Abstract
Research and financial efforts spent on biodefense technologies highlight the current concern for biothreat event preparedness. Nonhazardous but relevant “simulant” microorganisms are typically used to simplify technological developments, testing, and staff training. The bacteriophage MS2, a small RNA virus, is classically used as the reference simulant for biothreat viruses within the biodefense community. However, variola virus, considered a major threat, displays very different features (size, envelope, and double-stranded DNA genome). The size parameter is critical for aerosol sampling, detection, and protection/filtration technologies. Therefore, a panel of relevant simulants should be used to cover the diversity of biothreat agents. Thus, we investigated a new virus model, the Cydia pomonella granulovirus (baculovirus), which is currently used as a biopesticide. It displays a size similar to that of poxviruses, is enveloped, and contains double-stranded DNA. To provide a molecular tool to detect and quantify this model virus, we developed an assay based on real-time PCR, with a limit of detection ranging from roughly 10 to a few tens of target copies per μl according to the sample matrix. The specificity of the assay against a large panel of potential cross-reactive microorganisms was checked, and the suitability of the assay for environmental samples, especially aerosol studies, was determined. In conclusion, we suggest that our PCR assay allows Cydia pomonella granulovirus to be used as a simulant for poxviruses. This assay may also be useful for environmental or crop treatment studies.
The use of biological agents as weapons in warfare and terrorism has been recorded throughout history and was highlighted after the deliberate release of anthrax through contaminated letters. Multiple pathogenic viruses such as poxviruses, hemorrhagic fever viruses, and agents of viral encephalitis are potentially serious risks against military troops and civilian populations because they are highly infectious and relatively easy to produce (3). For operational countermeasures to address these risks, in addition to medical treatments, biodefense systems integrating detection, protection, and decontamination functions are needed. In order to equip first responders with such devices, all stages of their developments from lab bench to operational use must be experimentally validated in representative conditions. Handling pathogenic agents is cumbersome due to cost and security problems and, therefore, these are restricted to few laboratories. Moreover, it is not conceivable to release aerosols of pathogenic agents outdoors in order to validate a complete detection system. Thus, nonpathogenic simulants for each class of typical biowarfare agents have been selected: ovalbumin to simulate protein toxins such as ricin, botulinum toxin, or staphylococcal enterotoxins; Bacillus atrophaeus (formerly called Bacillus globigii or Bacillus subtilis subsp. niger) spores in place of Bacillus anthracis spores; Escherichia coli or Pantoea agglomerans (formerly Erwinia herbicola) vegetative bacterial cells to simulate gram-negative agents such as plague; and the bacteriophage MS2 in place of pathogenic viruses (2, 20). Simulant agents should share common physical or biological characteristics (i.e., size, shape, genome, and taxonomy) with the “real pathogens.” Moreover, they should be harmless, qualify for approval by regulatory agencies for environmental test releases, and be easy to produce in a cost-effective manner. Because some simulant testing sometimes requires concentrated samples, the method of purification should be practical for large-scale processes. Basically, one must be confident that a biodefense technology is evaluated with a simulant agent that will display similar capabilities with the corresponding threat agent.
Current efforts are focused on the development of fully automated aerosol biodetectors that are capable of rapid (ideally real-time) sample collection and identification in the field (6, 14, 19). Viral biothreat agents are highly infectious and easily transmitted after airborne dissemination; however, the efficiency of aerosol sampling and filtration steps is strongly dependent on particle size (4, 11). Thus, the use of a very small virus such as MS2 (26 nm) is not suitable for simulating larger particles such as poxviruses (∼300 nm). Conversely, the use of larger biological particles (bacterial spores for instance displaying sizes above 1 μm) is not adequate either. The collection yield of some biocollectors rapidly declines for particles below 2 μm, which of course impacts the efficiency of a detection system for sample collection. The same considerations also apply for protection equipments as filters and suits materials that display variable efficiency as a function of particle size.
Although, for decades, MS2 has been used for the evaluation of detection equipments and allowed the development of immunoassays and array biosensors (18, 23, 25), it is unfortunately very different in shape and size from the presumed most serious viral biothreat agent, the smallpox virus. Moreover, MS2 has no envelope (whereas variola virus displays enveloped and nonenveloped virions) and has a single-stranded RNA genome (while poxviruses particles contain double-stranded DNA genomes). It therefore appears that although MS2 can be useful as a simulant of the smallest RNA viruses (such as some of arenaviruses, alphaviruses displaying a size between 70 to 130 nm), it is not sufficient for developing, testing, and validating biodefense systems for the detection of larger viruses, especially poxviruses. As in the case of bacterial threat agents for which several simulants are available (i.e., spores and gram-negative bacteria), it seems impractical to use a single viral simulant in order to cover the diversity of viral threat agents. Thus, the development of a simulant for poxviruses appeared to be a useful goal because of the high ranking of smallpox in the several priority lists of threat agents (i.e., category A in the Centers for Disease Control and Prevention classification [5]).
Among the DNA viruses, insect viruses from the baculovirus family are an interesting alternative to MS2. They share several key characteristics with poxviruses. Virions contain double-stranded DNA genomes, possess envelopes, and are closer in shape and size to smallpox (250 to 300 nm versus 220 to 450 nm long). Moreover, they have narrow host ranges among invertebrates and are therefore safe for humans and animals. This point is highlighted by their use as biopesticides in agriculture (North America, continental Europe, and Asia) and also as molecular biology tools in protein production processes. Therefore, it should be easier in these countries to obtain authorizations to use them for biodefense test and evaluation purposes, especially for field trials (7, 24). Baculoviruses thus display great potential as representative simulants of poxvirus biothreat agents.
The baculovirus family includes many different species, and we chose a granulovirus type of baculovirus, Cydia pomonella granulovirus (CpGV). This virus is specific for the codling moth, Cydia pomonella, a worldwide pest on apples, pears, chestnuts, and walnuts. In Europe, the presence of this moth leads to important crop damage, and CpGV is registered in several countries as a biopesticide.
As for all granuloviruses, late in infection, CpGV forms ovoid occlusion bodies called granules that generally contain a single virion which is named occlusion body-derived virus (ODV) (28). CpGV granules, which present a size varying between 400 and 600 nm in length, can be used as a simulant of biological particles presenting a size around 500 nm, whereas ODVs are considered a closer simulant of the variola virus.
Here, we describe the development of an assay based on real-time quantitative PCR that specifically detects a few copies of CpGV genomic DNA extracted from granules. Our results indicate that the sensitivity and specificity of our assay make possible the analysis of environmental samples. This study focuses on protocols for the production and the detection of CpGV and is a first step in the establishment of a new viral simulant agent for biodefense studies.
MATERIALS AND METHODS
Materials.
CpGV is the only granulovirus for which there exist permissive cell lines; however, the titers produced in cell culture are too low (106 PFU/ml) for testing and evaluating detection protocols, especially for aerosolization studies (28). For this reason, we decided to purchase partially purified but high-titer pellets (∼1 kg) containing ∼1014 granules/g from Natural Plant Protection (Pau, France), a subsidiary of Arysta Lifescience Corporation (Japan). This pellet was obtained from CpGV-infected larval suspensions after a three-step process (filtration and two centrifugations) removing heavy debris from isects. The determination of the concentration of the pellet (granules/g) was done by the supplier by infecting Cydia pomonella larvae with serial dilutions of the suspension.
This pellet was resuspended in 4 liters of sterile distilled water and stored at 4°C until use for further experiments.
Production of CpGV ODVs.
One milliliter of this solution of granules was loaded onto a 30 to 60% continuous sucrose gradient (wt/wt) and centrifuged in an ultracentrifuge at 90,000 × g for 1 h. The band of granules was then harvested from above the tube (not through the side of the tube) by using a Pasteur pipette, diluted in distilled water (approximately 1:10), and centrifuged at 10,000 × g for 30 min (L8 70 MR; Beckman). The final pellet was resuspended in 500 μl of sterile distilled water and stored at 4°C.
To release intact ODVs from granules, 500 μl of granule suspension was added to 500 μl of 0.05 M Na2CO3 in a final volume of 1 ml, followed by incubation for 30 min at 37°C with stirring (100 rpm). Different concentrations of Na2CO3 (0.05 M and 0.1 M) and incubation times (15, 30, and 45 min and 1 h) were tested (data not shown). Undissolved granules and other debris were removed by centrifugation (10,000 × g, 30 min) (L8 70 MR; Beckman). The supernatant fraction containing CpGV ODVs was centrifuged again at 90,000 × g for 1 h (L8 70 MR; Beckman). The pellet was resuspended in 200 μl of 1× TE (0.1 M Tris [pH 7.6], 0.05 M EDTA [pH 8]) and stored at −80°C. To check for the presence of CpGV ODVs and the absence of granules, the resulting suspensions were observed by electron microscopy.
CpGV DNA extraction.
To extract CpGV DNA from granules, occlusion bodies were disrupted by using a bead beater (PreCellys 24; Bertin Technologies, France) containing 500 mg of glass beads (100 nm in diameter) mixed with yeast RNA (100 μl of a 0.1-mg/ml solution) for 45 s at 6.5 rpm. The presence of yeast RNA on the surface of glass beads decreases the probability of binding between beads and CpGV DNA released from granules. Beads were removed by low-speed centrifugation (4,000 rpm, 1 min; Beckman Microfuge R). The DNA was then purified by using extraction columns according to the manufacturer's instructions (DNeasy tissue kit; Qiagen SA, Courtaboeuf, France). DNA was directly isolated from CpGV ODVs with the extraction columns previously mentioned.
Real-time PCR assay design and conditions.
Real-time PCR assays were designed with the PrimerExpress software version 2.0 (Applied Biosystems, Foster City, CA) by using as a template GenBank accession no. NC_002816, the complete genome sequence of CpGV (15). As a target sequence, we chose a part of CpGV ORF138 that encodes the VP1054 protein, which is implicated in the virion assembly. Although this target gene ORF138 is conserved in several baculoviruses, this sequence is specific to CpGV and does not present a significant homology with other baculovirus sequences of this structural protein available in GenBank.
The primers and TaqMan probe (Table 1) were designed with the following parameters: an amplicon length of 50 to 150 bp to optimize the PCR speed and a melting temperature (Tm) of 58 to 60°C to use classical temperature cycles. The primers and probe were purchased from Applied Biosystems (Courtaboeuf, France). The probe is labeled at the 5′ end with 6-carboxy-fluorescein (5′-FAM) and at the 3′ end with a nonfluorescent quencher/minor groove binder (3′-NFQ/MGB). PCR assays were performed by using a Quantitect Probe PCR MasterMix and RNase-free water (Qiagen SA, Courtaboeuf, France) according to the manufacturer's instructions. The primers and the probe were added to final concentrations of 500 and 200 nM, respectively.
TABLE 1.
Primer and probe sequences for real-time fluorogenic PCR assays for detection of CpGV
| Primer or probe | Positiona | Sequenceb | Tm (°C) |
|---|---|---|---|
| MGB Cpgp138 174forwardc | 118464 | AACCGGTGCACGTTGCA | 59 |
| MGB Cpgp138 232reversec | 118522 | AGCACAGTTCGTCCGATTGG | 60 |
| MGB Cpgp138 194 (TaqMan probe) | 118484 | AAGCAATGTCGACTGCAT | 70 |
That is, the position of the first base in each oligonucleotide sequence relative to the whole-genome sequence in GenBank accession number U53466.
The sequences are listed in the 5′-to-3′ direction. The TaqMan probe is labeled with 5′-FAM and 3′-NFQ/MGB.
The size of the resulting amplicon is 59 base pairs.
Assays were performed on a SmartCycler II (Cepheid Inc Europe, Toulouse, France) with a thermocycler profile of 95°C for 15 min, followed by 45 PCR cycles of 95°C for 15 s and 60°C for 30 s. Assays were prepared in 25-μl volumes containing 5 μl of DNA sample. Cepheid SmartCycler software (version 2.0c) was used for quantitative calculations.
Absolute quantification of unknown samples in real-time PCR.
For the quantification of unknown samples containing CpGV genomic DNA in real-time PCR, we decided to use a classical method to generate a standard curve by cloning the viral target sequence into a plasmid vector (8, 9, 26). The advantages of this technique are that large amounts of standard plasmid DNA can be produced on a reproducible basis, with a sequence-checked single target copy, and the DNA can easily be quantified by spectrophotometry.
We used the TOPO TA cloning kit (Invitrogen). First, PCR amplification of nucleotides 174 to 232 of ORF138 was performed using the primers MGB Cpgp138-174forward and MGB Cpgp138-232reverse (Table 1). The resulting 59-pb PCR product was purified by using a specific purification column (MinElute PCR purification kit; Qiagen SA) and then ligated into the vector plasmid pCR2.1-TOPO, supplied linearized according to the manufacturer's instructions. The resulting vector, pCR2.1-TOPO-CpGV, was analyzed by PCR and sequenced to confirm the presence of the amplicon. Its weight concentration was determined by measuring the absorbance at 260 nm in a spectrophotometer DU640 (Beckman). The concentration of pCR2.1-TOPO-CpGV copies was calculated using the appropriate equation.
DNA material for cross-reaction testing.
DNAs used to validate the specificity of our PCR assay were extracted from microorganisms of the CEB strain collections (Bombyx mori nuclear polyhedrosis virus, Autographa californica multiple nuclear polyhedrosis virus, Bacillus atrophaeus, Escherichia coli, Bacillus anthracis, Bacillus thuringiensis, Bacillus cereus, Bacillus mycoides, Bacillus stearothermophilus, Brucella abortus, Yersinia enterocolitica, Yersinia pestis, Micrococcus luteus, Francisella tularensis, Legionella pneumophila, and Staphylococcus aureus) except for human DNA (kindly provided by X. Coumoul, INSERM U707, Paris, France).
Preparation for electron microscopy. (i) Negative staining.
Portions (10 μl) of the suspension of biological agents were adsorbed onto the electron microscopy support grid for 3 min. The grid with adhering virus was transferred by using a forceps onto a 10-μl droplet of 1% sodium silicotungstate (Merck, Germany) for 1 min. The grid was removed, and excess stain was drained away with a strip of filter paper. The grid was then examined with a Philips CM120 electron microscope.
(ii) Thin sectioning.
Purified granules were fixed in 2% glutaraldehyde in 0.1 M cacodylate buffer at 4°C overnight. After centrifugation at 10,000 × g for 30 min, the resulting pellet containing CpGV granules was washed three times for 10 min each time with cacodylate buffer. Granules were then postfixed in 2% osmium tetroxide in the same buffer. After dehydration in 70% ethanol, granules were embedded in Epon 812 resin (Fluka, Switzerland). Thin sections were cut by using a diamond knife in an LKB Ultrotome III apparatus and were mounted on grids subsequently stained with uranyl acetate and lead citrate. All observations were performed with a Zeiss (Jena) EM 10C/CR electron microscope. All chemical products were purchased from Serva (Germany).
Air sample collection.
Air samples were collected by using an ICAS biocollector (BioTrace, United Kingdom) based on a wet cyclone technology, a technique that displays a d50 (diameter for which 50% of particles are retained) cutoff at about 0.45 to 2 μm (16). Sampling took place in several locations in France, both rural and semirural. The inlet of the biocollector was placed at about 1.5 m from ground level. In each case, sampling was performed for 15 min with an airflow of about 800 liters/min. Thus, each sample corresponds to about 12 m3 of air. The liquid output was set at a flow rate of 1 ml/min. Samples were then concentrated down to 1 ml by using Vivaspin filtration column with a 100-kDa cut diameter (Vivasciences, France). DNAs from environmental microorganisms and debris were extracted and eluted in 50 μl by using DNeasy tissue kit columns according to the manufacturer's instructions (Qiagen SA).
RESULTS
Preparation of CpGV virions.
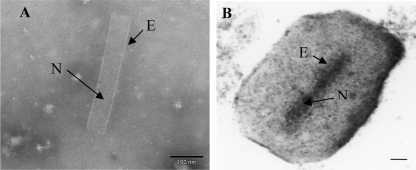
After the treatment of CpGV granules with alkali buffer to dissolve granulin, the presence of ODVs and the absence of undissolved granules and nonenveloped virions in the resulting suspension were observed by negative staining. Only CpGV virions were present, indicating that our protocol was reproducible and efficient (Fig. 1A). Purified CpGV granules were also observed by thin-section electron microscopy (Fig. 1B). Overall, fifty different preparations of purified granules and ODVs were observed.
FIG. 1.
(A) Negative staining. One enveloped virus (ODV) after alkali treatment of granules (magnification, ×200,000; the bar marker represents 100 nm). (B) Thin-section electron micrograph. CpGV granule containing a single enveloped virion after sucrose gradient purification (magnification, ×100,000; the bar marker represents 100 nm). N, nucleocapsid; E, envelope.
Sensitivity of the qPCR assay for CpGV.
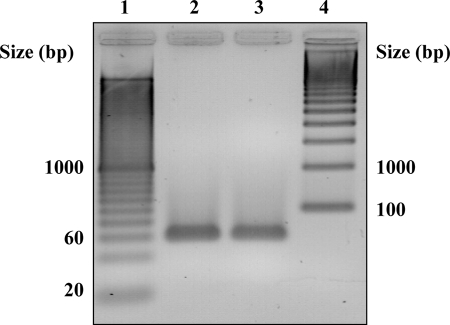
We designed PCR primers and a probe for a specific gene that encodes a structural protein of CpGV (see Materials and Methods). In a preliminary quantitative PCR (qPCR) assay, where no DNA standard was added, the success of this assay was assessed by checking with two different samples of CpGV DNA the presence of an amplicon with the expected size (59 bp, Fig. 2). Then, to determine the concentrations of unknown samples containing CpGV with our assay, we generated a standard curve by using a solution of pCR2.1-TOPO-CpGV (see Materials and Methods) serially diluted 10-fold from 1 to 106 genome copies/μl. The primers and probe provided sensitive fluorogenic PCR detection of the target sequence. The data from these experiments showed a linear correlation with a dynamic range of 7 orders of magnitude (R2 = 0.999, Fig. 3). This external standard curve was included and recalculated for each PCR run of unknown samples. The detection limit of our assay was approximately between 1 and 10 copies of cloned DNA (Table 2). Two assays using plasmid DNA were performed with each concentration in triplicate. The standard error to the mean calculated by comparing the CT values obtained for each concentration in six measures was found to vary between 0 and 0.24.
FIG. 2.
Agarose gel electrophoresis of amplicons produced by the qPCR assay for CpGV structural protein in two samples. Lane 1, 20-bp ladder; lanes 2 and 3, PCR products from two different template DNA samples; lane 4, 100-bp ladder.
FIG. 3.
External standard curve for the quantification of unknown samples containing CpGV genomic DNA by real-time PCR. (A) Different concentrations of the CpGV plasmid DNA were used and are associated with increasing CT values. The CT values represent the cycle number for which the fluorescence passes from background levels to clearly detectable (usually 10% of the maximum signal obtained with positive controls after 40 cycles [20]). (B) Correlation between sample concentration and CT values.
TABLE 2.
Performance of the real-time PCR assay for detection of CpGVa
| Sample concn (gene copies/μl) | Mean CT value ± SD
|
Sample quantity (pg or fg/PCR sample)e | ||
|---|---|---|---|---|
| Cloned CpGV DNA (plasmid)b | CpGV genomic DNA extracted from granules in a water samplec | CpGV genomic DNA, mixed with DNA extracted from a biocollection sampled | ||
| 1.00E+06 | 19.06 ± 0.09 | 22.9 1± 0.12 | 22.79 ± 0.10 | 135.4 pg |
| 1.00E+05 | 22.76 ± 0.05 | 26.38 ± 0.06 | 26.28 ± 0.06 | 13.54 pg |
| 1.00E+04 | 26.64 ± 0.07 | 29.84 ± 0.09 | 29.78 ± 0.07 | 1.354 pg |
| 1.00E+03 | 29.64 ± 0.07 | 33.20 ± 0.08 | 33.02 ± 0.08 | 135.4 fg |
| 1.00E+02 | 33.66 ± 0.08 | 36.66 ± 0.24 | 36.54 ± 0.23 | 13.54 fg |
| 1.00E+01 | 36.46 ± 0.24 | 38.50 ± 0.0 | NDf | 1.354 fg |
| 1.00E+00 | 39.05 ± 0.0 | |||
PCR runs were performed as described in Materials and Methods.
Detection of CpGV cloned DNA in six wells (three wells per experiment, two experiments).
Detection of CpGV genomic DNAs extracted from granules (and a PCR run in a water matrix sample) in seven wells (three and four wells per experiment, two experiments).
Detection of CpGV genomic DNA mixed with heterologous DNAs extracted from a biocollection sample in eight wells (four wells per experiment, two experiments).
Corresponding amounts of CpGV DNA are also indicated in pg or fg.
ND, no detection after 45 PCR cycles.
The detection sensitivity threshold for CpGV genomic DNA extracted from granules was then evaluated, using 10-fold dilutions in sterile water (from 10 to 106 target genomes/μl). The smallest amount significantly detected was a few tens of CpGV genome copies corresponding to a quantity between 1.3 to 13 fg (Table 2). We also verified that CpGV genomic DNA extracted from ODVs was detected with the same level of sensitivity (data not shown).
To examine the efficiency of the PCR assay in analyses of environmental samples, the influence of the sample matrix, especially biological environmental background, on the sensitivity of our assay was investigated. First, genomic CpGV DNA was mixed with DNA extracted from aerosol biocollection samples (see Materials and Methods), which contain a large panel of different microorganisms (13, 17; data not shown). We found that the detection limit was less than 100 genomes per μl, showing a slight loss in sensitivity (Table 2). In a second study, CpGV granules were added directly to 1-ml biocollection sample in increasing concentration (from 102 to 106 granules/ml), before the DNA extraction step (see Materials and Methods). These granules were taken from a purified suspension previously quantified by real-time PCR. The detection limit was between 100 and 1000 granules per spiked biocollection sample (1 ml) (Table 3). If we assume that the yield of DNA extraction was 100%, then 1,000 spiked granules within 1 ml should correspond to a PCR target concentration of 20 genome copies/μl (see Materials and Methods), which is consistent with Table 2. These results indicate that our PCR assay is reproducible and quite sensitive, even in the presence of complex environmental matrices.
TABLE 3.
Detection of CpGV DNA extracted from a matrix wherein granules were mixed with a biocollection sample in eight wells (four wells per experiment, six experiments)
| CpGV concn added in a biocollection sample (granules/ml) | Avg CT value for DNA extracted from granules mixed with a biocollection sample ± SD |
|---|---|
| 1.00E+06 | 27.70 ± 0.13 |
| 1.00E+05 | 30.71 ± 0.06 |
| 1.00E+04 | 34.28 ± 0.07 |
| 1.00E+03 | 37.55 ± 0.26 |
| 1.00E+02 | 38.21 ± 0.19 |
Specificity of the quantitative PCR assay.
To evaluate the specificity of the PCR assay, we first used DNAs extracted from two others baculoviruses: Bombix mori nuclear polyhedrosis virus and Autographa californica multiple nuclear polyhedrosis (AcMNPV), which are widely present in several countries. These DNAs did not give a signal when added in the assay sample with an amount equivalent to 1,000 times the limit amount necessary to detect CpGV (1.7 and 2 pg, respectively). Then, since other biological simulants, as well as biothreat agents, may be commonly used in the laboratories developing or evaluating biodefense technologies, we checked the absence of detection against whole-genomic DNA isolated from Bacillus atrophaeus, Escherichia coli, Bacillus anthracis, Bacillus thuringiensis, Bacillus cereus, Bacillus mycoides, Bacillus stearothermophilus, Brucella abortus, Yersinia enterocolitica, Yersinia pestis, Micrococcus luteus, Francisella tularensis, Legionella pneumophila, Staphylococcus aureus, and human DNA (in case of a contamination introduced from an operator). No cross-reaction was observed (data not shown), thus enabling the easy performance of CpGV detection and also its simultaneous use with other simulants if needed or its use as a control (either positive or negative) in biothreat agent assays.
Finally, to characterize the assay with environmental samples, we performed a rigorous specificity study. During 1 year, 241 air samples were collected in different places in France for the purpose of studying the natural airborne biological background. As a positive control, CpGV granules were spiked in one sample at a concentration of 105/ml. Samples were analyzed as previously described (see Table 3). CpGV granules were only significantly detected in the positive control. None of the air samples gave a significant fluorescent signal above the PCR detection threshold and are thus rated as negative, as is commonly described (12). More precisely, 90% of the samples gave a CT above 40, i.e., beyond the detection threshold of purified target in pure water (see Table 2). The remaining 10% of the samples gave a CT between 39 and 40, which is beyond the significant value for the detection of CpGV in complex environmental matrices (see Table 3). Conversely, the CT obtained for the positive control was 32 and was consistent with Table 3. These data indicate that the background levels of airborne CpGV cross-reactive species in the different places where air sampling was performed were below detection levels. Since the study was performed on a large number of different environmental samples, it can be assumed that significant cross-reactions should be scarcely if at all observed.
Altogether, these results suggest that our protocol of quantification could be used for the environmental detection of collected CpGV granules, even in concentrated air-sample matrices. The quantitative PCR assay described above is very specific to CpGV even in the presence of nontarget organisms that are found in environmental samples.
DISCUSSION
In this report, we suggest that CpGV could be a better candidate than bacteriophage MS2 for simulating large double-stranded DNA virus threat agents such as smallpox (10). It seems relevant to have several simulants close enough for each main viral biothreat agents group in the same way that there are several simulants for the main classes of bacterial agents (i.e., spores, gram negative). At least two groups of viral biowarfare agents can be discriminated: small RNA viruses and large DNA viruses. It seems to us and others that MS2 is not adequate as a viral simulant of variola virus (and more generally poxviruses), although it has long been used in the biological defense community. It is important to find a more suitable simulant of poxviruses for developing, testing, and validating biodefense technologies, especially biodetection devices.
The first step was to choose our candidate from the baculovirus family, which is not pathogenic and better approximates variola virus in genome, nucleic acid composition, and virion size. This virus family includes many different species, and our initial thought was to use AcMNPV, the most studied baculovirus (1). This virus is widely used for the high-level production of heterologous recombinant proteins in insect cells (21, 22). Unfortunately, this virus is not registered as a biopesticide in Europe, suggesting that it will not be easy to obtain authorization from competent administrations for nonconfined trials. Moreover, its production in insect cell lines yields large polyhedrons containing several viruses, and such particles cannot be used directly as a simulant because they are much larger (up to 15 μm) than poxviruses or any biothreat agents. Finally, the AcMNPV titers in Spodoptera frugiperda cells are ∼108 PFU/ml (21), which is too low to perform trials in aerosol facilities or in the field, where large quantities of viruses are usually required. For these reasons, we decided to turn to CpGV. This virus actually provides two kinds of simulant, virions named ODVs for poxvirus threat agents and granules for biological particles presenting a size of ∼500 nm and containing a double-stranded DNA genome. These granules could be used in different biodefense test and evaluation protocols, including aerosolization. We established a first protocol allowing the production of purified concentrated suspensions of CpGV granules. With regard to ODVs, we described a protocol for their production from CpGV granules that is reproducible, rapid, and easy to perform. In this way, concentrated suspensions of ODVs are produced from high-titer suspensions of granules, which makes the process easier than the use of an in vitro system.
The next step and main goal of the present study was to develop a real-time PCR assay for CpGV that can be adapted to various PCR technologies (27). The detection limit of our assay was optimized in order to detect only a few gene target copies with cloned DNA per PCR sample. Moreover, the very low deviation (R2 = 0.999) indicates that the assay is highly reproducible. In the case of CpGV genomic DNA extracted from granules or ODVs, the sensitivity of our assay ranged from 1.3 to 13 fg or approximately from 10 to 100 genome copies. We then investigated environmental samples and found that the detection limit, evaluated with genomic CpGV DNA extracted from granules mixed with DNA extracted from aerosol biocollection samples, was 13 fg or approximately 100 genome copies. Finally, for CpGV granules in environmental samples, the sensitivity of our protocol was between 100 and 1,000 granules per ml of samples (e.g., biocollected air samples). The determination of the yield of each step of the sample processing was beyond the scope of this study; however, it seems that the overall DNA extraction steps from an environmental sample are efficient and reproducible. When the assay was evaluated with the DNAs of two other baculoviruses and those of classical bacterial simulants and threat agents used in biodefense laboratories, the specificities of our assay proved to be very accurate (no cross-reaction was detected). Moreover, we found that our assay can be used for the environmental detection of CpGV even in concentrated air-sample matrices without false-positive detection. Altogether, these sensitivity and specificity results show that we have developed a robust assay and that our protocols of CpGV DNA extraction from granules and real-time quantification could be used in experimental test and evaluation procedures. These procedures may include the release of CpGV in aerosol facilities and in field trials or the analysis of spiked samples. According to our results, natural (environmental) cross-reactions are most unlikely and may only occur in the case of the agricultural spraying of a CpGV-based biopesticide in the vicinity.
The CpGV simulant and its related PCR assay described here can be used to evaluate and compare directly different integrated PCR technologies, using a simulant close enough to variola virus. Current PCR-based detection devices now include sample preparation. The efficiency of this process is dependent on the physical features of the target agent (size, structure, genome, etc.). Thus, a relevant simulant is required for the development and testing of such technologies in case the actual pathogenic target cannot be handled. Our PCR assay can also be used as a reference metrology to quantify CpGV challenges of any biodefense technology, including aerosol challenge.
Focusing on these challenges, using a biological particle of about 400 to 600 nm in long (such as CpGV granules) as a simulant for poxviruses allows for more accurate technologies development and testing, especially in the field of aerosol detection and protection. Indeed, biocollection efficiency often declines rapidly for particle sizes below 2 μm (16). In addition, biothreat detectors that analyze aerosol particles using a direct physical or optical signal (such as laser-induced fluorescence) are very dependent on the particle size as well. Challenging technologies with 1-μm particles (B. atrophaeus spores are the most classical simulant) is not sufficient since some biothreat agents such as viruses or some bacteria (such as Francisella tularensis) are smaller. Conversely, using MS2 bacteriophage is too great a challenge because of its very small size (26 nm). Besides, controlling the metrology of such small particles aerosols is most difficult and rarely available. Using CpGV granules would save the trouble to generate calibrated aerosols made up of nanometric particle clusters (e.g., simulating the size of a virus larger than MS2 using aggregates of MS2). Generating monodispersed aggregates is a very difficult process, and the metrology of nanometric particles requires special skills and costly specific apparatus and facilities. Thus, beyond its biological structure close to poxviruses, the CpGV model can be used for controlled submicronic bioaerosol challenge of equipments and emerging technologies. It would better simulate poxvirus aerosols than the classically used simulants MS2 (too small) or Bacillus atrophaeus (too large) for the evaluation of aerosol samplers and integrated field detection systems. More generally, it would thus expand the simulation tools available for the biodefense community.
In conclusion, we suggest using CpGV as a simulant of variola virus (and more generally poxviruses) for biodefense technological studies. We demonstrate the specific, real-time PCR identification of this virus, even in complex environmental samples. Thus, CpGV can be practically used as a simulant in test and evaluation protocols including field trials. We are currently testing polyclonal and monoclonal antibodies specific to granules and ODVs in order to develop immunoassays. The next step will be to perform calibrated aerosolization assays with this new viral simulant. Our PCR assay may also be useful for studies related with the crop treatments using CpGV as a biopesticide.
Acknowledgments
We are grateful to Iroudayanadin De La Manche and Marc Ravallec for assistance with the electron microscopy and to John W. Wills for critical reading of the manuscript.
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 2.Belgrader, P., W. Benett, D. Hadley, G. Long, R. Mariella, F. Milanovich, S. Nasarabadi, W. Nelson, J. Richards, and P. Stratton. 1998. Rapid pathogen detection using a microchip PCR array instrument. Clin. Chem. 44:2191-2194. [PubMed] [Google Scholar]
- 3.Bonze, M. S., M. M. Huycke, L. J. Machado, G. W. Voskuhl, and R. A. Greenfield. 2002. Viral agents as biological weapons and agents of bioterrorism. Am. J. Med. Sci. 323:316-325. [DOI] [PubMed] [Google Scholar]
- 4.Burton, N. C., S. A. Grinshpun, and T. Reponen. 2007. Physical collection efficiency of filter materials for bacteria and viruses. Ann. Occup. Hyg. 51:143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2000. Biological and chemical terrorism: strategic plan for preparedness and response. Recommendations of the CDC Strategic Planning Workgroup. MMWR Recomm. Rep. 49(RR-4):1-14. [PubMed] [Google Scholar]
- 6.Christensen, D. R., L. J. Hartman, B. M. Loveless, M. S. Frye, M. A. Shipley, D. L. Bridge, M. J. Richards, R. S. Kaplan, J. Garrison, C. D. Baldwin, D. A. Kulesh, and D. A. Norwood. 2006. Detection of biological threat agents by real-time PCR: comparison of assay performance on the R.A.P.I.D., the LightCycler, and the Smart Cycler platforms. Clin. Chem. 52:141-145. [DOI] [PubMed] [Google Scholar]
- 7.Cory, J. S., and D. H. L. Bishop. Use of baculoviruses as biological insecticides. Mol. Biotechnol. 7:303-313. [DOI] [PubMed]
- 8.Gunson, R. N., T. C. Collins, and W. F. Carman. 2006. Practical experience of high throughput real-time PCR in routine diagnostic virology settings. J. Clin. Virol. 35:355-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 10.Henderson, D. A., T. V. Inglesby, J. G. Bartlett, M. S. Ascher, E. Eitzen, P. B. Jahrling, J. Hauer, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. Perl, P. K. Russell, and K. Tonat. 1999. Smallpox as a biological weapon. JAMA 281:2127-2137. [DOI] [PubMed] [Google Scholar]
- 11.Hogan, C. J., E. M. Kettleson, M. H. Lee, B. Ramaswami, L. T. Angenent, and P. Biswas. 2005. Sampling methodologies and dosage assessment techniques for submicrometre and ultrafine virus aerosol particles. J. Appl. Microbiol. 99:1422-1434. [DOI] [PubMed] [Google Scholar]
- 12.Kurth, A., J. Achenbach, L. Miller, I. A. Mackay, G. Pauli, and A. Nitsche. 2008. Orthopoxvirus detection in environmental specimens during suspected bioterror attacks: inhibitory influences of common household products. Appl. Environ. Microbiol. 74:32-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lighthart, B., and B. T. Shaffer. 1995. Airborne bacteria in the atmospheric surface layer: temporal distribution above a grass seed field. Appl. Environ. Microbiol. 61:1492-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim, D. V., J. M. Simpson, E. A. Kearns, and M. F. Kramer. 2005. Current and developing technologies for monitoring of bioterrorism and biowarfare. Clin. Microbiol. Rev. 18:583-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luque, T., R. Finch, N. Crook, D. R. O'Reilly, and D. Winstanley. 2001. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 82:2531-2547. [DOI] [PubMed] [Google Scholar]
- 16.Mainelis, G., D. A. Masquelier, K. Willeke, A. Makarewicz, J. Dzenitis, and F. P. Milanovich. 2006. Performance of a compact air-to-liquid aerosol collector with high concentration rate. J. Aerosol Sci. 37:645-657. [Google Scholar]
- 17.Mancinelli, R., and W. A. Shulls. 1978. Airborne bacteria in an urban environment. Appl. Environ. Microbiol. 35:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride, M. T., S. Gammon, M. Pitesky, T. W. O'Brien, T. Smith, J. Aldrich, R. G. Langlois, B. Colston, and K. S. Venkateswaran. 2003. Multiplexed liquid arrays for simultaneous detection of simulants of biological warfare agents. Anal. Chem. 75:1924-1930. [DOI] [PubMed] [Google Scholar]
- 19.Molenkamp, R., A. Van der Ham, J. Schinkel, and M. Beld. 2007. Simultaneous detection of five different DNA targets by real-time TaqMan PCR using the Roche LightCycler480: application in viral molecular diagnostics. J. Virol. Methods 141:205-211. [DOI] [PubMed] [Google Scholar]
- 20.O'Connell, K. P., J. R. Bucher, P. E. Anderson, C. J. Cao, A. S. Khan, M. V. Gostomski, and J. J. Valdes. 2006. Real-time fluorogenic reverse transcription-PCR assays for detection of bacteriophage MS2. Appl. Environ. Microbiol. 72:478-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. W. H. Freeman & Co., New York, NY.
- 22.Possee, D. P. 1997. Baculoviruses as expression vectors. Cur. Opin. Biotechnol. 8:569-572. [DOI] [PubMed] [Google Scholar]
- 23.Rowe, C. A., L. M. Tender, M. J. Feldstein, J. P. Golden, S. B. Scruggs, B. D. MacCraith, J. J. Cras, and F. S. Ligler. 1999. Array biosensor for simultaneous identification of bacterial, viral, and protein analytes. Anal. Chem. 71:3846-3852. [DOI] [PubMed] [Google Scholar]
- 24.Szewczyk, B., L. Hoyos-Carvajal, M. Paluszek, I. Skrzecz, and M. Lobo de Souza. 2005. Baculoviruses-re-emerging biopesticides. Biotechnol. Adv. 24:143-160. [DOI] [PubMed] [Google Scholar]
- 25.Thomas, J. H., S. K. Kim, P. J. Hesketh, H. B. Halsall, and W. R. Heineman. 2004. Bead-based electrochemical immunoassay for bacteriophage MS2. Anal. Chem. 76:2700-2707. [DOI] [PubMed] [Google Scholar]
- 26.Vanguilder, H., K. Vrana, and W. Freeman. 2008. Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques 44:619-626. [DOI] [PubMed] [Google Scholar]
- 27.Watzinger, F., K. Ebner, and T. Lion. 2006. Detection and monitoring of virus infections by real-time PCR. Mol. Aspects Med. 27:254-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winstanley, D., and N. E. Crook. 1993. Replication of Cydia pomonella granulosis virus in cell cultures. J. Gen. Virol. 74(Pt. 8):1599-1609. [DOI] [PubMed] [Google Scholar]