determine whether proteins differentially expressed along the crypt-villus axis were enriched for specific biological processes, proteins were categorized according to their biological function and the percentage of proteins classified into various biological processes computed using the Panther data analysis tool (www.panther.org). For comparison, we repeated the analysis using an equal number of randomly selected proteins. During the course of this study we became aware of the limitation of DIGE in identifying low abundance proteins. Therefore, the 42 “random” proteins were selected from the highest intensity quartile from our corresponding cDNA microarray database of genes expressed in the mouse small intestine, as an unbiased means of selecting high abundance proteins for comparison. Enrichment of biological categories between the experimental and random group were identified using a Fisher Exact test. To discover transcription factors putatively involved in the regulation of proteins differentially expressed between the crypt and villus, we utilized the Metacore transcription regulation algorithm in the MetaCore analysis tool (GeneGo Inc). This is a manually curated interactions database for over 90% of human proteins of known function.
Western blot analysis and immunohistochemistry
Western blot analysis and immunohistochemistry were performed as described 28. Briefly, small intestinal tissues from 3 month old C57BL/6 mice were fixed overnight in 10% neutral buffered formalin and embedded in paraffin. After dewaxing and hydration, 5µm sections were pretreated with 3% H2O2 for 10 min at room temperature, and antigen retrieval was achieved by boiling in 10mM citrate buffer pH 6.0, for 30 min. All primary antibodies were diluted as recommended by the manufacturer, incubated with intestinal sections overnight at 4°C, following by detection using either Vectastain® ABC kit (Vector Laboratories, Burlingame, CA) and 3′,3′-diaminobenzidine hydrochloride as a peroxidase chromogen (Sigma, St Louis, MO) or FITC-conjugated secondary antibodies (Santa Cruz Biotechnology). The specificity of the reaction was tested by omitting the primary antibody. Nuclei were counterstained with either Mayer’s hematoxylin and/or DAPI. Anti-LFABP was a kind gift from Dr. Jeffrey Gordon 29. Anti-AGR2 was obtained from Imgenex (San Diego, CA), anti-cytokeratin 20 and anti-Ki67 were obtained from DakoCytomation (Carpinteria, CA), anti-SOX-9 was obtained from Chemicon (Temecula, CA), and anti-PCNA was obtained from Santa Cruz (Santa Cruz, CA).
For validation studies we also utilized immunohistochemical images generated by the Swedish Human Protein Atlas (HPA) database, established to enable the systematic exploration of the human proteome using Antibody-Based Proteomics (www.proteinatlas.org) 30, 31. Proteins identified in the present study which had been independently analyzed by immunohistochemistry by this consortium were downloaded, and areas of interest selected for presentation in accordance with the image use policy of protein atlas.
Measurement of Lactate and Pyruvate
Levels of lactate and pyruvate were measured in total cell lysates from cells isolated from the villus or crypt using lactate and pyruvate assay kits (Biovision, Mountain View, CA), respectively.
Computation of Codon Adaptation Index (CAI)
The codon adaptation index (CAI) was calculated as previously described by Sharp and Li 32, 33 using the Evolving code CAI calculator tool (http://www.evolvingcode.net/codon/cai/cais.php).
Microarray analysis
Changes in glycolytic gene expression were extracted from a cDNA microarray database in which gene expression changes following butyrate treatment of Caco-2 cells was determined using 27,000 feature cDNA microarrays generated by the AECOM microarray facility. Caco-2 cells were treated with 5 mM butyrate for 72 hours. RNA extraction and microarray analyses were performed as previously described 18.
Results
Validation of fractionation procedure
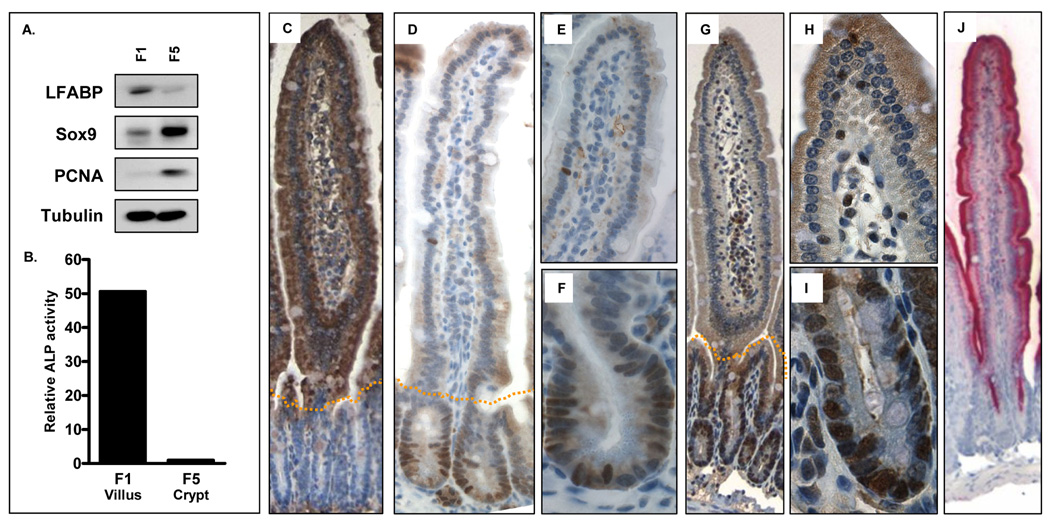
Intestinal epithelial cells that line the crypt-villus axis can be sequentially isolated beginning at the villus tip and culminating in the intestinal crypts by incubation in EDTA-buffer for progressive time periods 21, 24. While we have previously utilized a 10 cell fractionation protocol for this purpose 21, we reduced the number of fractions collected to 5 in the present study in order to maximize the number of cells available for subsequent proteomic analysis. To validate the fractionation efficiency of this modified protocol, activity or protein expression of the intestinal cell maturation markers, alkaline phosphatase and LFABP, and the proliferation markers SOX9 and PCNA, were assessed in villus (F1) and crypt (F5) fractions (Figure 2). Consistent with their immunohistochemical localization, activity and expression of the differentiation markers alkaline phosphatase and LFABP were maximal in the villus fraction. Conversely, expression of the proliferation markers SOX9 and PCNA was maximal in fraction 5, consistent with their immunohistochemical localization to the crypt base (Figure 2). These findings demonstrate efficient fractionation of differentiated villus and proliferating crypt cells using this method.
Figure 2.
Validation of cell fractionation efficiency. Cells were sequentially fractionated along the crypt-villus axis in mouse small intestine and fractionation efficiency determined by measurement of (A) LFABP, SOX9 and PCNA expression by western blot, and (B) Alkaline phosphatase (ALP) activity in villus (F1) and crypt (F5) fractions. Panels C–J show corresponding immunohistochemical staining in the jejunum of (C) LFABP, (D–F) Sox-9, (G–I) PCNA and (J) alkaline phosphatase activity.
Changes in the proteome during intestinal cell maturation along the crypt-villus axis
Proteins differentially expressed between the villus (F1) and crypt (F5), were identified by fluorescence labeling followed by 2D-fluorescence difference gel electrophoresis (Figure 1). Of a total of 932 spots analyzed, 123 (~13%) were differentially expressed (P<0.05, >1.5-fold, n=4) between the crypt and villus. Of these, 91 were successfully sequenced by analysis of a combination of Peptide Mass Fingerprinting and MALDI-TOF/TOF MS/MS spectra. Of these, 46 unique proteins differentially expressed between the crypt and villus were identified, with the remaining spots being multiple identifications of the same protein. Of these 46 proteins, 25 were upregulated in the villus and 17 were downregulated in the villus, while for 4 proteins contradictory findings were reproducibly observed depending upon the specific spot examined (Table 1) possibly reflecting different post-translational modification of the same protein. Notably, two of the 4 proteins showing contradictory findings belonged to the cytokeratin family (keratin 8 and keratin 19), and it is noteworthy that a previous proteomic analysis comparing colon tumor with adjacent normal mucosa also reported both up and downregulation of keratin 8 34. Representative spots of proteins up and downregulated in the crypt are shown in Figure 3.
Table 1.
Proteins identified by 2D-DIGE as being differentially expressed along the crypt-villus axis in mouse small intestine. Values shown are the mean fold change computed from the 8 gels. Score (Probability-based mascot score) and Coverage (Sequence percentage coverage which peptides matched to) were calculated from mascot based on combined mass and mass/mass spectra.
| Accession no. | Proteins Upregulated in the villus | Symbol | Biological Process | Fold change Villus/Crypt) | P Value | Score | # of peptides matched* | Coverage (%) |
|---|---|---|---|---|---|---|---|---|
| 56206029 | profilin 1a | PFN1 | Ctoskeletal org. biogenesis | 2 | 0.01 | 165 | 5,2 | 41 |
| 34980904 | keratin 20 | KRT20 | Cell structure | 1.84 | 0.00042 | 219 | 22,2 | 61 |
| 6678573 | villin 1 | VIL1 | Cell structure | 1.56 | 0.036 | 94 | 17,2 | 17 |
| 50881 | Ezrin | VIL2 | Cell structure | 1.61 | 0.0081 | 572 | 51,5 | 53 |
| 1703127 | actin, cytoplasmic | ACTB | Cell structure; Exo/Endocytosis;Cytokinesis; | 1.71 | 0.027 | 359 | 22,5 | 44 |
| 7548322 | aldolase 1, A isoformb | ALDOA | Glycolysis | 1.65 | 0.0078 | 91 | 16,1 | 45 |
| 21707669 | aldolase 2, B isoform | ALDOB | Glycolysis | 2.25 | 0.0029 | 288 | 22,5 | 37 |
| 54673814 | enolase 1, alpha non-neuron | ENO1 | Glycolysis | 1.61 | 0.019 | 541 | 33,8 | 53 |
| 28436836 | triosephosphate isomerase 1 | TPI1 | Glycolysis;Other metabolism | 1.56 | 0.02 | 103 | 10,1 | 33 |
| 31982229 | ketohexokinase | KHK | Monosaccharide metabolism | 1.8 | 0.019 | 80 | 11,2 | 33 |
| 56206644 | malate dehydrogenase 1, NAD (soluble) | MDH1 | Tricarboxylic acid pathway | 1.88 | 0.0013 | 251 | 14,5 | 35 |
| 58037546 | isocitrate dehydrogenase 1 (NADP+),soluble | IDH1 | Tricarboxylic acid pathway | 1.65 | 0.0049 | 438 | 35,7 | 54 |
| 19484047 | malate dehydrogenase 2, NAD (mitochondrial) | MDH2 | Tricarboxylic acid pathway | 1.54 | 0.049 | 251 | 16,3 | 43 |
| 16741459 | ATP synthase, H+ transp, mitochon.F0 complex, subunit d | ATP5H | ATP synthesis coupled proton transport | 1.91 | 0.0033 | 118 | 10,2 | 54 |
| 2894517 | fatty acid binding protein 1, livera | FABP1 | Lipid and fatty acid transport | 2 | 0.01 | 165 | 14,5 | 67 |
| 15215161 | microsomal triglyceride transfer protein | MTTP | Lipid and fatty acid transport;Transport | 1.54 | 0.0428 | 112 | 26,2 | 27 |
| 2492905 | annexin A4 | ANXA4 | Lipid, fatty acid and steroid metabolism | 1.56 | 0.0012 | 143 | 17,2 | 48 |
| 9845263 | dopa/tyroine sulfotransferase | SULT1B1 | Steroid hormone metabolism;Sulfur metab. | 1.87 | 0.00063 | 115 | 11,3 | 47 |
| 2257955 | cytochrome b-5 | CYB5A | Steroid metab. Electr. transp. | 1.54 | 0.049 | 120 | 6,1 | 80 |
| 397351 | mCRBPII | RBP2 | Steroid hormone metabolism | 2.53 | 0.003 | 670 | 21,8 | 32 |
| 71059735 | pyrophosphatase (inorganic) 1 | PPA1 | Polyphosphate catabolism | 1.69 | 0.049 | 72 | 9,2 | 32 |
| 71059839 | adenosine deaminase | ADA | Purine metabolism | 6.43 | 0.0061 | 266 | 23,5 | 42 |
| 19526926 | unnamed protein | C11orf54 | unclassified | 1.52 | 0.02 | 141 | 19,2 | 48 |
| 19484063 | arginase type II | ARG2 | Amino acid catabolism | 1.63 | 0.0051 | 104 | 12,6 | 34 |
| 52789310 | archain 1 | ARCN1 | Constitutive exocytosis | 1.99 | 0.0045 | 118 | 22,3 | 26 |
| Accession no. | Proteins downregulated in the villus | Symbol | Biological process | Fold change (Villus/Crypt) | P Value | Score | # of peptides matched* | Coverage (%) |
| 53237015 | peptidylprolyl isomerase A | Ppia | Protein folding;Nuclear transp;Immunity/defense | −1.61 | 0.03 | 105 | 8,2 | 43 |
| 76779273 | heat shock protein 1 (chaperonin) | Hspd1 | Protein folding/complex assembly | −1.93 | 0.019 | 359 | 24,6 | 39 |
| 38303969 | heat shock 70kD protein 5 (glucose-regulated protein) | HSPA5 | Protein folding/complex assembly; Stress resp. | −1.6 | 0.047 | 449 | 38,10 | 43 |
| 435839 | mortalin | HSPA9 | Protein folding/complex assembly; Stress resp. | −1.56 | 0.00044 | 674 | 32,8 | 61 |
| 123681 | heat shock protein 1, beta | HSP90AB1 | Protein folding; Stress resp. | −1.55 | 0.015 | 182 | 24,4 | 41 |
| 12860388 | ribosome binding protein 1 | Rrbp1 | Protein localization; Cell structure and motility | −1.86 | 0.038 | 82 | 19,0 | 20 |
| 8393866 | ornithine aminotransferase | OAT | Amino acid metab. Vision | −2.24 | 0.00081 | 691 | 35,10 | 50 |
| 20987331 | eukaryotic translation initiation factor 3, subunit 2b | EIF3S2 | Protein biosynthesis | −1.93 | 0.00028 | 197 | 17,2 | 46 |
| 9790219 | destrin | DSTN | Cell structure | −1.52 | 0.0013 | 61 | 4,2 | 15 |
| 13542680 | tubulin, beta 2c | TUBB2C | Protein traffic;C'some seg; Cell structure/motility | −1.56 | 0.025 | 287 | 32,5 | 40 |
| 20381452 | Lgals2 protein | LGALS2 | Cell adhesion; immune/defense; apoptosis | −1.5 | 0.016 | 384 | 15,6 | 69 |
| 37747847 | heterogeneous nuclear ribonucleoprotein A2/B1 | HNRPA2B1 | mRNA splicing | −1.62 | 0.033 | 121 | 8,3 | 44 |
| 73918911 | carbamoyl-phosphate synthetase 1 | CPS1 | Arginin, pyrimidine biosynthesis, urea cycle | −1.68 | 0.0057 | 476 | 58,7 | 48 |
| 31542333 | hypoxia up-regulated 1 | HYOU1 | Stress response | −1.57 | 0.016 | 489 | 55,7 | 43 |
| 51770420 | unknown protein | LOC433141 | Biological process unclassified | −1.59 | 0.029 | 118 | 10,3 | 30 |
| 31560560 | laminin receptor 1 | LAMR1 | Biological process unclassified | −1.68 | 0.0014 | 126 | 10,2 | 30 |
| 15426464 | anterior gradient 2 (Xenopus laevis) | AGR2 | Miscellaneous | −1.74 | 0.00042 | 128 | 9,4 | 42 |
| Accession no. | Contradictory findings | Symbol | Biological process | Fold change (Villus/Crypt) | P Value | Score | # of peptides matched* | Coverage (%) |
| 13624315 | keratin complex 2, basic, gene 8 | KRT8 | Cell structure, Ectoderm development | 2.11/−2.72 | 0.0085/0.028 | 343/344 | 43,8/42,5 | 52/59 |
| 6680606 | keratin complex 1, acidic, gene 19 | KRT19 | Cell structure | 2.82/−1.54 | 0.006/0.021 | 148/133 | 24,4/15,1 | 47/39 |
| 15488625 | fatty acid binding protein 2, intestinal | FABP2 | Lipid and fatty acid transport | 2.41/−2.54 | 0.013/0.013 | 393/149 | 15,5/10,2 | 79/52 |
| 42542422 | heat shock protein 8 | Hspa8 | Protein folding/complex assembly; Stress resp. | 1.56/−1.57 | 0.028/0.021 | 508/258 | 40,7/22,5 | 47/43 |
Numbers shown in the “ # of peptides matched column” represent the number of peptides matched with either MS or MS/MS and with MS/MS, respectively.
indicates multiple proteins present in one spot."
Figure 3.
(A) Representative 2D-DIGE Gel image. (B) Representative proteins upregulated in villus (F1) relative to crypt (F5) fractions. (C) Representative proteins upregulated in crypt (F5) relative to villus (F1) fractions.
Validation of proteomic changes
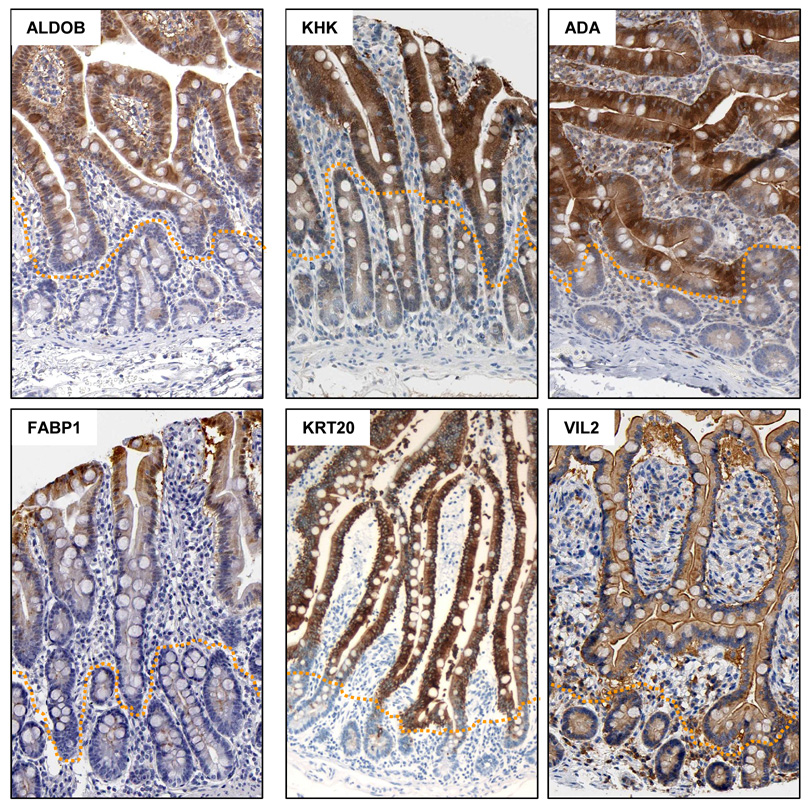
To validate the proteomic analysis, the expression patterns of a subset of the differentially expressed proteins were examined by immunohistochemistry in cross sections of mouse (AGR2) and human (KRT20) small intestinal tissue. In order to maximize the number of targets for validation, we also utilized the protein atlas database which contains immunohistochemical analyses of multiple human tissues utilizing 3014 antibodies (www.proteinatlas.org, version 3.1 2-15-2008). Collectively, immunohistochemical images for 6 of the proteins upregulated in the villus, and for 8 of the proteins downregulated in the villus were obtained for analysis. Of the proteins upregulated in the villus, 5/6 (ALDOB, KHK, ADA, FABP1, KRT20) showed staining patterns consistent with higher expression in the villus. No clear gradient of expression was evident for Villin 2 (Figure 4).
Figure 4.
Immunohistochemical validation of six proteins, aldolase 2, B isoform (ALDOB), Ketohexokinase (KHK), adenosine deaminase (ADA), Fatty acid binding protein 1 (FABP1), Keratin 20 (KRT20), and Ezrin/Villin 2 (VIL2), upregulated in the villus relative to the crypt. All images, with the exception of KRT20, were obtained from the protein atlas database (www.proteinatlas.org).
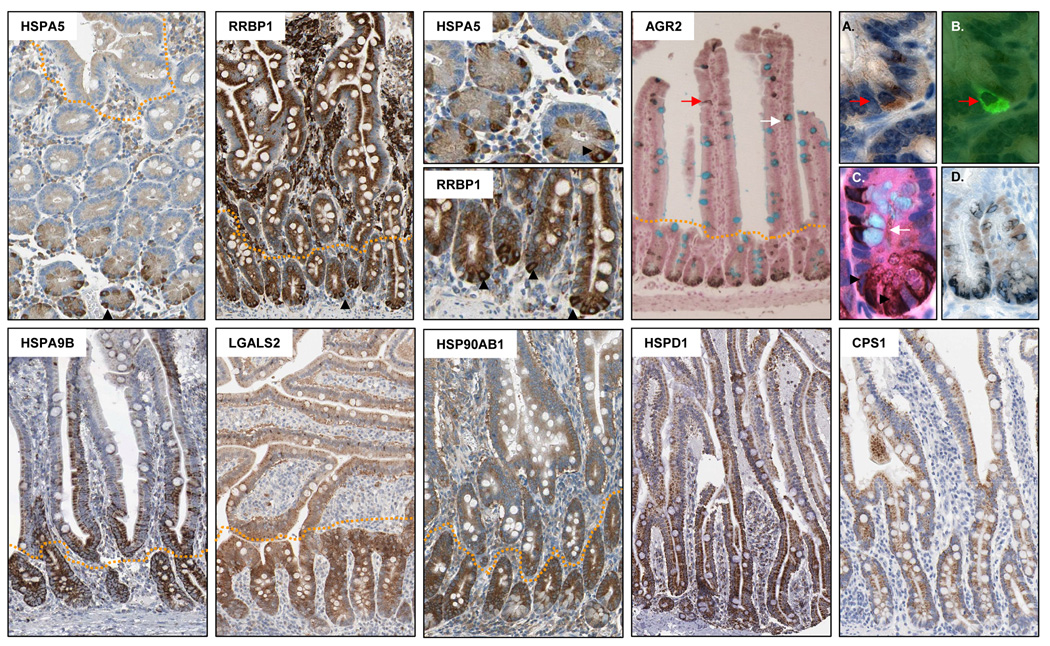
Likewise, immunohistochemical staining patterns consistent with the proteomic data were observed for 6/8 proteins maximally expressed in the crypt (HSPA5, RRBP1, HSP90AB1, HSPA9, LGALS2 and AGR2). Notably, for three of these proteins, HSPA5, RRBP1 and AGR2, staining patterns consistent with maximal expression in Paneth cells located at the base of the crypts was observed, identifying these proteins as novel markers of this differentiated cell lineage (Figure 5).
Figure 5.
Immunohistochemical validation of eight proteins maximally expressed in the crypt relative to the villus: heat shock 70kD protein 5 (HSPA5), ribosome binding protein 1 (RRBP1), anterior gradient 2 (AGR2), mortalin (HAPA9), Lgals2 protein (LGALS2), heat shock protein 1 beta (HSP90AB1), heat shock protein 1 (HSPD1), carbamoyl-phosphate synthetase 1 (CPS1). All images, with the exception of AGR2, were obtained from the protein atlas database (www.proteinatlas.org). Red arrow in the AGR2 panel indicates AGR2 expression in an enteroendocrine cell. White arrow indicates AGR2 expression in a goblet cell. (A–B) Overlap of Agr2 staining (brown, panel A) with the enteroendocrine cell marker, chromogranin (green, panel B, red arrow). (C) Co-staining with Agr2, alcian blue to identify goblet cells, and hematoxylin and eosin to identify paneth cells. Paneth cells are indicated by a black arrow head, goblet cells by a white arrow and enteroendocrine cells by a red arrow. (D) Dual staining of Agr2 (black) with the cell proliferation marker, Ki67 (brown).
To directly confirm the overlap in expression of AGR2 with Paneth cells, sections were counterstained with eosin which stains eosinophillic cytoplasmic granules present in this cell type. As shown in Figure 5, clear overlap between granulated Paneth cells in the crypt base and AGR2 staining was evident. In addition to paneth cells, occasional cells in both the crypt and the villus which stained positively for AGR2 were also evident. Alcian blue counterstaining confirmed that the majority of these cells were mucus containing goblet cells, while dual staining with AGR2 and the enteroendocrine marker, chromogranin, also demonstrated clear localization of AGR2 to enteroendocrine cells. Finally, dual staining of AGR2 with the proliferation marker Ki67 demonstrated minimal overlap between these two proteins, collectively identifying AGR2 as a bone-fide marker of differentiated cells of the secretory cell lineage (Figure 5A–D). No clear gradient of expression was evident for HSPD1 and CPS1 (Figure 5).
Proteins involved in lipid and steroid metabolism and in cell structure are upregulated in the villus compartment
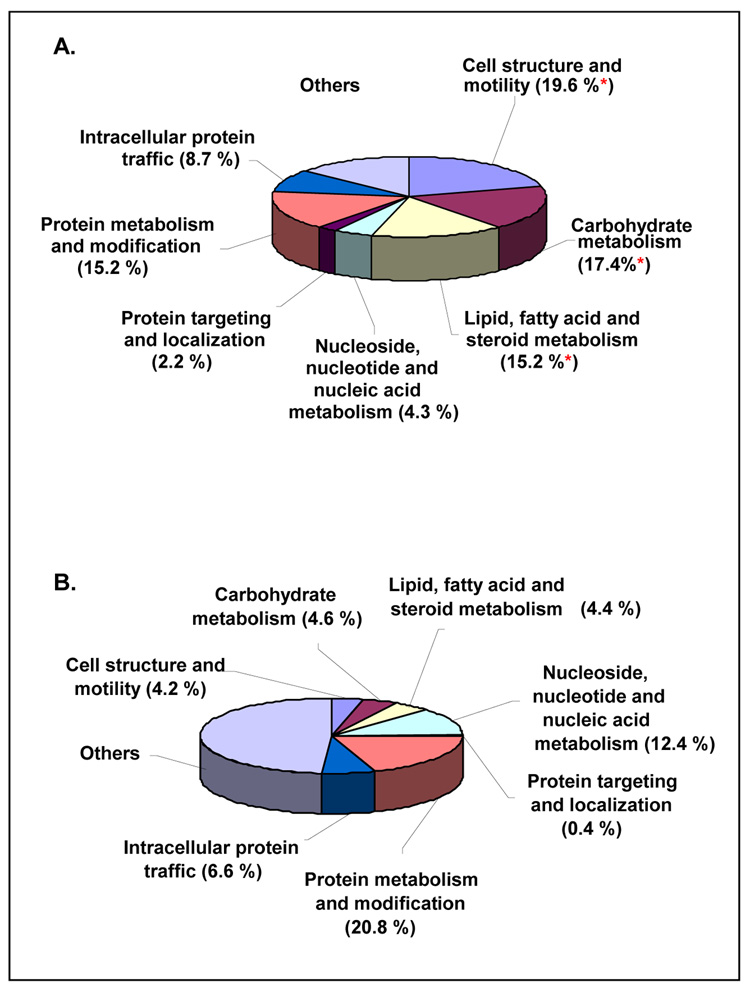
To determine whether the proteins differentially expressed along the crypt-villus axis were statistically enriched for particular biological processes, we performed an enrichment analysis using the Panther analysis tool 35. As a control, the enrichment analysis was repeated using 46 randomly selected proteins (Figure 6). This analysis identified that proteins differentially expressed along the crypt-villus axis were significantly enriched for proteins with a role in lipid/fatty acid/steroid metabolism, cell structure and motility and carbohydrate metabolism (P<0.005, Fisher exact test). Importantly, detailed examination of these categories revealed a high degree of consistency in terms of the directionality of change of the individual proteins comprising each category (Table 1). This included the co-ordinate upregulation in the villus of proteins involved in lipid/fatty acid (FABP1 and II, and MTTP), vitamin A (RBP2), and steroid hormone (SULT1B1) uptake and metabolism, a finding consistent with an established role of the intestinal epithelium in the absorption of these compounds 36, 37 (Table 1). Likewise, several of the cell structure and motility proteins were upregulated in the villus, including Villin I and II, actin, keratin 20, and profilin, which is consistent with the formation of microvilli during intestinal cell differentiation.
Figure 6.
(A) Functional categorization of proteins differentially expressed between crypt and villus. Classification of protein function was performed using the Panther analysis tool (www.pantherdb.org). (B) As a control, the enrichment analysis was repeated on 46 randomly selected proteins. Fisher exact tests identified significant enrichment of differentially expressed proteins between the crypt and villus relative to that expected by chance in the categories of cell structure and motility (p=0.0002), carbohydrate metabolism (p=0.0014), and lipid, fatty acid, and steroid metabolism (p=0.0045), denoted by *.
Proteins involved in glycolysis are upregulated in the villus compartment
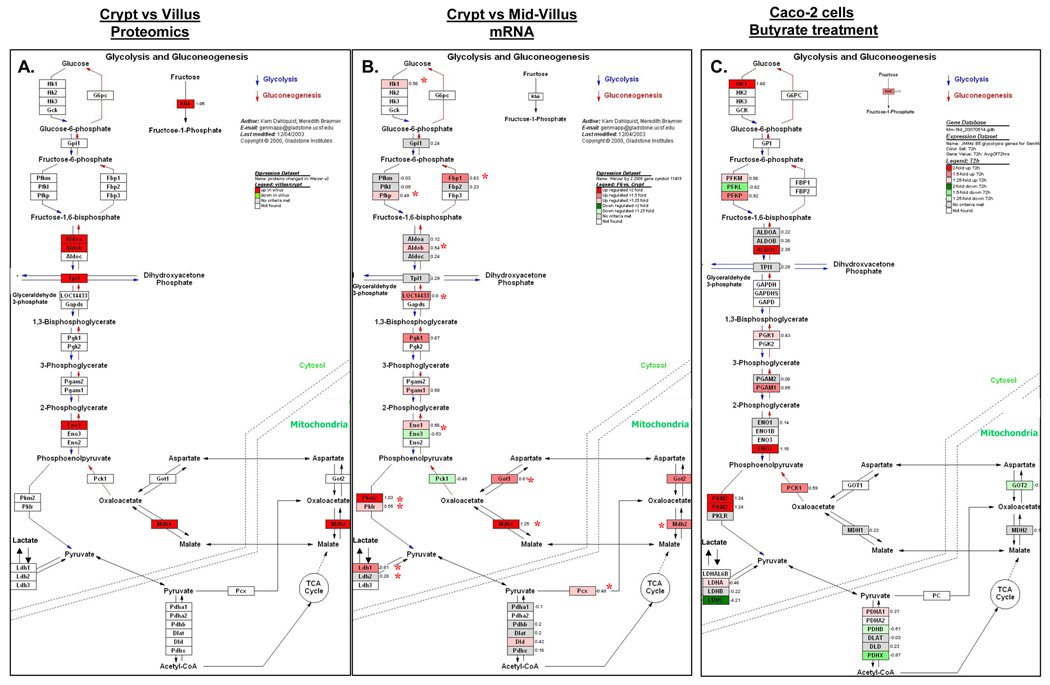
Notably, a number of components of the glycolytic pathway (ketohexokinase, aldolase A and B, enolase, malate dehydrogenase 1 and 2, and isocitrate dehydrogenase 1) were coordinately upregulated in the villus (Figure 7). The upregulation of ketohexokinase and aldoB in the villus was supported by the immunohistochemical localization of these proteins (Figure 4). Increased rates of glycolysis have been closely linked with cell proliferation 38, and is a classical feature of transformation 38, whereas the present study demonstrated increased expression of glycolytic enzymes in the non proliferating differentiated villus compartment relative to the proliferating crypt compartment. Therefore, to confirm this finding we examined parallel mRNA changes in glycolytic enzymes along the crypt-villus axis utilizing our previously published microarray database 21. In support of the proteomic changes, coordinated upregulation of mRNA expression of several glycolytic enzymes was evident in the villus fractions (Hk1, Pfkb, Aldob, LOC14433, Eno1, Pkm2, Pklr, Mdh1, Mdh2, Got1), with maximal elevation observed in the mid villus region (Figure 7B). To further address this, we also examined changes in glycolytic gene expression following butyrate treatment of the Caco-2 colon cancer cell line, an established in vitro model of intestinal cell maturation 39. Changes in gene expression following butyrate treatment were determined by cDNA microarray analysis. As shown in Figure 7C, >1.5 fold induction of mRNA expression of multiple glycolytic enzymes was evident 72h post butyrate treatment, including Hk1, Khk, Pfkp, Aldoc, Pgam1, Eno2, and Pkm2), suggesting induction of glycolytic enzymes is a consistent feature of intestinal cell maturation.
Figure 7.
(A) Pathway maps generated using GenMapp depicting increased glycolytic protein expression in differentiated intestinal epithelial cells relative to crypt cells. The intensity of red indicates the extent to which protein expression is elevated in the villus fraction. (B) Corresponding mRNA levels of glycolytic genes are upregulated in the villus fraction in mouse small intestine (n=4 mice). Red asterix indicates statistical significance (P<0.05) as assessed using a paired t-test. (C) Increased glycolytic gene expression following butyrate induced differentiation of Caco-2 colon cancer cells. Caco-2 cells were treated with 5 mM butyrate for 72 hours and changes in gene expression determined using 27,000 feature cDNA microarrays. For B and C, the intensity of red indicates the extent to which gene expression is elevated in the villus fraction, as indicated in the figure legend.
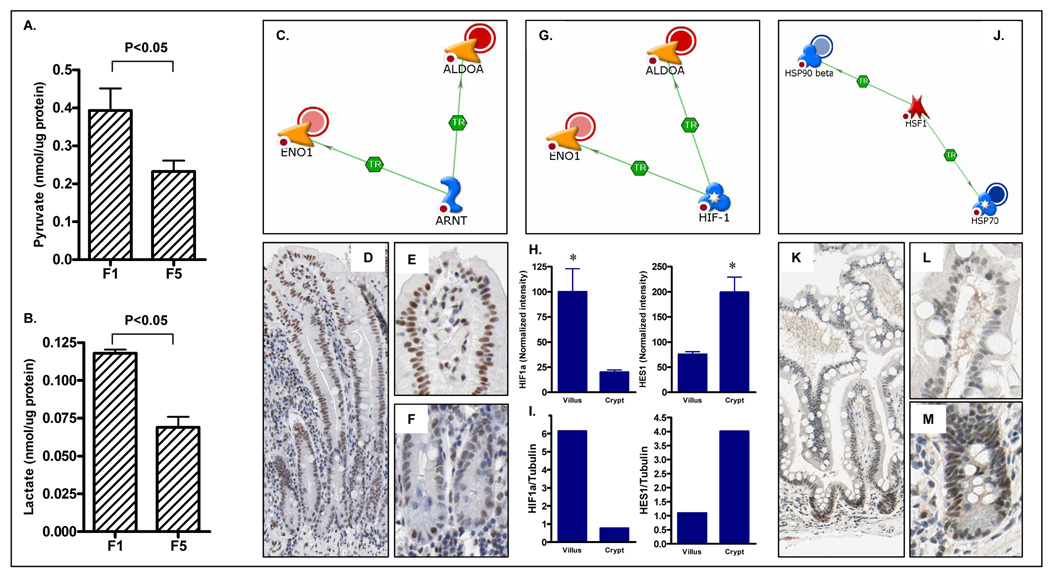
To determine whether the upregulation of glycolytic enzymes in the villus fraction resulted in a parallel increase in the concentrations of the metabolic products of glycolysis, we measured the levels of pyruvate and lactate in cell fractions isolated from the villus and the crypt. As shown in Figure 8A–B, and consistent with the higher expression of glycolytic enzymes, levels of pyruvate and lactate were increased by 41.3% and 40.9%, respectively, in the villus fraction (P<0.05).
Figure 8.
Increased levels of glycolytic metabolites (A) pyruvate and (B) lactate in villus fractions (mean + SD, n=4). (C,G, H) Transcription factors previously linked to the regulation of 2 or more differentially expressed proteins were identified using the transcription regulation algorithm in MetaCore. Among the transcription factors identified were (C) ARNT, (G) HIF1α and (J) HSF1. (D–F) Immunohistochemical localization demonstrating maximal expression of ARNT in the villus epithelium, and expression of (K–M) HSF1 in crypt cells. (H, I) Expression of HIF1α mRNA as assessed by (H) microarray analysis or (I) QPCR is maximal in the villus fraction. Shown for comparison is expression of HES1 an established marker of crypt progenitor cells.
Finally, to identify possible common regulators of glycolytic and other proteins differentially expressed along the crypt-villus axis, we performed a network analysis using the Metacore pathway analysis tool. As shown in Figure 8, this analysis suggested that the heterodimeric transcription factors HIF1α and ARNT might function as positive regulators of two of the glycolytic enzymes upregulated in the villus, namely Eno1 and ALDOA. To validate these network analysis predictions, we once again utilized the protein atlas database. As shown in figure 8, immunohistochemical staining of ARNT revealed maximal expression in the villus. To determine the expression pattern of HIF1α, we measured HIF1α mRNA expression in crypt and villus fractions by both microarray and QPCR. As shown in figure 8, HIF1α mRNA expression was significantly higher in the villus compared to the crypt fraction. The upregulation of ARNT and HIF1α in the villus fraction is therefore consistent with the parallel upregulation of glycolytic enzymes in the villus, suggesting a possible mechanism of regulating glycolytic gene expression along the crypt villus axis.
Proteins involved in protein processing and folding are maximally expressed in the crypt compartment
Among the differentially expressed proteins were several with a role in protein processing and folding, including several heat shock proteins, mortalin, peptidylpropyl isomerase A, and eIF32B. While this category was not significantly enriched when compared to the random set, the majority of these proteins were maximally expressed in the crypt, likely reflecting the increased rate of protein synthesis that accompanies cell proliferation.
As shown in Figure 8, the network analysis identified the transcription factor HSF1 as an established regulator of two of these proteins, hsp90AB1 (HSP90beta) and HSPA5 (Hsp70). To determine the expression pattern of HSF1 in the small intestine, we utilized the protein atlas database. As shown in Figure 8K–M, consistent with the expression pattern of its target genes, HSF1 expression was maximal in the crypt compartment.
Proteomic changes correlate significantly with corresponding gene expression changes
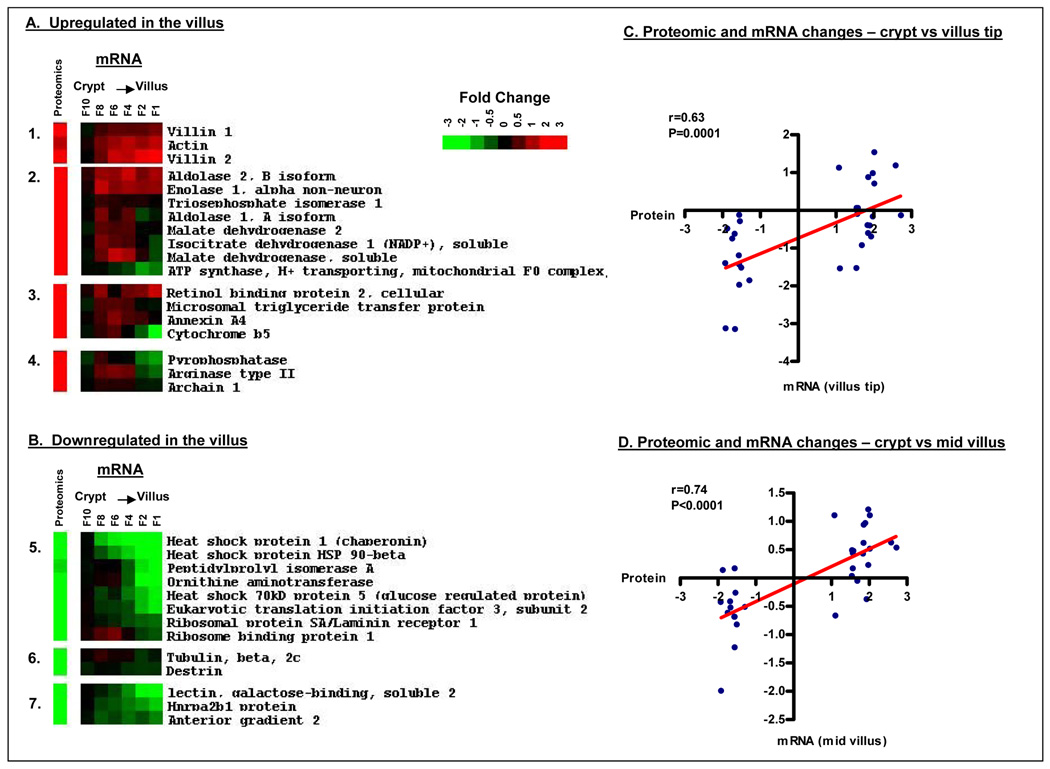
To determine the degree to which the identified changes in protein expression correlated with our previous study of gene expression changes, we performed a correlation analysis of the sequences present in both datasets. Of the 46 proteins identified as differentially expressed by DIGE, we obtained corresponding mRNA expression data for 31 sequences (Figure 9). Regression analysis demonstrated a moderate although statistically significant correlation between proteomic and gene expression changes between the crypt base and the villus tip (r=0.63, P=0.0001, Figure 9D). Interestingly, a stronger correlation (r=0.74, P<0.0001) between proteomic changes in the villus tip and corresponding mRNA changes in the mid-villus region was observed, consistent with the interpretation that transcriptional changes precede changes in protein level and that proteins synthesized during maturation are likely more stable than the mRNAs that code for them.
Figure 9.
Proteomic changes along the crypt-villus axis correlate significantly with corresponding gene expression changes. Corresponding gene expression data were available for 31 of the 46 differentially expressed proteins. Shown in panels A and B are proteomic and corresponding microarray data for proteins up and downregulated in the villus respectively. (C–D) Pearson’s correlation of proteomic and corresponding gene expression changes. (C) Correlation of proteomic changes between the crypt and villus with corresponding degree of mRNA change between these two fractions (F1 vs F10; r=0.67, p<0.0001). (D) Correlation of proteomic changes between the between crypt and villus with corresponding degree of mRNA change between the crypt and mid-villus region (F1 vs F4; r=0.71, p<0.0001).
Discussion
This study provides a novel profile of the proteomic changes that occur during intestinal cell maturation along the crypt-villus axis in mouse small intestine in vivo. Demonstrating the validity of this approach, several proteins previously shown to be altered in expression along the crypt-villus axis were similarly identified in the present study. These included villin 21, ezrin 40, CRBPII 29 and FABP1 29, each of which were maximally expressed in the villus compartment.
Analyses to identify functionally enriched categories of proteins demonstrated that proteins with a role in lipid/fatty acid/steroid metabolism and cell structure were significantly enriched among the differentially expressed proteins identified. The proteins involved in each of these processes were coordinately upregulated in the villus compartment, consistent with the absorption and metabolism of dietary lipids and cell polarization, respectively, being important features of differentiated intestinal epithelial cells. Specific examples included the upregulation of microsomal triglyceride transfer protein (MTTP) in the villus epithelium, which plays a central role in chylomicron assembly in the intestine by lipidating apo B-48 and apo A-IV with newly synthesized triglycerides 36.
The analysis also identified a coordinate induction of multiple proteins involved in glycolysis in the villus compartment. Consistent with the increased expression of glycolytic enzymes, levels of 2 metabolites of glycolysis, lactate and pyruvate, were elevated in the villus compartment, suggesting that although differentiated, villus epithelial cells have a higher energetic demand than their proliferating crypt counterparts. This demand may be driven in part by the major role played by differentiated intestinal epithelial cells in nutrient uptake, which is predominantly an active process, likely requiring significant energy expenditure 41. Additionally, the high glycolytic rates may reflect the high rates of protein synthesis and degradation which have been observed in the gastrointestinal tracts of several animals, a process proposed to permit abrupt changes in gut size to occur in order to match absorptive capacity with nutrient delivery 41. Consistent with these processes, oxygen utilization studies indicate that approximately 20% of all incoming energy is consumed in the intestinal tract as part of the absorption and digestion process 41.
The upregulation of glycolytic proteins may also be linked to the intraluminal absorption of monosaccharides, primarily glucose, fructose and galactose, which is a key function of the differentiated intestinal epithelium 42. Reflecting this, the brush border hydrolases sucrase-isomaltase and lactase-phloridzin hydrolase, which generate monosaccharides from disaccharides 43, and the hexose transporters SGLT1 44, and GLUT5 45–47, which actively transport glucose and fructose respectively, are maximally expressed in enterocytes present in the differentiated villus compartment. What is less clear is the fate of these absorbed sugars in enterocytes following absorption. Some studies indicate that the majority of intraluminally absorbed glucose is transported intact into the portal vein 48, which has been shown to occur primarily via the basolateral GLUT2 transporter 47. However, Stumpel et al demonstrated that the kinetics of glucose uptake in mice with intestinal-specific deletion of the GLUT2 transporter was normal, and demonstrated the existence of an alternate transcellular glucose transport pathway which required partial glucose metabolism by enterocytes 49. Other studies have also demonstrated that 50–60% of absorbed sugars, in particular fructose, are metabolized via glycolysis to lactate within the intestinal epithelium, and that the intestine is a major producer of portal-venous lactate 48. Finally, a recent proteomic analysis demonstrated that a number of glycolytic proteins were among those downregulated in the intestine following starvation-induced glucose deprivation in mice 50. The upregulation of glycolytic enzymes in the villus epithelium is therefore consistent with several observations indicating enterocytic metabolism of absorbed sugars is a component of the uptake process.
Notably, of the differentially expressed proteins identified the majority were membrane or cytoplasmic proteins with a role in cell structure, metabolism and nutrient transport. To formally address whether this reflected a limitation of the DIGE technique for the preferential identification of high abundance proteins, we used our complementary gene expression database to determine the corresponding gene expression intensity for each of the differentially expressed proteins. This analysis demonstrated that the mRNA expression intensity of 92% of the differentially expressed proteins fell into the uppermost intensity quartile of the overall distribution of mRNA expression intensity. In addition, several studies have demonstrated a bias in codon usage that correlates with the abundance of a protein 51 52. The codon adaptation index (CAI), which is one method of computing codon bias within sequence data, was therefore calculated for the 46 genes whose proteins expression was changed along the crypt villus axis, as an alternative method of estimating protein abundance. The CAI for each protein was calculated as previously described by Sharp and Li 32, 33 , using the Evolving code CAI calculator tool (http://www.evolvingcode.net/codon/cai/cais.php). In order to place these values into the context of the entire proteome, the distribution of CAI values of all cDNA sequences in the mouse genome for which CAI could be computed was also determined (12,550 sequences). The CAI for 85% of these genes (39 of 46 genes) fell within the 4th quartile of the overall distribution, while 93% of genes (43 of 46 genes) fell within the 3rd and 4th quartiles, implying that the differentially expressed proteins identified by DIGE are biased towards high abundance proteins.
To overcome this limitation, a pathway analysis using a bioinformatic approach was performed to obtain insight into possible transcriptional regulators of the differentially expressed proteins. Based upon prior reports, the network analysis identified the HIF1α/ANRT transcriptional heterodimer as a potential regulator of glycolytic gene expression in the villus, and the HSF1 transcription factor as a potential regulator of heat shock gene expression in the crypt. Importantly, immunohistochemical and QPCR validation studies demonstrated upregulation of ARNT and HIF1α in the villus compartment, and maximal expression of HSF1 in the crypt, consistent with the expression patterns of their target genes. Notably, mice with targeted deletion of HIF1α or ARNT in the intestinal epithelium have recently been generated 53. While the histological appearance of the intestinal epithelium in these mice appears normal, intestinal-specific disruption of HIF1α results in increased epithelial permeability and increased sensitivity to experimentally induced colitis 53. Whether disruption of glycolytic gene expression contributes to this process remains to be determined. Therefore, the approach of leveraging proteomic data, which by its nature has limited coverage, with network analysis tools, clearly provided important insights.
Never-the-less proteomic methodologies which can successfully interrogate low abundance proteins are clearly needed. In this regard, newer techniques are emerging for fractionating proteins based upon their isoelectric properties. One advantage of combining pre-fractionation by isoelectric point and using a narrower pH gradient is to reduce protein complexity, thus enabling the loading of higher amounts of protein with increased resolution. An alternative approach for improving the resolution of low abundant proteins is the prefractionation of proteins based upon their subcellular localization, or fractionation using columns with affinity for specific classes of proteins. While such approaches reduce the overall scope of the study, they increase sensitivity by increasing the amount of low abundant proteins relative to other proteins. For example, a recent study demonstrated that a priori isolation and purification of mitochondria helped to overcome the masking of mitochondrial proteins by more abundant cytosolic proteins 54.
This analysis extends our previous gene expression profiling experiments of the transcriptional changes that define intestinal cell maturation by providing a complementary profile of the parallel proteomic changes. We observed a significant correlation between the changes in mRNA and protein expression during differentiation along the crypt-villus axis, with the correlation stronger when mid-villus mRNA expression was compared to villus tip proteome expression. This provides a high throughput validation of the publicly available gene expression database of transcriptional changes along the crypt-villus 21. Collectively, these observations emphasize the linear and coordinated nature in which intestinal cell maturation is regulated along the crypt-villus axis with transcriptional changes driving most protein expression changes.
This study also emphasizes a number of important complements of proteomic profiling to RNA expression profiling. First, the integration of gene expression data with parallel proteomic profiling studies suggested that a subset of proteins induced during maturation is likely more stable than the encoding mRNAs. Focusing exclusively on mRNA levels would therefore have failed to identify this subset of differentiation markers. Second, for another subset of differentially expressed proteins, parallel gene expression data were not available either because corresponding probes were not represented on the array or because they failed to provide reliable signal. With regards to the former, proteomic profiling has the important advantage of not being limited to pre-determined targets.
In conclusion, this study characterizes the proteomic changes that occur during intestinal cell maturation along the crypt-villus axis in mouse small intestine. This database complements existing databases that have examined the changes in gene expression that take place during intestinal / colonic cell maturation and transformation, including studies profiling the differences in expression between normal and tumor tissue in both humans 55, 56 and mice 57; that occur in response to drug treatment 18, 58, 59 or manipulation of specific genes 4; that occur in models of intestinal cell differentiation in vitro 19, 60, or which occur along the anterior-posterior axis of the gastrointestinal tract 61. Integration of these databases will provide a powerful method of identifying novel candidate genes involved in the regulation of normal intestinal cell maturation and in disease.
Acknowledgements
We thank Dr. Kathryn Tanaka for her assistance with immunohistochemical analyses.
Grants: Supported by NIH grants NIH U54-100926, CA100823, P30-CA043703 and P30-CA013330.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon JI, Hermiston ML. Differentiation and self-renewal in the mouse gastrointestinal epithelium. Curr. Opin. Cell Biol. 1994;6:795–803. doi: 10.1016/0955-0674(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 2.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 3.Mariadason JM, Bordonaro M, Aslam F, Shi L, Kuraguchi M, Velcich A, Augenlicht LH. Down-regulation of beta-catenin TCF signaling is linked to colonic epithelial cell differentiation. Cancer Res. 2001;61:3465–3471. [PubMed] [Google Scholar]
- 4.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 5.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 6.Batts LE, Polk DB, Dubois RN, Kulessa H. Bmp signaling is required for intestinal growth and morphogenesis. Dev. Dyn. 2006;235:1563–1570. doi: 10.1002/dvdy.20741. [DOI] [PubMed] [Google Scholar]
- 7.He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, Mishina Y, Li L. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 8.van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, Nielsen C, Gaffield W, van Deventer SJ, Roberts DJ, Peppelenbosch MP. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- 9.Gao X, Sedgwick T, Shi Y-B, Evans T. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol. Cell. Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shie JL, Chen ZY, O'Brien MJ, Pestell RG, Lee ME, Tseng CC. Role of gut-enriched Kruppel-like factor in colonic cell growth and differentiation. Am. J. Physiol. Gastrointest.Liver Physiol. 2000;279:G806–G814. doi: 10.1152/ajpgi.2000.279.4.G806. [DOI] [PubMed] [Google Scholar]
- 11.Burgess AW. Growth control mechanisms in normal and transformed intestinal cells.Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1998;353:903–909. doi: 10.1098/rstb.1998.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clatworthy JP, Subramanian V. Stem cells and the regulation of proliferation,differentiation and patterning in the intestinal epithelium: emerging insights from gene expression patterns, transgenic and gene ablation studies. Mech. Dev. 2001;101:3–9. doi: 10.1016/s0925-4773(00)00557-8. [DOI] [PubMed] [Google Scholar]
- 13.Wang TC, Dockray GJ. Lessons from genetically engineered animal models. I.Physiological studies with gastrin in transgenic mice. Am. J. Physiol. 1999;277:G6–G11. doi: 10.1152/ajpgi.1999.277.1.G6. [DOI] [PubMed] [Google Scholar]
- 14.Hermiston ML, Gordon GI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science. 1995;270:1203–1207. doi: 10.1126/science.270.5239.1203. [DOI] [PubMed] [Google Scholar]
- 15.Kedinger M, Lefebvre O, Duluc I, Freund JN, Simon-Assmann P. Cellular and molecular partners involved in gut morphogenesis and differentiation. Philos. Trans. R. Soc. Lond.B. Biol. Sci. 1998;353:847–856. doi: 10.1098/rstb.1998.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 17.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 18.Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–4572. [PubMed] [Google Scholar]
- 19.Mariadason JM, Arango D, Corner GA, Aranes M, Hotchkiss KA, Yang W, Augenlicht LH. A gene expression profile that defines colon cell maturation in vitro. Cancer Res. 2002;62:4791–4804. [PubMed] [Google Scholar]
- 20.Velcich A, Corner GA, Paul D, Zhuang M, Mariadason JM, Laboisse C, Augenlicht LH. Qantitative rather than qualitative differences in gene expression predominate in intestinal cell maturation along distinct cell lineages. Exp. Cell Res. 2004 doi: 10.1016/j.yexcr.2004.10.014. In Press. [DOI] [PubMed] [Google Scholar]
- 21.Mariadason JM, Nicholas C, L'Italien KE, Zhuang M, Smartt HJM, Heerdt BG, Yang W, Corner GA, Wilson AJ, Klampfer L, Arango D, Augenlicht LH. Gene Expression Profiling of intestinal Epithelial Cell Maturation along the Crypt-Villus Axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 22.Ferraris RP, Villenas SA, Diamond J. Regulation of brush-border enzyme activities and enterocyte migration rates in mouse small intestine. Am. J. Physiol. 1992;262:G1047–G1059. doi: 10.1152/ajpgi.1992.262.6.G1047. [DOI] [PubMed] [Google Scholar]
- 23.Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis. II.Glycosyltransferases and endogenous acceptors of the undifferentiated cell surface membrane. J. Biol. Chem. 1973;248:2542–2548. [PubMed] [Google Scholar]
- 24.Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J. Biol. Chem. 1973;248:2536–2541. [PubMed] [Google Scholar]
- 25.Rabilloud T, Adessi C, Giraudel A, Lunardi J. Improvement of the solubilization of proteins in two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1997;18:307–316. doi: 10.1002/elps.1150180303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Friedman DB, Wang SE, Whitwell CW, Caprioli RM, Arteaga CL. Multivariable difference gel electrophoresis and mass spectrometry: a case study on transforming growth factor-beta and ERBB2 signaling. Mol Cell Proteomics. 2007;6:150–169. doi: 10.1074/mcp.D600001-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Wilson AJ, Byun DS, Popova N, Murray LB, L'Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 29.Rubin DC, Ong DE, Gordon JI. Cellular differentiation in the emerging fetal rat small intestinal epithelium: mosaic patterns of gene expression. Proc. Natl. Acad. Sci. U S A. 1989;86:1278–1282. doi: 10.1073/pnas.86.4.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, Steen J, Stenvall M, Sterky F, Stromberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Walden A, Wan J, Wernerus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Ponten F. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson P, Paavilainen L, Larsson K, Odling J, Sundberg M, Andersson AC, Kampf C, Persson A, Al-Khalili Szigyarto C, Ottosson J, Bjorling E, Hober S, Wernerus H, Wester K, Ponten F, Uhlen M. Towards a human proteome atlas: high-throughput generation of mono-specific antibodies for tissue profiling. Proteomics. 2005;5:4327–4337. doi: 10.1002/pmic.200500072. [DOI] [PubMed] [Google Scholar]
- 32.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu G, Culley DE, Zhang W. Predicted highly expressed genes in the genomes of Streptomyces coelicolor and Streptomyces avermitilis and the implications for their metabolism. Microbiology. 2005;151:2175–2187. doi: 10.1099/mic.0.27833-0. [DOI] [PubMed] [Google Scholar]
- 34.Polley AC, Mulholland F, Pin C, Williams EA, Bradburn DM, Mills SJ, Mathers JC, Johnson IT. Proteomic analysis reveals field-wide changes in protein expression in the morphologically normal mucosa of patients with colorectal neoplasia. Cancer Res. 2006;66:6553–6562. doi: 10.1158/0008-5472.CAN-06-0534. [DOI] [PubMed] [Google Scholar]
- 35.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Black DD. Development and physiological regulation of intestinal lipid absorption. I.Development of intestinal lipid absorption: cellular events in chylomicron assembly and secretion. Am J Physiol Gastrointest Liver Physiol. 2007;293:G519–G524. doi: 10.1152/ajpgi.00189.2007. [DOI] [PubMed] [Google Scholar]
- 37.Levin MS, Li E, Ong DE, Gordon JI. Comparison of the tissue-specific expression and developmental regulation of two closely linked rodent genes encoding cytosolic retinol-binding proteins. J Biol Chem. 1987;262:7118–7124. [PubMed] [Google Scholar]
- 38.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Mariadason JM, Rickard KL, Barkla DH, Augenlicht LH, Gibson PR. Divergent phenotypic patterns and commitment to apoptosis of Caco-2 cells during spontaneous and butyrate-induced differentiation. J. Cell. Physiol. 2000;183(3):347–354. doi: 10.1002/(SICI)1097-4652(200006)183:3<347::AID-JCP7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 40.Barila D, Murgia C, Nobili F, Perozzi G. Transcriptional regulation of the ezrin gene during rat intestinal development and epithelial differentiation. Biochim Biophys Acta. 1995;1263:133–140. doi: 10.1016/0167-4781(95)00090-4. [DOI] [PubMed] [Google Scholar]
- 41.Cant JP, McBride BW, Croom WJ., Jr The regulation of intestinal metabolism and its impact on whole animal energetics. J Anim Sci. 1996;74:2541–2553. doi: 10.2527/1996.74102541x. [DOI] [PubMed] [Google Scholar]
- 42.Cheeseman CI, O'Neill D. Basolateral D-glucose transport activity along the crypt-villus axis in rat jejunum and upregulation induced by gastric inhibitory peptide and glucagon-like peptide-2. Exp Physiol. 1998;83:605–616. doi: 10.1113/expphysiol.1998.sp004142. [DOI] [PubMed] [Google Scholar]
- 43.Drozdowski LA, Thomson AB. Intestinal sugar transport. World J Gastroenterol. 2006;12:1657–1670. doi: 10.3748/wjg.v12.i11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang ES, Hirayama BA, Wright EM. Distribution of the SGLT1 Na+/glucose cotransporter and mRNA along the crypt-villus axis of rabbit small intestine. Biochem Biophys Res Commun. 1991;181:1208–1217. doi: 10.1016/0006-291x(91)92067-t. [DOI] [PubMed] [Google Scholar]
- 45.Davidson NO, Hausman AM, Ifkovits CA, Buse JB, Gould GW, Burant CF, Bell GI. Human intestinal glucose transporter expression and localization of GLUT5. Am J Physiol. 1992;262:C795–C800. doi: 10.1152/ajpcell.1992.262.3.C795. [DOI] [PubMed] [Google Scholar]
- 46.Jiang L, David ES, Espina N, Ferraris RP. GLUT-5 expression in neonatal rats: crypt-villus location and age-dependent regulation. Am J Physiol Gastrointest Liver Physiol. 2001;281:G666–G674. doi: 10.1152/ajpgi.2001.281.3.G666. [DOI] [PubMed] [Google Scholar]
- 47.Ferraris RP. Dietary and developmental regulation of intestinal sugar transport. Biochem J. 2001;360:265–276. doi: 10.1042/0264-6021:3600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rich-Denson C, Kimura RE. Evidence in vivo that most of the intraluminally absorbed glucose is absorbed intact into the portal vein and not metabolized to lactate. Biochem J. 1988;254:931–934. doi: 10.1042/bj2540931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stumpel F, Burcelin R, Jungermann K, Thorens B. Normal kinetics of intestinal glucose absorption in the absence of GLUT2: evidence for a transport pathway requiring glucose phosphorylation and transfer into the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:11330–11335. doi: 10.1073/pnas.211357698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lenaerts K, Sokolovic M, Bouwman FG, Lamers WH, Mariman EC, Renes J. Starvation induces phase-specific changes in the proteome of mouse small intestine. J Proteome Res. 2006;5:2113–2122. doi: 10.1021/pr060183+. [DOI] [PubMed] [Google Scholar]
- 51.Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- 53.Karhausen J, Furuta GT, Tomaszewski JE, Johnson RS, Colgan SP, Haase VH. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest. 2004;114:1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, Cheung KH, Miller P, Gerstein M, Roeder GS, Snyder M. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 56.Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, Charafe-Jauffret E, Loriod B, Bachelart L, Montfort J, Victorero G, Viret F, Ollendorff V, Fert V, Giovaninni M, Delpero JR, Nguyen C, Viens P, Monges G, Birnbaum D, Houlgatte R. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 57.Paoni NF, Feldman MW, Gutierrez LS, Ploplis VA, Castellino FJ. Transcriptional profiling of the transition from normal intestinal epithelia to adenomas and carcinomas in the APCMin/+ mouse. Physiol. Genomics. 2003;15:228–235. doi: 10.1152/physiolgenomics.00078.2003. [DOI] [PubMed] [Google Scholar]
- 58.Clarke PA, George ML, Easdale S, Cunningham D, Swift RI, Hill ME, Tait DM, Workman P. Molecular pharmacology of cancer therapy in human colorectal cancer by gene expression profiling. Cancer Res. 2003;63:6855–6863. [PubMed] [Google Scholar]
- 59.Maxwell PJ, Longley DB, Latif T, Boyer J, Allen W, Lynch M, McDermott U, Harkin DP, Allegra CJ, Johnston PG. Identification of 5-fluorouracil-inducible target genes using cDNA microarray profiling. Cancer Res. 2003;63:4602–4606. [PubMed] [Google Scholar]
- 60.Fleet JC, Wang L, Vitek O, Craig BA, Edenberg HJ. Gene expression profiling of Caco-2 BBe cells suggests a role for specific signaling pathways during intestinal differentiation. Physiol. Genomics. 2003;13:57–68. doi: 10.1152/physiolgenomics.00152.2002. [DOI] [PubMed] [Google Scholar]
- 61.Bates MD, Erwin CR, Sanford LP, Wiginton D, Bezerra JA, Schatzman LC, Jegga AG, Ley-Ebert C, Williams SS, Steinbrecher KA, Warner BW, Cohen MB, Aronow BJ. Novel genes and functional relationships in the adult mouse gastrointestinal tract identified by microarray analysis. Gastroenterology. 2002;122:1467–1482. doi: 10.1053/gast.2002.32975. [DOI] [PubMed] [Google Scholar]