Abstract
Muscle regeneration following injury is dependent on the ability of muscle satellite cells to activate, proliferate and fuse with damaged fibres. This process is controlled by the myogenic regulatory factors (MRF). Little is known about the temporal relation of the MRF with the expression of known myogenic growth factors (i.e. IGF-1) in humans following muscle damage. Eight subjects (20.6 ± 2.1 years; 81.4 ± 9.8 kg) performed 300 lengthening contractions (180 deg s−1) of their knee extensors in one leg on a dynamometer. Blood and muscle samples were collected before and at 4 (T4), 24 (T24), 72 (T72) and 120 h (T120) post-exercise. Mechano growth factor (MGF), IGF-1Ea and IGF-1Eb mRNA were quantified. Serum IGF-1 did not change over the post-exercise time course. IGF-1Ea and IGF-1Eb mRNA increased ∼4- to 6-fold by T72 (P < 0.01) and MGF mRNA expression peaked at T24 (P = 0.005). MyoD mRNA expression increased ∼2-fold at T4 (P < 0.05). Myf5 expression peaked at T24 (P < 0.05), while MRF4 and myogenin mRNA expression peaked at T72 (P < 0.05). Myf5 expression strongly correlated with the increase in MGF mRNA (r2= 0.83; P = 0.03), while MRF4 was correlated with both IGF-1Ea and -Eb (r2= 0.90; r2= 0.81, respectively; P < 0.05). Immunofluorescence analysis showed IGF-1 protein expression localized to satellite cells at T24, and to satellite cells and the myofibre at T72 and T120; IGF-1 was not detected at T0 or T4. These results suggest that the temporal response of MGF is probably related to the activation/proliferation phase of the myogenic programme as marked by an increase in both Myf5 and MyoD, while IGF-1Ea and -Eb may be temporally related to differentiation as marked by an increase in MRF4 and myogenin expression following acute muscle damage.
Muscle repair/regeneration following injury is dependent on the action of muscle satellite (stem) cells to activate, proliferate and terminally differentiate. The paired box transcription factor Pax7 is expressed in quiescent and activated satellite cells and directs the induction of target genes such as Myf5 that regulate entry of satellite cells into the myogenic programme (Kuang et al. 2006; McKinnell et al. 2008). Myogenesis is controlled by a group of transcriptional networks known as the myogenic regulatory factors (MRF) (Rudnicki et al. 1993; Holterman & Rudnicki, 2005). There are four MRF involved in adult myogenesis, MyoD, Myf5, MRF4 and myogenin. MyoD and Myf5 are transcription factors known to regulate myoblast proliferation while MRF4 and myogenin are transcription factors that regulate terminal differentiation (for review see Holterman & Rudnicki, 2005). The time course for MRF expression is well defined in cell culture; however, data from humans are less clear (Psilander et al. 2003; Bickel et al. 2005). The myogenic commitment of satellite cells is marked by the presence of Myf5, which is expressed early after mitogenic activation. The proliferative phase of the myogenic programme is associated with the coexpression of Myf5 and MyoD (Cooper et al. 1999; Sabourin et al. 1999; Yablonka-Reuveni et al. 1999). The subsequent down-regulation of Myf5 and MyoD is associated with the induction of differentiation, while the up-regulation of myogenin and MRF4 is known to direct terminal differentiation.
The MRF expression time course in human tissue is not well defined, in part due to the varying models used to examine MRF expression, but also due to the fact that MRF expression profiles are confounded by non-satellite cell myofibrillar gene expression (Psilander et al. 2003; Kadi et al. 2004; Bickel et al. 2005; Yang et al. 2005; Crameri et al. 2007). Although exercise has been used to examine MRF gene expression in humans (Psilander et al. 2003; Bickel et al. 2005; Yang et al. 2005; Crameri et al. 2007), there is a paucity of information on the time course of MRF expression following contraction-induced muscle damage. There is even less information on the temporal association between potential satellite cell regulators, such as insulin-like growth factor-1 (IGF-1), and MRF expression in human tissue in the context of muscle repair and regenesis.
The expression of the MRF appears to be regulated, in part by the mitotic and myogenic activity of locally produced isoforms of IGF-1 (Chakravarthy et al. 2000; Mourkioti & Rosenthal, 2005). IGF-1 is a growth factor found in the circulation (produced in the liver) and locally in muscle. Three specific isoforms of IGF-1, IGF-1Ea, IGF-1Eb and IGF-1Ec have been described in muscle and each may contribute in some extent to muscle regeneration (Rotwein et al. 1986; Yang et al. 1996; Chakravarthy et al. 2000). IGF-1Eb may be involved in hypertrophy or muscle regeneration; however, the physiological role of IGF-1Eb in human skeletal muscle is currently unknown. IGF-1Ec or mechano growth factor (MGF) is a splice variant of IGF-1Ea that may have both autocrine and paracrine functions stimulating satellite cell proliferation and muscle hypertrophy following muscle stretch or muscle damage (Yang et al. 1996; Hill & Goldspink, 2003). IGF-1Ea and MGF expression have been shown to increase as early as 24 h after exercise and injury in rats (Hill & Goldspink, 2003; Hill et al. 2003); however, results from human studies are conflicting (Bickel et al. 2005; Greig et al. 2006). Information regarding the response of IGF-1Ea to exercise is equivocal (Hill & Goldspink, 2003; Hill et al. 2003; Hameed et al. 2003; Psilander et al. 2003; Bickel et al. 2005; Greig et al. 2006). There is little evidence regarding the expression of IGF-1Eb in human muscle following exercise, and there is no information on how the IGF-1 isoforms relate to MRF expression in humans.
The aims of the current study were to: (1) investigate the time course of expression for IGF-1Ea, IGF-1Eb and MGF in human muscle following intense damaging muscle contractions; (2) investigate the time course of expression for MyoD, Myf5, MRF4 and myogenin in human muscle following damaging exercise; and (3) determine if there were any relationships between the temporal expression of the IGF-1 isoforms and MRF in the post-exercise period. Based on available data from humans we hypothesized that both MyoD and Myf5 would be expressed early following injury (within 24 h) and that MRF4 and myogenin expression would occur after Myf5 and MyoD. Further, MGF and IGF-1 expression would increase early after exercise and remain elevated for a number of days post-exercise.
Methods
Subjects
Eight healthy males (age 20.6 ± 2.1 years, height 180.5 ± 5.2 cm, weight 81.4 ± 9.8 kg) were recruited from the McMaster University community. Subjects underwent a routine medical screening, completed a health questionnaire and were not involved in any lower body resistance training for at least 6 months prior to beginning the study. Subjects were told to refrain from exercising throughout the time course of the study. All subjects were informed of the procedures and potential risks associated with the study and gave their written informed consent to participate. This study was approved by the Hamilton Health Sciences Research Ethic Board and conforms to the Declaration of Helsinki concerning the use of human subjects as research participants.
Muscle damage protocol
Maximal isokinetic unilateral muscle-lengthening contractions of the quadriceps femoris were performed using the Biodex dynamometer (Biodex-System 3, Biodex Medical Systems, Inc., USA) at 180 deg s−1. For each subject, one leg was selected randomly to perform the exercise protocol described below. Movement at the shoulders, hips and thigh were restrained with straps in order to isolate the knee extensors during the protocol.
Immediately prior to the intervention, subjects underwent a brief familiarization with the equipment, involving 5–10 submaximal lengthening contractions of the leg to be exercised. Subjects were required to perform 30 sets of 10 maximal knee extensions with 1 min rest between sets, for a total of 300 lengthening contractions. During each set, investigators provided verbal encouragement for the subjects to complete and exert maximal force during each contraction. This protocol has been previously shown to induce a significant level of skeletal muscle damage (Beaton et al. 2002).
Muscle biopsies
Five percutaneous needle biopsies were obtained from the mid-portion of the vastus lateralis under local anaesthetic (1% lidocaine (lignocaine)). One muscle biopsy was obtained prior to the intervention. Baseline measures were generated from the pre-intervention biopsy (PRE) taken in the morning prior to beginning the exercise session. Four biopsies were obtained from the working leg at different time points following the intervention. Muscle biopsies and blood samples were taken concurrently at 4 h (T4), 24 h (T24), 72 h (T72) and 120 h (T120) post-intervention from a new incision 3–5 cm proximal to the last biopsy site. Approximately 150 mg of muscle tissue was collected from each biopsy using manual suction. Following collection of the sample, the muscle was dissected free of adipose and connective tissue and flash-frozen in liquid nitrogen, and stored at −80°C for later analysis. For immunohistochemistry, a fresh piece of muscle (approximately 20 mg) was dissected from the biopsies, orientated in cross-section, mounted in OCT compound (Tissue-Tek, Sakura Finetek, USA) and frozen in isopentane cooled with liquid nitrogen. The mounted samples were stored at −80°C and then sectioned (7 μm) at −20°C. The cross-sections were mounted on slides and stored at −80°C for immunohistochemical analysis.
Immunohistochemistry/immunofluorescence
Immunohistochemistry/immunofluorescent staining procedures were modified from previously published methods (Singh et al. 1999; Reimann et al. 2004). Briefly, 7 μm sections were fixed with 2% paraformaldehyde (PFA, Sigma, USA) for 10 min followed by several washes in 1 × PBS. Sections were then covered for 30 min in a blocking solution containing: 2% bovine serum albumin, 5% fetal bovine serum, 0.2% Triton X-100, 0.1% sodium azide followed by incubation in the Pax7 antibody (DSHB, USA) overnight at 4°C. Sections were washed in 1 × PBS and incubated in the secondary antibody (AlexaFluor 594 goat anti-mouse; 1: 200, Invitrogen, Molecular Probes Inc., USA) for 2.5 h at room temperature. After washing, sections were re-fixed with 2% PFA to prevent migration of the secondary antibody. Sections were then incubated for 30 min in a blocking solution containing 10% goat serum (Sigma, USA) and 0.2% Triton X-100. Sections were then incubated with an anti-human IGF-1 antibody (1: 50, Chemicon, Millipore, USA) for 1.5 h at room temperature. After washing in PBS, sections were incubated for 1 h with a polyclonal goat anti-mouse immunoglobulin biotinylated secondary antibody (Dako Canada, Inc., Canada). After washing in PBS, sections were then incubated for 1 h with a streptavidin–FITC fluorochrome (Biosource, USA). Sections were then washed with PBS and DAPI for nuclear staining. Sections were mounted with a fluorescent mounting medium (Dako Canada, Inc., Canada) to preserve the fluorescence signal. Stained slides were viewed with the Nikon Eclipse 90i Microscope (Nikon Instruments, Inc., USA) and images were captured using the Nikon NIS Elements 3.0 software (Nikon). Stains were compared to secondary-only negative controls to ensure the staining patterns observed were not due to non-specific binding of the secondary antibodies.
Blood measures
A resting blood sample was obtained from the antecubital vein immediately prior to the intervention. Blood was also drawn at T4, T24, T72 and T120. Approximately 20 ml of blood was collected and separated into one heparinized and one non-heparinized vacutainer tube at each time-point. Samples were separated into 50 μl aliquots, flash-frozen in liquid nitrogen, and stored at −80°C for analysis at a later date. Serum samples were analysed for IGF-1 protein using commercially available Enzyme-Linked ImmunoSorbant Assay (ELISA) kits following the manufacturer's instructions (R & D Systems, Inc., USA).
RNA isolation
RNA was isolated from homogenized muscle samples using the TRIzol method. Briefly, a total of 1.0 ml of TRIzol Reagent (Invitrogen Corporation, Canada) was added to the homogenizer. Approximately 25 mg (average weight 25.33 ± 1.67 mg) of each muscle sample was cut individually in liquid nitrogen using an RNase-free razor blade treated with Ambion RNaseZap (Ambion Biosystems, USA). Each muscle sample was ground and the muscle homogenate was centrifuged at 12 000 g and 4°C for 10 min. The clear supernatant was allowed to incubate at room temperature for 15 min. Chloroform (200 μl) was mixed with the supernatant and incubated at room temperature for 2–3 min, then centrifuged at 12000 g for 15 min at 4°C. The aqueous phase was transferred to a fresh tube, allowed to incubate with 500 μl isopropyl alcohol for 10 min at room temperature, then centrifuged at 12000 g and 4°C for 10 min. The supernatant was removed and the resulting RNA pellet was washed in 1.0 ml of 75% ethanol, resuspended, and centrifuged at 7500 g for 5 min. The pellet was air-dried and dissolved in 10 μl of sterile double-distilled water (ddH2O) treated with diethylpyrocarbonate.
The resulting RNA solution was quantified for RNA purity and concentration using a spectrophotometer (Ultrospec 3000 pro UV, Amersham Biosciences, USA). RNA quality was assessed on a subset of random samples using denaturing gel electrophoresis. After this, quantification samples were stored at −80°C for later analysis.
Reverse transcription (RT)
In 0.2 ml Eppendorf tubes, 20 μl RT reactions were set up on ice for each individual sample using a commercially available kit (Applied Biosystems High Capacity cDNA Reverse Transcription Kit; Applied Biosystems, USA) according to the manufacturer's instructions. Briefly, 10 μl of RNA pre-diluted to 100 ng μl−1 was added to 10 μl of a master mixture containing 2.0 μl of 10 × RT buffer, 0.8 μl of 25 × dNTP, 2.0 μl of 10 × RT random primers, 1 μl of MultiScribe reverse transcriptase and 4.2 μl of nuclease-free H2O. The cDNA synthesis reaction was carried out using an Eppendorf Mastercycle epgradient thermal cycler (Eppendorf Canada). Following RT, samples were stored at −20°C until further analysis.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Reactions (25 μl) were set up in 0.2 ml Stratagene PCR tubes (Stratagene, USA) for each individual reaction. Reactions were run in duplicate for each time-point. Primers were custom-made using published sequences (Table 1) and were re-suspended in 1 × TE buffer (10 mm Tris-HCl, 0.11 mm EDTA) and stored at −20°C prior to use. In each PCR tube, 1.0 μl of cDNA and 7.5 μl of ddH2O were added to 16.5 μl of a master mix containing 12.5 μl of RT2 Real-Time SYBR Green/Rox PCR master mix (SuperArray Bioscience Corp., USA), 2 μl of the specific forward primer and 2 μl of the specific reverse primer. qRT-PCR reactions were carried out using a Stratagene Mx3000P real-time PCR system (Stratagene, USA). Reactions were set up using Stratagene MxPro QPCR Software version 3.00 (Stratagene, USA). Fold changes in gene expression were calculated using the delta-delta Ct method (Livak & Schmittgen, 2001). Briefly, Ct values were first normalized to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Table 1). Ct values normalized to GAPDH are expressed as delta-Cts (ΔCt). ΔCt values were then normalized to PRE values, expressed as delta-delta Cts (ΔΔCt). Values were then transformed out of the logarithmic scale using the formula: fold change = 2−ΔΔCt (Livak & Schmittgen, 2001). Thus, mRNA values are expressed as a fold change from PRE (mean ±s.e.m.). GAPDH expression was not different from PRE at any of the post-intervention time-points.
Table 1.
qRT-PCR primer sequences
| Gene name | Forward sequence | Reverse sequence |
|---|---|---|
| IGF-1Ea | 5′-GACATGCCCAAGACCCAGAAGGA-3′ | 5′-CGGTGGCATGTCACTCTTCACTC-3′ |
| IGF-1Eb | 5′-GCCCCCATCTACCAACAAGAACAC-3′ | 5′-CAGACTTGCTTCTGTCCCCTCCTTC-3′ |
| MGF | 5′-GCCCCCATCTACCAACAAGAACAC-3′ | 5′-CGGTGGCATGTCACTCTTCACTC-3′ |
| MyoD | 5′-GGTCCCTCGCGCCCAAAAGAT-3′ | 5′-CAGTTCTCCCGCCTCTCCTAC-3′ |
| Myf5 | 5′-ATGGACGTGATGGATGGCTG-3′ | 5′-GCGGCACAAACTCGTCCCCAA-3′ |
| MRF4 | 5′-CCCCTTCAGCTACAGACCCAA-3′ | 5′-CCCCCTGGAATGATCGGAAAC-3′ |
| Myogenin | 5′-CAGTGCACTGGAGTTCAGCG-3′ | 5′-TTCATCTGGGAAGGCCACAGA-3′ |
| GAPDH | 5′-CCTCCTGCACCACCAACTGCTT-3′ | 5′-GAGGGGCCATCCACAGTCTTCT-3′ |
IGF-1, insulin-like growth factor-1; MGF, mechano growth factor (IGF-1Ec); MyoD, myogenic determination factor; Myf5, myogenic factor-5; MRF4, myogenic regulatory factor-4; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Statistical analysis
Statistical analysis was performed using SigmaStat 3.0 analysis software (Systat, SPSS Inc., USA). Serum IGF-1 concentrations, MRF mRNA and IGF-1 splice variant mRNA were analysed using a one-way repeated measures analysis of variance (ANOVA). Correlations were analysed using the Pearson product moment correlation. Statistical significance was accepted at P < 0.05. Differences between the expressions of the different IGF-1 splice variants at a given time-point was analysed with a t test. Significant interactions and main effects were analysed using Tukey's HSD post hoc test. All results are presented as means ±s.e.m.
Results
Serum IGF-1
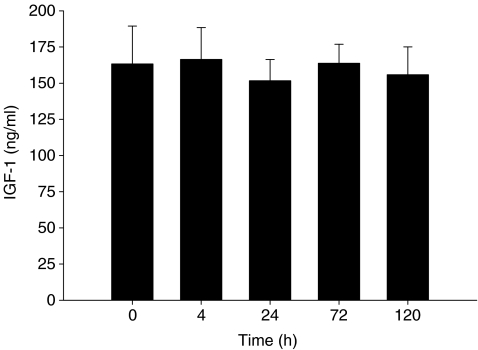
Serum IGF-1 concentration was not different from rest at any time-point following the exercise intervention (Fig. 1A).
Figure 1.
Average serum insulin-like growth factor-1 (IGF-1) concentration. Time 0 h corresponds to pre-exercise serum concentrations; all other time-points correspond to post-exercise time (h). Values are reported as mean ±s.e.m.
MRF mRNA
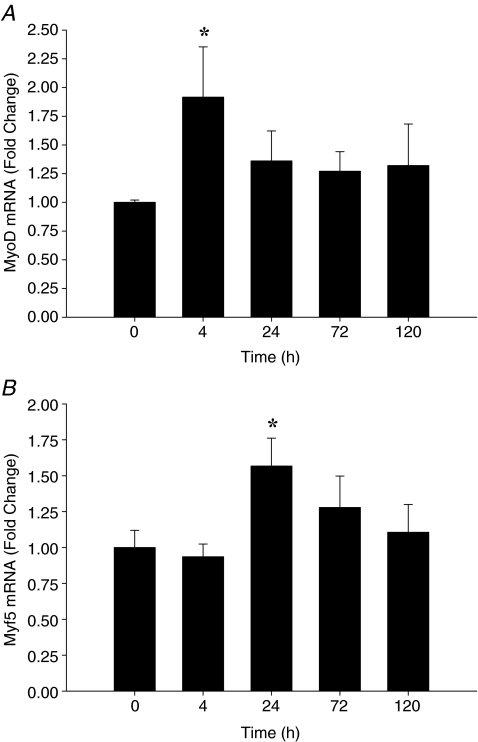
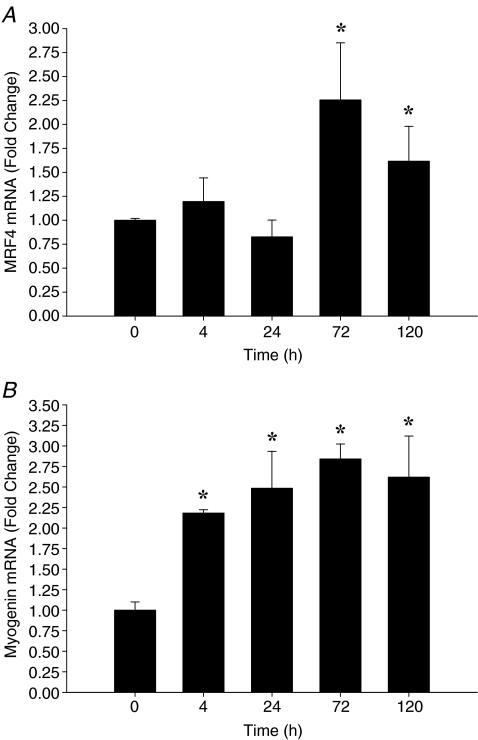
Following muscle-lengthening contractions, gene expression of MyoD and Myf5 increased early following exercise, while MRF4 and myogenin increased at later time-points following the intervention. MyoD mRNA expression increased approximately 2-fold by T4 (P = 0.001) (Fig. 2A) with a trend to remain elevated at T24 (P = 0.07). There were no significant differences between MyoD mRNA expression from PRE at T72 or T120. Myf5 mRNA expression peaked at T24 (P < 0.05), followed by a gradual return to PRE expression levels by T120 (Fig. 2B). MRF4 mRNA expression increased ∼2.3-fold at T72 (P = 0.039) and remained ∼1.5-fold higher than PRE at T120 (Fig. 3A). Myogenin mRNA expression increased at T4 and T24 (P < 0.05) and peaked at T72 (∼2.8-fold, P = 0.009). Myogenin expression remained ∼2.5-fold higher than PRE (P = 0.015) at T120 (Fig. 3B).
Figure 2.
Relative expression of MyoD mRNA (A) and Myf5 mRNA (B) expression following exercise, expressed as fold change from 0 h (pre-exercise). Data are normalized to GAPDH and reported as mean ±s.e.m.*P < 0.05 versus 0 h.
Figure 3.
Relative expression of MRF4 mRNA (A) and myogenin mRNA (B) expression following exercise, expressed as fold change from 0 h (pre-exercise). Data are normalized to GAPDH and reported as mean ±s.e.m.*P < 0.05 versus 0 h.
IGF-1 mRNA
IGF-1Ea mRNA expression increased ∼4-fold at T72 (P = 0.004) and began to decrease by T120 (Fig. 4A). Interestingly, the change in IGF-1Eb mRNA expression from baseline was greater than the change in IGF-1Ea expression from baseline at T72 and T120 (P < 0.05), increasing ∼5.8-fold at T72 (P < 0.001), and remained elevated (∼3.5-fold above PRE, P = 0.035) at T120 (Fig. 4B). MGF mRNA expression increased 48 h before increases in IGF-1Ea or IGF-1Eb were detected, increasing ∼3.3-fold at T24 (P = 0.005), which was also significantly greater than T4 (P = 0.009). MGF expression decreased by T72; however, there was a trend towards MGF remaining elevated above PRE levels at both T72 and T120 (Fig. 4C).
Figure 4.
Relative expression of IGF-1Ea mRNA (A), IGF-1Eb mRNA (B) and mechano growth factor (MGF) mRNA (C) expression following exercise, expressed as fold change from 0 h (pre-exercise). Data are normalized to GAPDH and reported as mean ±s.e.m.*P < 0.05 versus 0 h; †P < 0.05 versus 4 h; ‡P < 0.05 versus IGF-1Ea at T72 and T120.
Correlations
In an attempt to determine the potential temporal relation between MRF and the IGF-1 isoforms, linear Pearson correlations were performed between variables. The significant correlations are described below.
MGF expression and the MRF
MGF expression showed a positive correlation (r = 0.91, r2= 0.83, P = 0.03) with the expression of Myf5 following muscle damage (Fig. 5A). MGF expression occurred earlier than the other IGF-1 splice variants suggesting that MGF may have a distinct role in muscle repair following contraction-induced damage.
Figure 5.
A, Pearson correlation of the time course of MGF mRNA expression (fold change) versus the time course of Myf5 mRNA expression (fold change) (r2= 0.83; P = 0.03). B, Pearson correlation of the time course of IGF-1Ea mRNA expression (fold change) versus the time course of MRF4 mRNA expression (fold change) (r2= 0.90; P = 0.015). C, Pearson correlation of the time course of IGF-1Eb mRNA expression (fold change) versus the time course of MRF4 mRNA expression (fold change) (r2= 0.81; P = 0.037). Correlations are representative of the individual data points and are presented as mean values (•) ±s.d. (error bars).
IGF-1Ea and IGF-1Eb expression and the MRF
Muscle IGF-1Ea and -Eb expression positively correlated with the expression of MRF4 (r = 0.95, r2= 0.90, P = 0.015 versus r = 0.90, r2= 0.81, P = 0.037; IGF-1Ea and IGF-1Eb, respectively; Fig. 5B and C).
IGF-1/Pax7 immunofluorescence
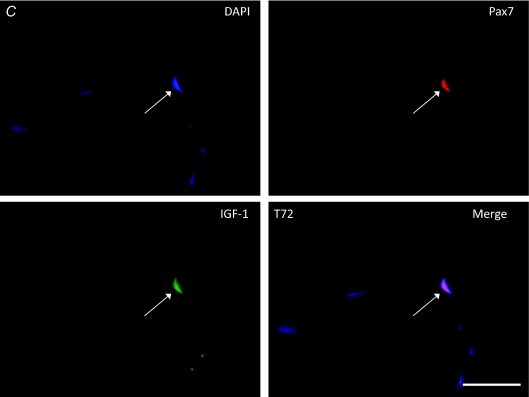
Baseline muscle sections (T0) and sections from T4 co-stained for IGF-1 and Pax7 had no detectable IGF-1 present in the fibre, or the satellite cell (as evaluated by Pax7/DAPI co-staining). Figure 6A shows a representative cross-section at T0 lacking IGF-1 expression. At T24, IGF-1 was detected in the majority of Pax7-positive cells (Fig. 6B); however, IGF-1 was undetectable in the myofibre. By T72 (Fig. 6C) and T120 (not shown) IGF-1 was detected in the majority of Pax7-positive cells examined and also diffusely present in the myofibre.
Figure 6.
Immunofluorescent staining of muscle cross-sections triple-stained for nuclei (DAPI), satellite cells (Pax7; TRITC), and IGF-1 protein expression (FITC) at T0 (A; scale bar = 20 μm, arrows denote Pax7+/DAPI+ satellite cells), T24 (B; scale bar = 20 μm), T72 (C; scale bar = 20 μm). For A–C, arrows denote satellite cells (Pax7+/DAPI+) (B and C) expressing IGF-1 protein. Note: no IGF-1 present at T0 (and T4, not shown) and diffuse IGF-1 staining in the myofibre at T72 and T120 (not shown).
Discussion
This study examined the temporal relation of all three IGF-1 splice variants and the expression of the MRF, in vivo, following an intense muscle-lengthening protocol designed specifically to elicit damage in human skeletal muscle. Although IGF-1Ea and MGF mRNA expression have been examined following exercise in humans (Psilander et al. 2003; Bickel et al. 2005; Greig et al. 2006), no study has yet examined all of the individual splice variants of IGF-1 in the context of muscle stem cell regulation over such an extended time course. Furthermore, this is the first study to our knowledge to directly show the expression of IGF-1 in Pax7-positive cells in human muscle in vivo.
MRF expression following exercise
The differential expression of the myogenic regulatory factors following an acute muscle-lengthening protocol in the present study was similar to the expression profiles observed in animal and cell culture studies (Smith et al. 1994; Cooper et al. 1999; Yablonka-Reuveni et al. 1999), with both MyoD and Myf5 up-regulated early (∼2-fold) following exercise (4–24 h) followed by the induction of MRF4 (72 h) and the peak expression of myogenin (72 h). Although the time-line of MRF expression in the present study is similar to observations made in animal and cell culture studies (Cooper et al. 1999; Smith et al. 1994; Yablonka-Reuveni et al. 1999), MRF data from human exercise studies are equivocal. In addition, few data exist with regards to Myf5 expression and the post-exercise response in humans. Yang et al. (2005) measured Myf5 following heavy resistance exercise, but they did not observe any changes in Myf5 mRNA expression. Previous studies measuring MRF expression following exercise have reported increases in MyoD immediately following exercise, returning to baseline at 2 h post-exercise; to an increase at 12 h which is sustained after 24 h (Psilander et al. 2003; Bickel et al. 2005). Furthermore, other studies have reported increased MRF4 and myogenin expression as early as 2–6 h following exercise, returning to baseline levels by 48 h post-exercise (Psilander et al. 2003; Bickel et al. 2005). In agreement with rodent and in vitro data, our data show MRF4 mRNA expression increased ∼72 h after exercise (Smith et al. 1994). MRF4 has been shown to act upstream of myogenin, with an increase in MRF4 mRNA leading to an increased myogenin expression (Rhodes & Konieczny, 1989). Our data support the temporal association of the expression of MRF4 and myogenin, with myogenin being significantly elevated 72–120 h post-exercise.
Although we reported an increase in myogenin expression 4–24 h following the intervention it is important to note that unlike data from in vitro studies, myogenin may be expressed in post-mitotic fibres, in vivo, following exercise (Kadi et al. 2004). Our data may differ from previous findings due to the nature of the intervention. This study employed a muscle-lengthening model, which has been previously shown to induce significant muscle damage (Beaton et al. 2002). Thus the stimulus for inducing satellite cell activation and MRF expression may have been much greater in this study compared to the lower-volume mixed concentric–eccentric protocols performed in other studies (Psilander et al. 2003; Bickel et al. 2005; Greig et al. 2006).
In addition our intervention employed a dynamometer which allowed us to isolate the quadriceps group and limit the contribution from other muscles, as opposed to the free movement that occurs during regular resistance exercise.
IGF-1 splice variants following exercise
In the present study the IGF-1 splice variants were differentially expressed following the intervention. MGF mRNA increased significantly 24 h after the intervention, while the expression of both IGF-1Ea and IGF-1Eb mRNA were not elevated until 72 h post-intervention. In addition, IGF-1Eb was elevated 120 h following the intervention, while IGF-1Ea was not, suggesting that IGF-1Ea and IGF-1Eb may have specific roles following muscle injury. Immunofluorescent staining of muscle cross-sections with a pan-IGF-1 antibody showed IGF-1 detectable in the satellite cell compartment at 24 h, with IGF-1 becoming detectable in the myofibre (diffuse staining) and satellite cell at 72 and 120 h, the timing of which is in line with the IGF-1 splice variant mRNA expression. Unfortunately, there is no specific antibody for MGF, thus we are unable to discern the different splice variant protein expressions from our immunofluorescent staining. Our IGF-1Ea and MGF data are in agreement with the expression of IGF-1Ea and MGF following exercise in a mouse model, where MGF increased ∼24 h after exercise and IGF-1Ea was increased 5 days after exercise (Hill & Goldspink, 2003; Hill et al. 2003). The authors suggested that MGF was related to the activation of satellite cells, while IGF-1Ea was related to the increased need for protein synthesis in late differentiation (Hill & Goldspink, 2003). Our results agree with this notion since MGF expression was positively correlated with the expression of Myf5, which is robustly expressed in proliferating myoblasts (Cooper et al. 1999). Furthermore, the time-line of IGF-1Ea expression correlated strongly with the expression of MRF4 which marks the commitment of proliferating myoblasts to terminal differentiation (Rhodes & Konieczny, 1989; Smith et al. 1994). This suggests that the expression of MRF4 and the IGF-1 isoforms Ea and Eb follow similar temporal profiles following muscle damage, the time-line of which is similar to the beginning of terminal differentiation of satellite cells after activation (Holterman & Rudnicki, 2005). Although our results agree with animal studies, they differ significantly from other work published on exercise and IGF-1 in human skeletal muscle (Hameed et al. 2003; Psilander et al. 2003; Greig et al. 2006).
MGF expression has been reported to have a high degree of variability in expression following various contraction protocols in humans (Hill & Goldspink, 2003; Psilander et al. 2003; Bickel et al. 2005; Greig et al. 2006). The conflicting results within the literature may be attributed to the differences in the intensity of the exercise stimuli as well as the post-exercise time course examined. In the present study, we chose an intense muscle-lengthening protocol that has been previously shown to induce significant muscle damage (Beaton et al. 2002). Furthermore, we chose an extensive post-contraction time course aimed at capturing satellite cell activation, expansion and differentiation (Rhodes & Konieczny, 1989; Smith et al. 1994; Cooper et al. 1999; Yablonka-Reuveni et al. 1999).
IGF-1Eb expression was up-regulated 72 h after damaging exercise and remained elevated at 120 h post-exercise. The correlation with MRF4 was robust suggesting a temporal relationship between IGF-1Eb expression and myoblast differentiation. Furthermore, the change in expression of IGF-1Eb was greater compared to IGF-1Ea at T72 (∼5.8-fold versus∼4-fold, P < 0.05) and remained significantly elevated longer (i.e. up to 120 h post-exercise). Immunofluorescence analysis shows a visible increase in IGF-1 protein in the myofibre and the satellite cell compartment at T72 and T120 compared to pre-exercise staining. At T0 and T4 there was no overt or diffuse IGF-1 staining within the fibre or the satellite cell. There was a discernible increase in diffuse IGF-1 staining in the myofibre at T72 and T120. These findings are of particular interest as they suggest that IGF-1Eb may play an important role in late differentiation, in which the preferential splicing of IGF-1 is directed from Ea to Eb. The physiological significance of IGF-1Eb is unknown at this time; however, it may be induced due to the increased need for protein and mitochondrial biosynthesis required by terminally differentiating cells. IGF-1 has been shown to regulate differentiation in L6A1 myoblasts through the activation of the phosphotidylinositol 3-kinase (PI3-K) signalling (Coolican et al. 1997). Although the temporal expression of IGF-1Ea and IGF-1Eb was not correlated with the increase in myogenin expression, the IGF-1 activation of PI3-K/Akt signalling has been shown to stimulate the expression of myogenin in vitro (Xu & Wu, 2000; Wilson et al. 2004). The elevated expression of IGF-1Ea and -Eb at 72 h post-intervention coincides with the peak myogenin expression (T72). It is possible that IGF-1Ea and -Eb may influence myogenin expression in the proliferating myoblasts during the post-intervention time course; however, we are not able to discern this from our whole-muscle data. Since myogenin is expressed in post-mitotic muscle following acute exercise (Kadi et al. 2004), it is possible that the unexpected expression profile of myogenin and lack of a significant correlation with IGF-1 was due to the expression of myogenin in the post-mitotic tissue which may have a different expression profile as compared to myogenin in the myoblasts. At this time, more work is needed to examine the effect of IGF-1Eb expression on the activation of the PI3-K/Akt pathway and myoblast differentiation in isolated myoblasts.
IGF-1 has also been shown to stimulate the proliferation of satellite cells in vitro through the activation of mitogen-activated protein kinase (MAPK) (Coolican et al. 1997). The results of the present study suggest that the IGF-1Ea and -Eb isoforms were not related to proliferation as the expression of both isoforms were not elevated before T72. Furthermore, circulating IGF-1 was not elevated at any time-point following exercise, suggesting that activation/proliferation of satellite cells was not due to an increased blood IGF-1 concentration. IGF-1 has been shown to increase differentiation in primary human myoblasts in culture (Ates et al. 2007). In that investigation, the expression of MGF was shown to block myoblast differentiation, thus enhancing proliferation (Ates et al. 2007). Furthermore, the ectopic addition of MGF-ct24E (a 24 amino acid peptide corresponding to the c-terminal part of the E-domain of MGF) enhanced proliferation of transplanted human myoblasts in vitro (Mills et al. 2007). The authors suggest that the enhanced proliferation was due to a mechanism different from IGF-1 receptor binding. Although it has been shown that MGF does not enhance proliferation through the IGF-1 receptor (Yang & Goldspink, 2002; Mills et al. 2007), it is possible that MGF may activate the MAP kinase pathway via another mechanism yet to be determined. The expression of MGF following resistance exercise has been shown to increase concomitantly with an increase in cyclin D1 mRNA expression (Kim et al. 2005). Cyclin D1 is essential to cell cycle initiation and is up-regulated in response to MAP kinase signalling (Coolican et al. 1997). Further work must be done to understand the mechanism of MGF-stimulated myoblast activation/proliferation, specifically identifying the cellular signalling machinery in isolated human satellite cells in order to understand the role of MGF in regulating the myogenic programme.
From histological analysis of muscle cross-sections, IGF-1 protein was undetectable in the myofibre and satellite cell compartment at baseline. IGF-1 protein began to be coexpressed with Pax7 at T24 with no IGF-1 detected in the myofibre, suggesting the initial appearance of IGF-1 may be confined to the satellite cell compartment. The timing of the initial IGF-1 expression coincides with the peak in MGF mRNA expression and may be related to the proposed role of MGF in increasing myoblast proliferation (Hill & Goldspink, 2003). Interestingly, IGF-1 protein was detectable in both the satellite cell compartment and in the myofibre (diffusely) at T72 and T120 which would agree with the idea that IGF-1 is partially responsible for preparing and directing proliferating satellite cells into terminal differentiation (Hill & Goldspink, 2003; Ates et al. 2007). The coexpression of IGF-1 with Pax7 shows that IGF-1 is present in satellite cells following contraction-induced muscle injury and the correlation of IGF-1 isoforms with MRF expression further confirms the important role of IGF-1 in regulating the satellite cell response to muscle injury.
In conclusion, these results suggest that the temporal response of MGF expression is related to the activation and proliferative phase of the myogenic programme, as marked by a significant increase in MyoD and Myf5 mRNA which is strongly correlated with MGF expression and the timing of which is in line with the coexpression of IGF-1 protein with Pax7 in muscle cross-sections. Additionally, the expression of IGF-1Ea and IGF-1Eb are temporally related to myogenic differentiation as marked by increased myogenin and MRF4 expression which is strongly correlated with increased IGF-1Ea and -Eb expression. The exact mechanisms of MGF and IGF-1Eb signalling are currently unknown. By employing ex vivo methods to study the actions of IGF-1 splice variants in isolated human primary myoblasts after damaging muscle contractions, we may be able to provide key insights into the role of these factors and the regenerative response following muscle damage.
References
- Ates K, Yang SY, Orrell RW, Sinanan AC, Simons P, Solomon A, Beech S, Goldspink G, Lewis MP. The IGF-I splice variant MGF increases progenitor cells in ALS, dystrophic, and normal muscle. FEBS Lett. 2007;581:2727–2732. doi: 10.1016/j.febslet.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Beaton LJ, Tarnopolsky MA, Phillips SM. Contraction-induced muscle damage in humans following calcium channel blocker administration. J Physiol. 2002;544:849–859. doi: 10.1113/jphysiol.2002.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. J Appl Physiol. 2005;98:482–488. doi: 10.1152/japplphysiol.00895.2004. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Abraha TW, Schwartz RJ, Fiorotto ML, Booth FW. Insulin-like growth factor-I extends in vitro replicative life span of skeletal muscle satellite cells by enhancing G1/S cell cycle progression via the activation of phosphatidylinositol 3′-kinase/Akt signaling pathway. J Biol Chem. 2000;275:35942–35952. doi: 10.1074/jbc.M005832200. [DOI] [PubMed] [Google Scholar]
- Coolican SA, Samuel DS, Ewton DZ, McWade FJ, Florini JR. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J Biol Chem. 1997;272:6653–6662. doi: 10.1074/jbc.272.10.6653. [DOI] [PubMed] [Google Scholar]
- Cooper RN, Tajbakhsh S, Mouly V, Cossu G, Buckingham M, Butler-Browne GS. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J Cell Sci. 1999;112:2895–2901. doi: 10.1242/jcs.112.17.2895. [DOI] [PubMed] [Google Scholar]
- Crameri RM, Aagaard P, Qvortrup K, Langberg H, Olesen J, Kjaer M. Myofibre damage in human skeletal muscle: effects of electrical stimulation versus voluntary contraction. J Physiol. 2007;583:365–380. doi: 10.1113/jphysiol.2007.128827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig CA, Hameed M, Young A, Goldspink G, Noble B. Skeletal muscle IGF-I isoform expression in healthy women after isometric exercise. Growth Horm IGF Res. 2006;16:373–376. doi: 10.1016/j.ghir.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Hameed M, Orrell RW, Cobbold M, Goldspink G, Harridge SD. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. J Physiol. 2003;547:247–254. doi: 10.1113/jphysiol.2002.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M, Goldspink G. Expression and splicing of the insulin-like growth factor gene in rodent muscle is associated with muscle satellite (stem) cell activation following local tissue damage. J Physiol. 2003;549:409–418. doi: 10.1113/jphysiol.2002.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M, Wernig A, Goldspink G. Muscle satellite (stem) cell activation during local tissue injury and repair. J Anat. 2003;203:89–99. doi: 10.1046/j.1469-7580.2003.00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman CE, Rudnicki MA. Molecular regulation of satellite cell function. Semin Cell Dev Biol. 2005;16:575–584. doi: 10.1016/j.semcdb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Kadi F, Johansson F, Johansson R, Sjöström M, Henriksson J. Effects of one bout of endurance exercise on the expression of myogenin in human quadriceps muscle. Histochem Cell Biol. 2004;121:329–334. doi: 10.1007/s00418-004-0630-z. [DOI] [PubMed] [Google Scholar]
- Kim JS, Cross JM, Bamman MM. Impact of resistance loading on myostatin expression and cell cycle regulation in young and older men and women. Am J Physiol Endocrinol Metab. 2005;288:E1110–E1119. doi: 10.1152/ajpendo.00464.2004. [DOI] [PubMed] [Google Scholar]
- Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le GF, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills P, Dominique JC, Lafreniere JF, Bouchentouf M, Tremblay JP. A synthetic mechano growth factor E Peptide enhances myogenic precursor cell transplantation success. Am J Transplant. 2007;7:2247–2259. doi: 10.1111/j.1600-6143.2007.01927.x. [DOI] [PubMed] [Google Scholar]
- Mourkioti F, Rosenthal N. IGF-1, inflammation and stem cells: interactions during muscle regeneration. Trends Immunol. 2005;26:535–542. doi: 10.1016/j.it.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Psilander N, Damsgaard R, Pilegaard H. Resistance exercise alters MRF and IGF-I mRNA content in human skeletal muscle. J Appl Physiol. 2003;95:1038–1044. doi: 10.1152/japplphysiol.00903.2002. [DOI] [PubMed] [Google Scholar]
- Reimann J, Brimah K, Schroder R, Wernig A, Beauchamp JR, Partridge TA. Pax7 distribution in human skeletal muscle biopsies and myogenic tissue cultures. Cell Tissue Res. 2004;315:233–242. doi: 10.1007/s00441-003-0833-y. [DOI] [PubMed] [Google Scholar]
- Rhodes SJ, Konieczny SF. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 1989;3:2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- Rotwein P, Pollock KM, Didier DK, Krivi GG. Organization and sequence of the human insulin-like growth factor I gene. Alternative RNA processing produces two insulin-like growth factor I precursor peptides. J Biol Chem. 1986;261:4828–4832. [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary MyoD–/– myogenic cells derived from adult skeletal muscle. J Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MA, Ding W, Manfredi TJ, Solares GS, O'Neill EF, Clements KM, Ryan ND, Kehayias JJ, Fielding RA, Evans WJ. Insulin-like growth factor I in skeletal muscle after weight-lifting exercise in frail elders. Am J Physiol Endocrinol Metab. 1999;277:E135–E143. doi: 10.1152/ajpendo.1999.277.1.E135. [DOI] [PubMed] [Google Scholar]
- Smith CK, Janney MJ, Allen RE. Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J Cell Physiol. 1994;159:379–385. doi: 10.1002/jcp.1041590222. [DOI] [PubMed] [Google Scholar]
- Wilson EM, Tureckova J, Rotwein P. Permissive roles of phosphatidyl inositol 3-kinase and Akt in skeletal myocyte maturation. Mol Biol Cell. 2004;15:497–505. doi: 10.1091/mbc.E03-05-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wu Z. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J Biol Chem. 2000;275:36750–36757. doi: 10.1074/jbc.M005030200. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev Biol. 1999;210:440–455. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Alnaqeeb M, Simpson H, Goldspink G. Cloning and characterization of an IGF-1 isoform expressed in skeletal muscle subjected to stretch. J Muscle Res Cell Motil. 1996;17:487–495. doi: 10.1007/BF00123364. [DOI] [PubMed] [Google Scholar]
- Yang Y, Creer A, Jemiolo B, Trappe S. Time course of myogenic and metabolic gene expression in response to acute exercise in human skeletal muscle. J Appl Physiol. 2005;98:1745–1752. doi: 10.1152/japplphysiol.01185.2004. [DOI] [PubMed] [Google Scholar]
- Yang SY, Goldspink G. Different roles of the IGF-I Ec peptide (MGF) and mature IGF-I in myoblast proliferation and differentiation. FEBS Lett. 2002;522:156–160. doi: 10.1016/s0014-5793(02)02918-6. [DOI] [PubMed] [Google Scholar]