Abstract
Arthrobacter aurescens TC1, Arthrobacter chlorophenolicus A6, Arthrobacter crystallopoietes, and Arthrobacter oxydans produce long-chain monoalkenes, predominantly cis-3,25-dimethyl-13-heptacosene. Four other Arthrobacter strains did not form alkenes. The level of cis-3,25-dimethyl-13-heptacosene in Arthrobacter chlorophenolicus A6 remained proportional to cell mass during growth. cis-3,25-Dimethyl-13-heptacosene did not support growth of A. chlorophenolicus A6.
Bacterial hydrocarbon biosynthesis has garnered renewed interest in the context of generating new biofuels that are superior to ethanol (8, 12). A number of bacteria make long-chain, nonisoprenoid hydrocarbons that are being explored for biofuel and specialty chemical applications (23). An unusual class of unsaturated C22 to C31 alkenes produced by Micrococcus and Stenotrophomonas species was first described more than 40 years ago, but the biological function and mechanism of formation of these alkenes have not yet been elucidated (4, 18, 21). In addition, the assignment of the structure for specific compounds has differed in different reports (19, 20). The precise structures of the compounds are relevant to understanding the biosynthetic mechanism and biological utility of these compounds. Micrococcus and Stenotrophomonas make a complex mixture of alkenes; identifying new organisms with a simpler product profile should facilitate mechanistic and biological experiments. Moreover, discovering alkenes in new bacteria that have been subjected to complete genome sequencing should advance efforts to identify the relevant biosynthetic genes and enzymes.
Arthrobacter spp. are high-G+C-content gram-positive bacteria (10) for which several genome sequences are currently available (NCBI sequence accession number NC_008541 and NCBI sequence accession number ABKU00000000 [13]). In the present study, we screened cultures of divergent Arthrobacter species for the presence of hydrocarbons. Long-chain alkenes, reminiscent of a subset of those previously found in Micrococcus, were observed in several Arthrobacter species. Arthrobacter strains tested here were observed to produce a more uniform alkene chain length, predominantly C29. To positively identify the products, C29 alkenes with different methyl branching patterns and with cis or trans stereochemistry were prepared by chemical synthesis (see Fig. S1 in the supplemental material). The corresponding alkanes were also synthesized. This set of 11 chemical standards allowed the identification of the products as specific dimethyl-13-heptacosenes with an unambiguous demonstration of a cis relative stereochemistry at the double bonds.
Demonstration of alkenes in cultures of Arthrobacter spp.
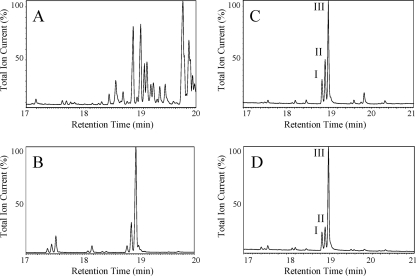
Nonpolar material extracted from Arthrobacter strains was compared to the hydrocarbons produced by Micrococcus luteus ISU and Stenotrophomonas (formerly Pseudomonas) maltophilia ATCC 17674 (17) (Fig. 1). Four-day-old cultures were extracted by using a modified Bligh and Dyer protocol (5) as described previously (24). Evaporated extracts were dissolved in 1 ml chloroform and applied to a 3.5-g silica gel column, eluted with 30 ml hexanes, concentrated, and subjected to gas chromatography-mass spectrometry (GC-MS) analysis using an HP6890 gas chromatograph connected to an HP5973 mass spectrometer (Hewlett Packard, Palo Alto, CA). The GC conditions were helium gas, 1 ml/min; HP-1ms column (100% dimethylpolysiloxane capillary; 30 m by 250 μm by 0.25 μm); temperature ramp, 100 to 320°C; and 10°C/min, with a 5-min hold at 320°C. The MS was run in electron impact mode at 70 eV and 35 μA.
FIG. 1.
Gas chromatograms of nonpolar extracts from Stenotrophomonas, Micrococcus, and Arthrobacter strains showing the variety of alkenes produced by each strain. (A) Stenotrophomonas maltophilia ATCC 17674. (B) Micrococcus luteus ISU. (C) Arthrobacter chlorophenolicus A6. (D) Arthrobacter crystallopoietes ATCC 15481. The numbers I, II, and III refer to the major C29 alkene products, and details of the their structures are described in the text.
Unlike the more-complex Stenotrophomonas (Fig. 1A) and Micrococcus (Fig. 1B) hydrocarbon profiles, the large majority of the Arthrobacter hydrocarbons were apparent in three readily resolvable peaks that eluted in the narrow range of 18.8 to 19.1 min. The major hydrocarbons extracted from Arthrobacter strains, represented in Fig. 1C and D, were identified by MS as C29 monoalkenes (m/z = 406; C29H58). Treatment of the alkenes with hydrogen gas over a palladium catalyst produced saturated alkanes, and subsequent GC-MS gave slightly longer retention times and mass spectra that were consistent with a gain of two mass units for each (m/z = 408; C29H60).
Table 1 shows the relative distribution of different alkene chain lengths in Stenotrophomonas, Micrococcus, and eight Arthrobacter species. The Arthrobacter strains were obtained from other researchers (9, 14) or the American Type Culture Collection (ATCC) or were isolated in this laboratory (7, 16). The Arthrobacter strains generally showed a narrower distribution of chain lengths (Table 1). This trend was most pronounced with A. oxydans ATCC 14358, which produced only C29 alkenes. Four other Arthrobacter strains did not yield any detectable alkenes. One of those not producing alkenes, Arthrobacter sp. strain FB24, has been subjected to genome sequencing (NCBI sequence accession number CP000454). Two Arthrobacter strains producing alkenes have had their genomes sequenced (NCBI sequence accession numbers ABKU00000000 and NC_008711 [13]).
TABLE 1.
Distribution of alkene chain lengths detected by GC-MS for a variety of Arthrobacter strains and comparison to their distribution in two previously studied bacteria, S. maltophilia and M. luteus
| Organism | % of indicated chain length in straina
|
||||||
|---|---|---|---|---|---|---|---|
| C25 | C26 | C27 | C28 | C29 | C30 | C31 | |
| Stenotrophomonas maltophilia ATCC 17674 | − | − | 1.15 | 5.15 | 36.3 | 39.4 | 18.0 |
| Micrococcus luteus ISU ATCC 27141 | 4.0 | 1.2 | 14.0 | 5.2 | 75.5 | − | − |
| Arthrobacter aurescens TC1 | − | − | − | − | 79.7 | 12.7 | 7.6 |
| Arthrobacter chlorophenolicus A6 | − | − | 1.5 | 4.5 | 80.3 | 11.6 | 2.1 |
| Arthrobacter crystallopoietes ATCC 15481 | − | − | 3.9 | 2.9 | 93.2 | − | − |
| Arthrobacter oxydans ATCC 14358 | − | − | − | − | 100 | − | − |
| Arthrobacter nicotianae ATCC 15236 | − | − | − | − | − | − | − |
| Arthrobacter sp. strain FB24 | − | − | − | − | − | − | − |
| Arthrobacter sp. strain 1-NP | − | − | − | − | − | − | − |
| Arthrobacter globiformis ATCC 35698 | − | − | − | − | − | − | − |
−, not detected.
Rigorous assignment of structures to resolved C29 isomers.
The mass spectra of the major alkenes produced by Arthrobacter bacteria were consistent with their assignment as dimethylheptacosenes (m/z = 406; C29H58). Arthrobacter strains produce predominantly C15 methyl-branched fatty acids (6, 22), and thus, if a head-to-head fatty acid condensation mechanism were operative as previously proposed (2, 3), dimethylheptacosenes (C29H58) would be the anticipated products. Arthrobacter bacteria produce both iso and anteiso methyl-branched C15 fatty acids, and so the alkenes could be iso-iso, iso-anteiso, anteiso-iso, anteiso-anteiso, or some mixture of the different isomers (see Fig. S1 in the supplemental material). Hydrogenation of the alkenes provided preliminary evidence from fragmentation patterns (M-15 and M-43) that the 18.8-min peak (I) was iso-iso branched, or 2,26-dimethylheptacosene (data not shown). The 19.0-min (major) peak (III) showed fragmentation (M-29 and M-59) consistent with an anteiso-anteiso branching that identifies the compound as 3,25-dimethylheptacosene (data not shown). The middle (18.9 min) peak (II) showed characteristics of iso-anteiso branching and thus could be identified as 2,25-dimethylheptacosene, 3,26-dimethylheptacosene, or a mixture of the two.
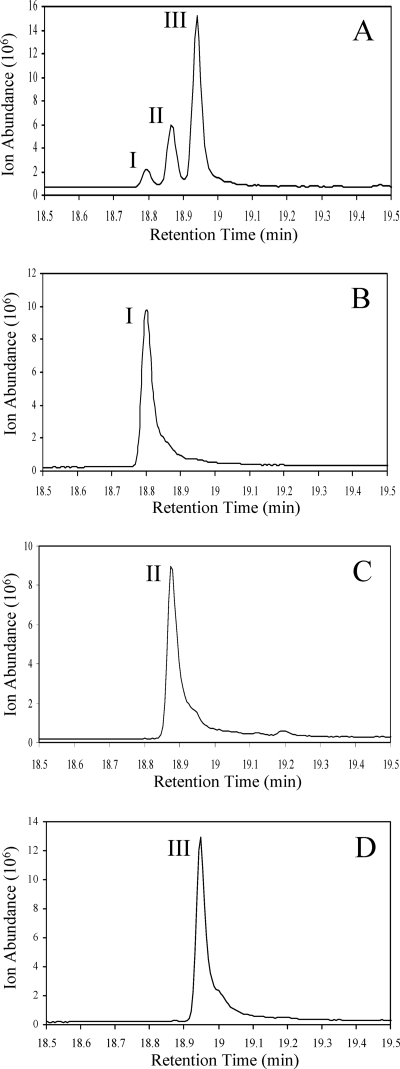
To help resolve these issues, and to determine the precise structures of the products, synthetic standards were prepared. Previous reports of alkenes biosynthesized by Micrococcus luteus indicated that the double bond of the alkenes was near the middle of the chain, based on their chemical degradation to fatty acids (1). The stereochemistry of the double bond was proposed to be cis based on their retarded migration on silica gel impregnated with silver (1). However, no synthetic chemical standards were available in those previous studies to compare the properties of cis- and trans-alkenes. Moreover, the same peak was assigned different structures in different studies (19, 20). In that context, cis- and trans-13-dimethylheptacosenes with all combinations of iso and anteiso branching patterns were synthesized (detailed synthetic conditions will be described elsewhere). The selective synthesis of cis or trans isomeric standards could be controlled by the synthetic methods used and was confirmed by nuclear magnetic resonance spectroscopy. Hydrogenation of different dimethylheptacosynes to the corresponding alkenes by using a Lindlar catalyst produced predominantly cis-olefins with trace amounts of the trans isomer, as shown in Fig. 2B, C, and D. The trans isomers eluted as a shoulder on the tail end of each cis-isomer peak. A more-aggressive hydrogenation reaction using 5% Pd/C catalyst produced predominately trans isomers. Long-term hydrogenation with 5% Pd/C led to complete reduction, yielding the corresponding alkanes. In this manner, eight different dimethyl-13-heptacosene standards and three dimethylheptacosane standards were synthesized. All of these were used as GC standards to determine retention times and mass spectra by GC-MS. This allowed the identification of the three major peaks as (Fig. 2A, left to right) cis-2,26-dimethyl-13-heptacosene (I), either cis-2,25-dimethyl-13-heptacosene or cis-3,26-dimethyl-13-heptacosene or a mixture of the two (II), and cis-3,25-dimethyl-13-heptacosene (III).
FIG. 2.
Gas chromatograms of the C29 alkene region from an Arthrobacter chlorophenolicus A6 extract shown in comparison with synthetic standards. (A) Arthrobacter chlorophenolicus A6 extract. (B to D) Synthetic standards: cis-2,26-dimethyl-13-heptacosene (B), a mixture of cis-2,25-dimethyl-13-heptacosene and cis-3,26-dimethyl-13-heptacosene (C), and cis-3,25-dimethyl-13-heptacosene (D).
The structural identifications made via separate injections were further confirmed by coinjection of standards in admixture with biological material on a GC. Coinjection of the respective standards gave uniform peaks, thus confirming the identity of the biological material eluting in peaks I and III as described above. Synthetic cis-2,25-dimethyl-13-heptacosene and cis-3,26-dimethyl-13-heptacosene had identical retention times and similar mass spectra, consistent with the conclusion that peak II could be either one of the compounds by itself or a mixture of the two.
Growth studies.
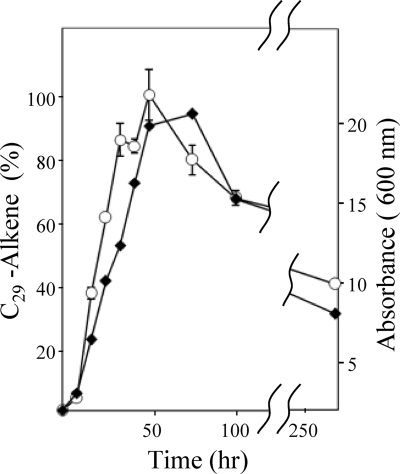
Micrococcus bacteria are spherical cells at all growth stages, whereas Arthrobacter species grow as rod-shaped cells during the exponential phase and become spherical during stationary phase (10). Thus, it was considered that alkenes might be preferentially formed by Arthrobacter bacteria during stationary phase. To test this hypothesis, a 50-μl preculture was used to inoculate 50 ml tryptic soy broth in a 125-ml Erlenmeyer flask. Cultures were grown at 28°C with shaking at 225 rpm. Duplicate cultures for growth studies were extracted at the time points of 0 h, 8 h, 16 h, 24 h, 32 h, 48 h, 96 h, and 288 h. The cells underwent a characteristic rod-to-coccus transition at 48 h. Prior to extraction, 62.5 μmol of cis-9-tricosene was added as an internal standard to quantitatively determine the levels of 3,25-dimethylheptacosene. The data obtained from these extractions showed that the alkene levels closely paralleled culture optical density (Fig. 3). Thus, both rod and spherical cell forms contain similar levels of 3,25-dimethylheptacosene.
FIG. 3.
Time course of culture optical density and C29 alkene accumulation in Arthrobacter chlorophenolicus A6. Alkene accumulation was determined by quantifying cis-3,25-dimethyl-13-heptacosene and normalizing the maximum concentration, found at 48 h, to 100%. The open circles denote alkene percentage. Closed diamonds represent absorbance. Error bars indicate standard deviations divided by n.
Arthrobacter chlorophenolicus A6 was tested for growth on S,S-cis-3,25-dimethyl-13-heptacosene, the major isomer thought to be formed biosynthetically by that strain. A. chlorophenolicus A6 was grown in M9 minimal medium (15) containing either glucose or S,S-cis-3,25-dimethyl-13-heptacosene, added to a concentration equivalent to 130 mM of carbon. Phenol was also used as a positive control, at 2 mM, 10 mM, and 20 mM in different cultures (14). Growth studies with each carbon source were set up in triplicate. For the inoculum, cells were grown overnight in tryptic soy broth and washed twice with M9 minimal medium without a carbon source. The medium was inoculated to an optical density of 0.07 at 600 nm and grown in capped test tubes at 28°C with shaking at 255 rpm. Optical density measurements were taken at 600 nm (Beckman DU 7400) after 17 days of growth.
No discernible cell growth was supported by cis-3,25-dimethyl-13-heptacosene. The average optical density at 600 nm in the test cultures was 0.049 ± 0.001 (mean ± standard deviation). That compared to an average of 0.042 ± 0.001 in a control without alkene. With phenol substituted for the alkene, the optical density at the same time point was 1.04 ± 0.03. With d-glucose as the carbon source, the optical density was 1.70 ± 0.18. These data suggested that the long-chain alkenes are not produced for the function of storing carbon or energy. The observation in this study that some Arthrobacter strains produce long-chain alkenes and others do not (Table 1) indicated that these compounds are not essential under the laboratory growth conditions used.
Conclusions.
This study has shown that some Arthrobacter strains produce C29 olefinic hydrocarbons, the structures of which were rigorously established by comparison with synthetic standards. In one previous study of Micrococcus bacteria, it was noted that one Arthrobacter strain, now identified as Arthrobacter citreus, also produced alkenes and that other Arthrobacter strains did not (11). No data were shown. However, that report, coupled with the present study, suggests that alkene formation is not ubiquitous amongst Arthrobacter species, in contrast to Micrococcus strains, which appear to uniformly produce olefinic hydrocarbons. Complete genome sequences are available for Arthrobacter aurescens TC1 (13) and Arthrobacter chlorophenolicus A6 (NCBI sequence accession numbers NC_00871 and ABXU00000000), which produced alkenes, and for Arthrobacter sp. strain FB24 (NCBI sequence accession number NC_008541), which did not. These observations pave the way to use comparative genomic analysis to identify alkene-biosynthetic genes in these microorganisms.
Supplementary Material
Acknowledgments
We thank Cindy Nakatsu and Janet Jansson for providing Arthrobacter sp. strain FB24 and Arthrobacter chlorophenolicus A6, respectively.
This work was supported by NIH Training Grant T32 GM08347 and NIH Training Grant 5 T32 GM008700 (to J.A.F.) and by grant LG-B13 from the Institute for Renewable Energy and the Environment and a Discovery Grant from the Institute of the Environment (to L.P.W.).
Footnotes
Published ahead of print on 23 January 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain nonisoprenoid hydrocarbons. I. Characterization of the hydrocarbons of Sarcina lutea and the isolation of possible intermediates of biosynthesis. Biochemistry 8:394-404. [DOI] [PubMed] [Google Scholar]
- 2.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain nonisoprenoid hydrocarbons. III. The metabolic relationship of long-chain fatty acids and hydrocarbons and other aspects of hydrocarbon metabolism in Sarcina lutea. Biochemistry 8:1913-1918. [DOI] [PubMed] [Google Scholar]
- 3.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain nonisoprenoid hydrocarbons. IV. Characteristics of synthesis by a cell-free preparation of Sarcina lutea. Biochemistry 8:3317-3324. [DOI] [PubMed] [Google Scholar]
- 4.Albro, P. W., and C. K. Huston. 1964. Lipids of Sarcina lutea. II. Hydrocarbon content of the lipid extracts. J. Bacteriol. 88:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bligh, E. G., and W. J. Dyer. 1959. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 6.Caudales, R., C. Forni, and J. M. Wells. 1998. Cellular fatty acid composition of rod and coccus forms of Arthrobacter globiformis, A. crystallopoietes and A. nicotianae isolated from the water fern Azolla. J. Appl. Microbiol. 84:784-790. [Google Scholar]
- 7.Dodge, A. G. 2008. Ph.D. thesis. University of Minnesota, Twin Cities, St. Paul.
- 8.Huber, G. W., and A. Corma. 2007. Synergies between bio- and oil refineries for the production of fuels from biomass. Angew. Chem. Int. Ed. Engl. 46:7184-7201. [DOI] [PubMed] [Google Scholar]
- 9.Jerke, K., C. H. Nakatsu, F. Beasley, and A. Konopka. 2008. Comparative analysis of eight Arthrobacter plasmids. Plasmid 59:73-85. [DOI] [PubMed] [Google Scholar]
- 10.Keddie, R. M., and D. Jones. 2006. The genus Arthrobacter, p. 945-960. In Martin Dworkin (ed.), Archaea, Bacteria: Firmicutes, Actinomycetes. The prokaryotes, 3rd ed., vol. 3. Springer-Verlag, New York, NY. [Google Scholar]
- 11.Kloos, W. E., T. G. Tornabene, and K.-H. Schleifer. 1974. Isolation and characterization of micrococci from human skin, including two new species: Micrococcus lylae and Micrococcus kristinae. J. Syst. Bacteriol. 24:79-101. [Google Scholar]
- 12.Lee, S. K., H. Chou, T. S. Ham, T. S. Lee, and J. D. Keasling. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19:556-563. [DOI] [PubMed] [Google Scholar]
- 13.Mongodin, E. F., N. Shapir, S. C. Daugherty, R. T. DeBoy, J. Emerson, A. Shvartzbeyn, D. Radune, J. Vamathevan, V. Grinberg, H. Khouri, L. P. Wackett, K. E. Nelson, and M. J. Sadowsky. 2006. Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet. 2:20214-20220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordin, K. 2004. Ph.D. thesis. Stockholm University, Stockholm, Sweden.
- 15.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Strong, L. C., C. Rosendahl, G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2002. Arthrobacter aurescens TC1 metabolizes diverse S-triazine ring compounds. Appl. Environ. Microbiol. 68:5973-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suen, Y., G. U. Holzer, J. S. Hubbard, and T. G. Tornabene. 1988. Biosynthesis of acyclic methyl branched poly-unsaturated hydrocarbons in Pseudomonas maltophilia. J. Ind. Microbiol. 2:337-348. [Google Scholar]
- 18.Tornabene, T. G., E. O. Bennett, and J. Oro. 1967. Fatty acid and aliphatic hydrocarbon composition of Sarcina lutea grown in three different media. J. Bacteriol. 94:344-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tornabene, T. G., E. Gelphi, and J. Oro. 1967. Identification of fatty acids and aliphatic hydrocarbons in Sarcina lutea by gas chromatography and combined gas chromatography-mass spectrometry. J. Bacteriol. 94:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tornabene, T. G., and S. P. Markey. 1971. Characterization of branched monounsaturated hydrocarbons of Sarcina lutea and Sarcina flava. Lipids 3:190-195. [DOI] [PubMed] [Google Scholar]
- 21.Tornabene, T. G., and S. L. Peterson. 1978. Pseudomonas maltophilia: identification of the hydrocarbons, glycerides, and glycolipoproteins of cellular lipids. Can. J. Microbiol. 254:525-532. [PubMed] [Google Scholar]
- 22.Unell, M., N. Kabelitz, J. K. Jansson, and H. J. Heipieper. 2007. Adaptation of the psychrotroph Arthrobacter chlorophenolicus A6 to growth temperature and the presence of phenols by changes in the anteiso/iso ratio of branched fatty acids. FEMS Microbiol. Lett. 266:138-143. [DOI] [PubMed] [Google Scholar]
- 23.Wackett, L. P. 2008. Microbial-based motor fuels: science and technology. Microb. Biotechnol. 1:211-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wackett, L. P., J. A. Frias, J. L. Seffernick, D. J. Sukovich, and S. M. Cameron. 2007. Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1. Appl. Environ. Microbiol. 73:7192-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.