Abstract
Lactococcus sp. strain QU 12, which was isolated from cheese, produced a novel cyclic bacteriocin termed lactocyclicin Q. By using cation-exchange chromatography, hydrophobic interaction chromatography, and reverse-phase high-performance liquid chromatography, lactocyclicin Q was purified from culture supernatant, and its molecular mass was determined to be 6,062.8 Da by mass spectrometry. Lactocyclicin Q has been characterized by its unique antimicrobial spectrum, high level of protease resistance, and heat stability compared to other reported bacteriocins of lactic acid bacteria. The amino acid sequence of lactocyclicin Q was determined chemically, and this compound is composed of 61 amino acid residues that have a cyclic structure with linkage between the N and C termini by a peptide bond. It showed no homology to any other antimicrobial peptide, including cyclic bacteriocins. On the basis of the amino acid sequences obtained, the sequence of the gene encoding the prepeptide lactocyclicin Q was obtained. This is the first report of a cyclic bacteriocin purified from a strain belonging to the genus Lactococcus.
Consumers demand a high level of safety, especially with regard to food. Empirical use of the antimicrobial agents produced by lactic acid bacteria (LAB) for the preservation of foods has been a common practice throughout history. Bacteriocins produced by LAB are antimicrobial proteins or peptides with antimicrobial activity that inhibits not only species closely related to the producer strain but also food-borne bacteria, such as food spoilage bacteria and pathogens. Bacteriocins are commonly divided into several groups (19, 27). Class I bacteriocins, termed lantibiotics and represented by nisin, are characterized by heat stability and their unusual amino acids, such as lanthionine and 3-methyllanthionine (13, 26, 37). Class II bacteriocins are heat-stable and nonlantibiotic peptides that are divided into three subgroups. Class IIa bacteriocins, including pediocin PA-1, are peptides with anti-Listeria activity and a consensus YGNGVXC sequence in the N-terminal region (8); class IIb bacteriocins, including lactococcin Q, comprise two peptides (39), both of which are required for full antimicrobial activity (12); and class IIc bacteriocins are other class II bacteriocins, such as lacticin Q (10).
Recently, several cyclic bacteriocins that are bound at the N and C termini have been discovered, and their classification is being discussed. Cotter et al. proposed a new classification of LAB bacteriocins that includes three main groups (6). Specifically, cyclic bacteriocins were defined as class IIc bacteriocins based on their criteria. On the other hand, Kemperman et al. classified cyclic bacteriocins in class V (18).
So far, enterocin AS-48 (11, 21), gassericin A (15, 16), reutericin 6 (17), and uberolysin (36) have been reported to be cyclic bacteriocins produced by LAB. Recently, several natural variants of these compounds have been reported. Enterocin AS-48 is the most extensively studied because of its cyclization characteristics and the fact that it shows great potential as a food preservative. It comprises 70 amino acid residues and is widely active against both related and unrelated bacteria, such as Listeria, Bacillus, Salmonella choleraesuis, and Escherichia coli. Several natural enterocin AS-48 variants produced by Enterococcus faecalis or Enterococcus faecium have been reported, including enterococcin EFS2 (22), enterocin 4 (14), bacteriocin 21 (33), and AS-48RJ (3). In particular, AS-48RJ differs at one amino acid residue, the residue at position 20 in the primary structure, where AS-48 contains Glu20 and AS-48RJ contains Val20. Acidocin B (20) is considered a potential natural variant of gassericin A. Gassericin A contains Met24, and acidocin B contains Val24. However, more structural analysis of acidocin B needs to be performed before this compound can be confirmed to be a cyclic bacteriocin.
Microorganisms belonging to the genus Lactococcus are the most important microorganisms in the dairy industry, as they are commonly used as starter organisms. In particular, Lactococcus lactis is generally recognized as a safe organism. Both the basic and application aspects of its bacteriocins, especially nisin, have been studied extensively. Additionally, nisin is the best bacteriocin so far because of its broad and strong inhibitory activity. However, using nisin for a long period of time against a specific target strain may cause the emergence of a nisin-resistant strain, even though this has not been reported. Therefore, to eliminate such a problem, various efficient bacteriocins other than nisin are required.
In this report, we describe identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12, which was isolated from cheese. Lactocyclicin Q is characterized by a unique antimicrobial spectrum and high heat stability compared to the other LAB bacteriocins. Lactocyclicin Q has a cyclic structure in which the N and C termini are linked to each other and comprises 61 amino acid residues that show no homology to the residues of any other antimicrobial peptide.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacteriocin-producing organism Lactococcus sp. strain QU 12 was isolated on an MRS agar plate containing 0.5% CaCO3 from overnight-aged milk of a cheese intermediate and was identified by 16S rRNA gene sequencing and sugar fermentation pattern analysis, as described in a previous report (10). Gram stain, motility, and catalase tests were performed as a preliminary step in the screening of LAB.
Lactococcus sp. strain QU 12 was stored at −80°C in M17 medium (Merck, Darmstadt, Germany) with 15% glycerol. Before use, it was propagated in M17 medium at 30°C for 18 h. Bacterial strains used as indicator strains for the bacteriocin assay were propagated for 18 h at the temperature (30°C or 37°C) recommended by the culture collections. MRS medium (Oxoid, Basingstoke, United Kingdom) was used to culture all indicator strains except Lactococcus sp. strain QU 12 and L. lactis subsp. cremoris ATCC 19275T, which were cultured in M17 medium.
Bacteriocin activity assay.
The bacteriocin assay was performed using the spot-on-lawn method (38) with some modifications, in which 10 μl of twofold dilutions of a bacteriocin preparation was spotted onto a double layer composed of 5 ml of MRS medium supplemented with 1% agar. After overnight incubation at temperatures appropriate for the indicator strains, the bacterial lawns were checked for inhibition zones.
The MICs of the bacteriocin for the various indicator strains were determined with purified bacteriocin solutions using the spot-on-lawn method described above. The MIC was defined as the minimum bacteriocin concentration that yielded a clear zone of growth inhibition in the indicator lawn.
Bacteriocin purification.
Using a three-step procedure, lactocyclicin Q was purified from a 1-liter culture grown until early stationary phase in M17 medium at 30°C for 18 h. The culture was adjusted to pH 3.0 with 1 M hydrochloric acid in order to recover bacteriocin peptides adsorbed on the cells, and then the cells were removed by centrifugation at 8,000 × g for 20 min at 4°C. The supernatant obtained was applied to an SP Sepharose Fast Flow cation-exchange chromatography column (length, 100 mm; internal diameter, 10 mm; GE Healthcare, Uppsala, Sweden) equilibrated with 50 mM sodium citrate buffer, pH 3.0 (buffer A). After the column was washed with 100 ml buffer A, the bacteriocin was eluted with 50 ml of 1 M NaCl in buffer A. Subsequently, the eluted active fraction was applied to an Octyl-Sepharose CL-4B column (length, 50 mm; internal diameter, 10 mm; GE Healthcare) equilibrated with 1 M ammonium sulfate in 50 mM sodium phosphate buffer, pH 5.6 (buffer B). Then an active fraction was eluted with 70% ethanol in buffer B. This fraction was applied to a 3-ml RESOURCE RPC column (Amersham Biosciences, Uppsala, Sweden) in an LC-2000 Plus high-performance liquid chromatography (HPLC) system (JASCO, Tokyo, Japan). The active fraction was eluted in the following manner with a gradient of MilliQ water-acetonitrile containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min: 0 to 5 min, 0 to 40% acetonitrile; 5 to 10 min, 40% acetonitrile; 10 to 35 min, 40 to 65% acetonitrile; 35 to 40 min, 100% acetonitrile; and 40 to 45 min, 100% acetonitrile. The activities of the fractions were examined by the spot-on-lawn method.
The purified lactocyclicin Q was stored at −30°C. The antibacterial activity of the fraction obtained in each purification step was determined as described above by using Bacillus coagulans JCM 2257T as an indicator strain. The protein concentration of each fraction was estimated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) according to the manufacturer's instructions.
For MIC determination and characterization of the bacteriocins, the solvent was removed from the purified fractions by using a SpeedVac concentrator (Savant, Farmingdale, NY), and the bacteriocins were dissolved at appropriate concentrations in distilled water supplemented with 0.1% Tween 80 unless otherwise mentioned.
MS and amino acid sequencing.
The molecular mass of the purified lactocyclicin Q was analyzed by electrospray ionization time of flight mass spectrometry (ESI-TOF MS) with a JMS-T100LC mass spectrometer (JEOL, Tokyo, Japan). The amino acid sequence was determined based on Edman degradation with a protein sequencer (model PPSQ-21; Simadzu, Kyoto, Japan).
Effects of enzymes, heat, and pH on the activity of lactocyclicin Q.
To examine the enzyme sensitivity of lactocyclicin Q, it was treated with the following enzymes (Sigma-Aldrich, St. Louis, MO): trypsin (1,000 U/mg), α-chymotrypsin (37 U/mg), proteinase K (0.6 U/mg), and papain (17 U/mg).
All enzymes were dissolved in appropriate buffers with optimal pHs and then were filter sterilized and added to a purified lactocyclicin Q solution at an enzyme-to-peptide molar ratio of 1:100 or 1:1. Following incubation at 37°C for 3 h, the enzymes were denatured by heating them at 100°C for 5 min. The residual bacteriocin activities were determined by using B. coagulans JCM 2257T as described above.
For evaluation of the heat and pH stability of lactocyclicin Q, the purified bacteriocin solution was adjusted to pH values between 3.0 and 10.0 using 1 M hydrochloric acid or 1 M sodium hydroxide. Then the preparations were heated at 80°C, 100°C, or 121°C for 30 or 15 min. The residual bacteriocin activities were determined as described above. The bacteriocin activity of the pH 3.0 sample without heat treatment was defined as 100%.
Fragmentation of lactocyclicin Q.
Two fragmentation procedures, 3-bromo-3-methyl-2-(2-nitrophenylthio)-3H-indole (BNPS-skatole) and hydrochloric acid treatments, were performed with lactocyclicin Q.
Purified lactocyclicin Q was dissolved in 700 μl acetic acid with a 100-fold molar excess of BNPS-skatole (Sigma-Aldrich). The presence of this chemical reagent leads to cleavage of the polypeptide at the C-terminal side of tryptophan residues. The reaction was performed at 37°C for 72 h. The product was purified by reverse-phase HPLC, and it was then subjected to ESI-TOF MS and amino acid sequencing as described above.
Purified lactocyclicin Q was also dissolved in 800 μl of 0.6 M and 0.03 M hydrochloric acid at a final concentration of 0.04 mg/ml and then incubated at 37°C for 24 h and at 110°C for 3 h, respectively.
DNA sequence analysis.
To determine the gene encoding lactocyclicin Q, oligonucleotide primers were designed on the basis of the amino acid sequences obtained. DNA manipulations were performed according to a previously described protocol (10). The primers used in this study are listed in Table 1. Degenerate primers (Q12.F3 and Q12.R1) and Lactococcus sp. strain QU 12 total DNA were employed to amplify part of the lactocyclicin Q structural gene by using Taq DNA polymerase (Promega, Madison, WI) according to the standard protocol (10). To amplify the upstream or downstream region of the structural gene, ligation-anchored PCR was performed as described previously, with some modifications (34, 35). Lactococcus sp. strain QU 12 total DNA was digested with BamHI, EcoRI, HindIII, KpnI, SacI, or XbaI (Nippon Gene, Tokyo, Japan), and the digested DNA was ligated into a dephosphorylated pUC18 cloning vector (Toyobo, Osaka, Japan) that was treated with the corresponding restriction enzyme. Each of the mixtures was then used as a template for PCR using a structural gene-specific primer (Lact.F1) and vector-specific primers (1stMup13-f and 1stMup13-r). The second (with primers Lact.F2, Mup13-f, and Mup13-r) and third (with Lact.F3, s-M13-f, and s-M13-r) PCRs were performed as described above. The fragments obtained were purified by using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced. Similarly, the DNA sequence of the downstream region was analyzed using the specific primers Lact.R1, Lact.R2, and Lact.R3 and vector-specific primers. A single fragment containing the entire lactocyclicin Q gene was amplified using the newly designed specific primers LycQ-F and LycQ-R. The product was purified and directly sequenced.
TABLE 1.
Oligonucleotide primers used to obtain the gene encoding lactocyclicin Q
| Primer | Corresponding amino acid sequence for degenerate primer | Sequence (5′-3′) |
|---|---|---|
| QU12.F1 | VKHQGK | GTNAARCAYCARGGNAA |
| QU12.R1 | KGQHKV | TTNCCYTGRTGYTTNAC |
| QU12.F2 | WLIDHL | TGGYTNATHGAYCAYYT |
| QU12.R2 | LHDILW | ARRTGRTCDATNARCCA |
| QU12.F3 | WAVDTI | TGGYTNATHGAYCAYYT |
| QU12.R3 | ITDVAW | ATNGTRTCNACNGCCCA |
| QU12.F4 | NLASWV | AAYYTNGCNWSNTGGGT |
| QU12.R4 | VWSALN | ACCCANSWNGCNARRTT |
| 1stMup13-f | TTAACTATGCGGCATCAGA | |
| 1stMup13-r | TAATGTGAGTTAGCTCACTC | |
| Mup13-f | AAGGCGATTAAGTTGGGTA | |
| Mup13-r | GTATGTTGTGTGGAATTGTG | |
| s-M13-f | GTAAAACGACGGCCAGT | |
| s-M13-r | TTCACACAGGAAACAGG | |
| Lact.F1 | AAATTAATTGATCATTTAGGTGC | |
| Lact.R1 | GCACCTAAATGATCAATTAATTT | |
| Lact.F2 | AGCTGGTTTAGCAACTGCT | |
| Lact.R2 | AGCAGTTGCTAAACCAGCT | |
| Lact.F3 | AACATCAAGGTAAAGCTGCC | |
| Lact.R3 | GGCAGCTTTACCTTGATGTT | |
| LycQ-F | GTGCACGGGTAATTAATAG | |
| LycQ-R | CTATGGATTACTCCTAAAGCG |
DNA sequencing was carried out by Macrogen Inc. (Seoul, Republic of Korea).
Computer analysis of DNA and amino acid sequences.
The DNA and amino acid sequences obtained were analyzed using GENETYX-WIN software (GENETYX, Tokyo, Japan). Database searches were performed using National Center for Biotechnology Information BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The secondary structure was predicted using PSIPRED, the Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/psiform.html).
Nucleotide sequence accession number.
The DNA sequence obtained has been deposited in the DDBJ database under accession number AB462499.
RESULTS
Identification of strain QU 12.
The 16S rRNA gene sequence of strain QU 12 showed a maximum level of identity of 94% to the Lactococcus raffinolactis accession no. EU091469 sequence (reference strains). In addition, an API50 CHL sugar fermentation pattern test of this strain revealed 70% identity to L. lactis subsp. lactis. Therefore, strain QU 12 can be classified as a member of the genus Lactococcus, but it cannot be classified at the species level.
Purification and structural analysis of lactocyclicin Q.
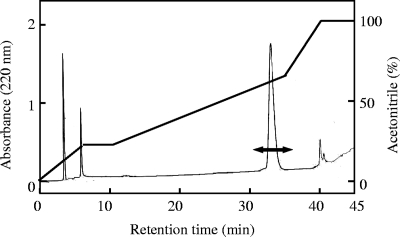
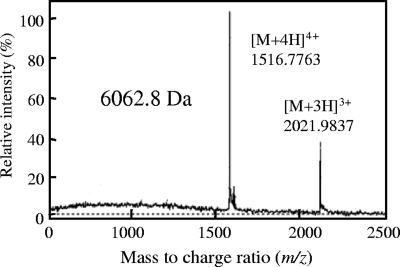
The bacteriocin from Lactococcus sp. strain QU 12 was purified by using three steps, as shown in Table 2. In the final reverse-phase HPLC step, antimicrobial activity was detected at a retention time of about 29 min (Fig. 1). The bacteriocin was purified with a yield of 25% and a concentration of 0.401 mg/ml, as shown in Table 2. The molecular mass of the purified lactocyclicin Q was determined by ESI-TOF MS analysis to be 6,062.8 Da (Fig. 2). Amino acid composition analysis proved that lactocyclicin Q was proteinaceous (data not shown). However, no signals were obtained when lactocyclicin Q was subjected to N-terminal amino acid sequencing by automated Edman degradation. This suggested that the N terminus of lactocyclicin Q was blocked.
TABLE 2.
Purification of lactocyclicin Q produced by Lactococcus sp. QU 12
| Step | Vol (ml) | Total activity (AU)a | Yield (%) | Total protein (mg)b | Purification (fold) |
|---|---|---|---|---|---|
| Supernatant | 1,000 | 128 × 105 | 100 | 16,100 | 1.00 |
| SP Sepharose | 50 | 102 × 105 | 80 | 4,530 | 2.84 |
| Octyl-Sepharose | 50 | 512 × 104 | 40 | 710 | 9.08 |
| Reverse-phase HPLC | 16 | 320 × 104 | 25 | 0.401 | 10,100 |
Antibacterial activity (expressed in arbitrary units [AU]) was assayed by the spot-on-lawn method using B. coagulans JCM 2257T as an indicator strain.
The protein concentration (in mg/ml) was estimated with a NanoDrop ND-1000 (A280).
FIG. 1.
Reverse-phase HPLC profile for purification of lactocyclicin Q produced by Lactococcus sp. strain QU 12. Lactocyclicin Q was isolated using a 3-ml RESOURCE RPC column (Amersham Biosciences, Uppsala, Sweden) at a flow rate of 1.0 ml/min under the gradient conditions indicated. Antibacterial activity was detected in the fractions indicated by the double-headed arrow.
FIG. 2.
ESI-TOF MS of purified lactocyclicin Q. The multiple charged molecular ions that were detected are indicated.
Fragmentation and amino acid sequence of lactocyclicin Q.
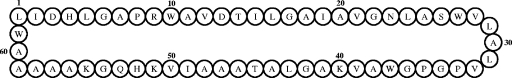
To obtain a partial amino acid sequence of lactocyclicin Q, various fragmentation procedures were performed. However, some treatments with enzymes and chemical reagents, such as lysyl-endopeptidase and cyanogen bromide, could not provide appropriate fragments of lactocyclicin Q (data not shown). BNPS-skatole treatment, which cleaves the C-terminal region of tryptophan residues, yielded three products with the molecular masses of 1,190.7, 2,290.1, and 1,022.3 Da. These fragment peptides were subjected to N-terminal amino acid sequencing, and the following sequences were obtained: LIDHIGAPR for the 1,190.7-Da fragment, AVKAGLATAAAIVKHQGKAAAAA for the 2,290.1-Da fragment, and VLALVPGPG for the 1,022.3-Da fragment. Tryptophan residues modified by the treatment were not detected, but it was expected that the fragments contained tryptophan at the ends of the C termini. In fact, the calculated molecular masses of the sequences obtained, including putative tryptophan at the ends of the C termini, were 1,176.43 Da, 2,275.66 Da, and 1,008.20 Da, respectively (Table 3). This indicated that the C-terminally modified tryptophan residues were oxidized by the treatment. Additionally, lactocyclicin Q was treated with 0.6 M or 0.03 M hydrochloric acid to obtain fragment peptides for amino acid sequence analysis. ESI MS revealed that the lactocyclicin Q treated with 0.6 M hydrochloric acid had a molecular mass of 6,080.8 Da. This suggested that the lactocyclicin Q combined with a molecule of water and was hydrolyzed. On the other hand, the peptide treated with 0.03 M hydrochloric acid had a molecular mass of 5,963.1 Da. These peptides were purified and sequenced, and the following sequences were obtained: GKAAAAAWLIDHLGAPRWAVDTILGAIAV for the peptide treated with 0.6 M hydrochloric acid and TILGAIAVGNLASWVLALVPGPGWAVKAG for the peptide treated with 0.03 M hydrochloric acid (Table 3). Consequently, the sequences obtained for the fragments were linked with each other, and all the amino acid sequences of lactocyclicin Q were chemically determined. Lactocyclicin Q is composed of 61 amino acid residues and forms a cyclic structure with a peptide bond (Fig. 3). The calculated molecular mass of the structure obtained was 6,080.8 Da, which was consistent with the observed peptides treated with 0.6 M hydrochloric acid. Lactocyclicin Q is the first cyclic bacteriocin purified from Lactococcus species, and it showed no significant homology to any other antimicrobial peptide, including cyclic bacteriocins.
TABLE 3.
Amino acid sequences of fragments obtained by treatment of lactocyclicin Q
| Treatment | Molecular mass of fragment obtained (Da) | Amino acid sequencea |
|---|---|---|
| BNPS-skatole | 1,190.7 | LIDHLGAPRW |
| 2,290.1 | AVKAGLATAAAIVKHQGKAAAAAW | |
| 1,022.3 | VLALVPGPGW | |
| 0.6 M HCl | 6,080.8 | GKAAAAAWLIDHLGAPRWAVDTILGAIAV |
| 0.03 M HCl | 5,963.1 | TILGAIAVGNLASWVLALVPGPGWAVKAG |
Underlined and double-underlined amino acid sequences indicate overlapping positions. Tryptophan residues at the C terminus of the peptide obtained by BNPS-skatole treatment were estimated by the reaction mechanism.
FIG. 3.
Structure of lactocyclicin Q. The cyclic structure of lactocyclicin Q is achieved through a dehydration reaction between L1 and W61. The numbers of the amino acid residues correspond to the sequence predicted by the DNA sequence of the gene encoding lactocyclicin Q.
DNA sequencing analysis for elucidation of the structure of lactocyclicin Q.
Degenerate primers were designed on the basis of the amino acid sequence obtained. When PCR was performed with the total DNA from Lactococcus sp. strain QU 12 and the degenerate primers, a 130-bp fragment was amplified. Then, based on the DNA sequence obtained, the specific primers were designed and employed for ligation-anchored PCRs to obtain the adjacent sequences. As a result, the DNA sequence of the structural gene encoding lactocyclicin Q, termed lycQ, was obtained (Fig. 4). A putative leader peptide (dipeptide) sequence, a ribosome-binding site, and a terminator sequence were also identified. The structural gene lycQ consisted of a 189-bp open reading frame encoding a primary translation product of 63 amino acid residues. This indicated that lactocyclicin Q was synthesized as a 63-amino-acid prepeptide, which was processed between lysine and leucine to produce a 61-amino-acid propeptide. It is expected that the putative leader peptide is cleaved off during cyclization. This suggests that the propeptide matures to yield cyclization of the N-terminal leucine to the C-terminal tryptophan with dehydration by a peptide bond. In addition, lactocyclicin Q has a K-X-X-X-X-X-W sequence at the end of the C terminus, like most other cyclic bacteriocins.
FIG. 4.
Nucleotide sequence of the region containing the lactocyclicin Q structural genes. Ribosome-binding sites (RBS) and terminator sequences are underlined. The putative leader peptide is indicated by italics. An asterisk indicates the stop codon. The sequence corresponding to the mature peptide is indicated by bold type.
Antimicrobial spectrum of lactocyclicin Q.
The inhibitory spectrum of lactocyclicin Q is shown in Table 4. Lactocyclicin Q showed a broad antimicrobial spectrum against gram-positive food spoilage bacteria, such as the genera Bacillus and Enterococcus and closely related bacteria in the genus Lactococcus. Furthermore, lactocyclicin Q had weak inhibitory activity against E. coli without other antimicrobial factors. However, the MICs for E. coli were at the same level of self-immunity as those for the lactocyclicin Q-producing strain. No activity was detected even with 312 μM of lactocyclicin Q against Salmonella enterica serovar Typhimurium NBRC 13245.
TABLE 4.
Antimicrobial spectrum of lactocyclicin Q
| Indicator straina | MIC (μM) |
|---|---|
| Lactococcus lactis subsp. lactis JCM 7638 | 0.141 |
| Lactococcus lactis subsp. lactis ATCC 19435T | 0.141 |
| Lactococcus lactis subsp. lactis IL1403 | 0.141 |
| Lactococcus lactis QU 1 | 0.141 |
| Lactococcus lactis subsp. cremoris ATCC 19275T | 0.283b |
| Lactococcus raffinolactis JCM 5706T | 0.710 |
| Lactobacillus sakei subsp. sakei JCM 1157T | 0.0150 |
| Lactobacillus casei subsp. casei JCM 1134T | 2.27 |
| Lactobacillus brevis JCM 1059T | 1.14 |
| Lactobacillus acidophilus JCM 1132T | 2.86 |
| Lactobacillus coryniformis subsp. coryniformis | |
| JCM 1164T | 2.86 |
| Lactobacillus kimchii JCM 10707T | 0.710 |
| Leuconostoc mesenteroides subsp. mesenteroides | |
| JCM 6124T | 1.03 |
| Weissella cibaria JCM 12495T | 0.710 |
| Pediococcus pentosaceus JCM 5885 | 0.55 |
| Pediococcus pentosaceus JCM 5890T | 2.86 |
| Pediococcus acidilactici JCM 8797T | 2.86 |
| Pediococcus dextrinicus JCM 5887T | 0.141 |
| Enterococcus faecium JCM5804T | 0.710 |
| Enterococcus hirae ATCC 10541 | 0.710 |
| Enterococcus durans NBRC 100479T | 0.710 |
| Enterococcus faecalis JCM 5803T | 0.260 |
| Streptococcus salivarius JCM 5707T | 22.9 |
| Streptococcus bovis JCM 5802T | 22.9 |
| Bacillus coagulans JCM 2257T | 0.0150 |
| Bacillus circulans JCM 2504T | 0.0310 |
| Bacillus subtilis JCM 1465T | 0.0640 |
| Bacillus cereus JCM 2152T | 11.4 |
| Micrococcus luteus IFO 12708 | 5.70 |
| Listeria innocua ATCC 33090T | 1.03 |
| Listeria monocytogenes ATCC BAA-679 | 1.03 |
| Staphylococcus aureus subsp. aureus ATCC 12600T | 91.6 |
| Salmonella enterica serovar Typhimurium NBRC 13245 | NAc |
| Escherichia coli JM109 | 34.3d |
| Escherichia coli NBRC 3301 | 17.3d |
| Lactococcus sp. strain QU 12 | 34.3b |
ATCC, American Type Culture Collection, Rockville, MD; IFO, Institute for Fermentation, Osaka, Japan; JCM, Japan Collection of Microorganisms, Saitama, Japan; NBRC, NITE Biological Resource Center, Chiba, Japan; QU, Laboratory of Microbial Technology, Kyushu University, Fukuoka, Japan.
M17 medium was used instead of MRS medium.
NA, no activity (>312 μM).
Observed after 48 h of incubation.
Stability of lactocyclicin Q with proteolytic enzymes, pH, and heat treatment.
The inhibitory activity of lactocyclicin Q was inactivated completely only by pepsin and proteinase K when the molar ratio of the enzyme to lactocyclicin Q was 1:1 (Table 5). In the other cases examined, especially at a molar ratio of 1:100, lactocyclicin Q retained its activity. The inhibitory activity was diminished by exposure to elevated temperatures and high pHs, but the activity was not decreased by autoclaving at 121°C for 15 min at pH 3.0 and 4.0 (Table 6). Lactocyclicin Q showed high resistance to proteases and a high level of stability against pH and heat compared to most LAB noncyclic bacteriocins.
TABLE 5.
Effect of enzyme treatment on lactocyclicin Q
| Enzyme | Relative activity (%) at an enzyme/protein molar ratio of:
|
|
|---|---|---|
| 1:100 | 1:1 | |
| α-Chymotrypsin | 80a | 60 |
| Trypsin | 80 | 40 |
| Pepsin | 40 | 40 |
| Papain | 40 | 20 |
| Proteinase K | 60 | 40 |
The activity of the control preparation without any treatment was defined as 100%.
TABLE 6.
Stability of purified lactocyclicin Q subjected to different pH and heat treatments
| pH | Relative activity (%) after incubation under the following conditionsa:
|
|||
|---|---|---|---|---|
| Room temperature, overnight | 80°C, 30 min | 100°C, 30 min | 121°C, 15 min | |
| 3.0 | 100 | 100 | 100 | 100 |
| 4.0 | 100 | 100 | 100 | 100 |
| 5.0 | 100 | 100 | 100 | 50.0 |
| 6.0 | 100 | 100 | 25.0 | 12.5 |
| 7.0 | 100 | 25.0 | 25.0 | 6.25 |
| 8.0 | 100 | 25.0 | 25.0 | 0 |
| 9.0 | 100 | 25.0 | 12.5 | 0 |
| 10.0 | 50 | 12.5 | 0 | 0 |
The activity of an untreated sample (pH 3.0) was defined as 100%.
DISCUSSION
This is the first report on a cyclic bacteriocin produced by Lactococcus spp. Here we describe identification and characterization of lactocyclicin Q that was purified from culture supernatant of Lactococcus sp. strain QU 12. Lactocyclicin Q had excellent characteristics, such as pH and heat stability, as well as protease resistance. The characteristics described above are likely due to lactocyclicin Q's cyclic structure. For example, the inhibitory activity of nisin Z was almost completely lost after exposure to an elevated temperature under autoclaving conditions (121°C for 15 min at pH 5.0) (28). In addition, the inhibitory activity of nisin Z was completely inactivated by treatments with proteinase K and α-chymotrypsin (28). In contrast, the inhibitory activity of enterocin AS-48 was retained with no reduction under similar conditions (120°C for 15 min at pH 5.0) (2). Enterocin AS-48 was also quite resistant to several proteolytic enzymes, including α-chymotrypsin (14, 15).
Lactocyclicin Q, which forms a cyclic structure like enterocin AS-48, was also resistant to heat and several enzymes, especially α-chymotrypsin (Table 5). Both enterocin AS-48 and lactocyclicin Q exhibited high stability in the presence of heat and protease, a common feature of cyclic peptides, compared to most LAB bacteriocins. In fact, cyclic structures have been proven to be responsible for high stability (5, 15, 23).
The primary structure of lactocyclicin Q was chemically determined, and the molecule was composed of 61 amino acid residues. The amino acid sequencing analysis showed that lactocyclicin Q had a cyclic structure bound at the N and C termini. In addition, lactocyclicin Q had the highest hydrophobic amino acid ratio (78.7%) among cyclic bacteriocins and showed no significant homology to any other bacteriocin. In addition, lactocyclicin Q showed high antimicrobial activity against the genera Bacillus, Lactococcus, and Enterococcus (Table 4). In a previous report, gassericin A showed inhibitory activity against Listeria monocytogenes, Bacillus cereus, Staphylococcus aureus, and other gram-positive bacteria (31). On the other hand, reutericin 6 had a narrower spectrum and weaker antimicrobial activity than gassericin A (32). Enterocin AS-48 showed inhibitory activity against not only the gram-positive genera Bacillus, Staphylococcus, and Listeria but also the gram-negative genus Salmonella, E. coli, and other gram-negative organisms (1, 2). The cyclic bacteriocins reported previously had antimicrobial spectra different than that of lactocyclicin Q, suggesting that lactocyclicin Q had an antimicrobial mechanism that was different from those of other cyclic bacteriocins. In addition, according to an analysis of the crystal structure, enterocin AS-48 is thought to have two different modes of molecular association for inhibitory activity. The model can be explained by the crystal containing chains of molecule pairs linked either by hydrophobic interactions (dimeric form I [water-soluble DF-I]) or by hydrophilic interactions (dimeric form II [membrane-bound DF-II]). It has been proposed that rearrangement from water-soluble DF-I to membrane-bound DF-II takes place at the membrane surface, which is followed by membrane insertion and molecular electroporation (30).
The antimicrobial mechanism of cyclic bacteriocins should be elucidated to develop these molecules as food preservatives. The structure of enterocin AS-48, which is deeply involved in antimicrobial activity, has been determined by nuclear magnetic resonance to consist of a globular arrangement of five α-helices (7, 30). The α1-helix (Ala9 to Glu21), α2-helix (Trp25 to Thr34), and α4-helix (Gly37 to Ala45) have a subtle amphiphilic character, and the overall structure shows high amphiphilicity (24).
On the other hand, four α-helices in the secondary structure of lactocyclicin Q (α1-helix [Ala11 to Ala18], α2-helix [Gly22 to Leu29], α3-helix [Ala41 to Ala48], and α4-helix [Ala57 to His4]), which may have subtle amphiphilic characteristics, have been predicted by using PSIPRED prediction. The molecular weights of the cyclic bacteriocins gassericin A, reutericin 6, acidocin B, and uberolysin determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis are significantly lower than the real molecular weights (15, 17). These bacteriocins are thought to form compact structures. However, the molecular weight of lactocyclicin Q was the same when ESI-TOF MS and sodium dodecyl sulfate-polyacrylamide gel electrophoresis were used (data not shown), like the molecular weight of enterocin AS-48 (2). This indicated that lactocyclicin Q and enterocin AS-48 had more elongated structures. The difference might be due to various factors, such as amino acid composition, charge, and/or number of α-helices.
The DNA sequence of the codons for a putative leader peptide, a ribosome-binding site, and a terminator sequence and the structural gene encoding lactocyclicin Q was obtained. Lactocyclicin Q has a short leader peptide composed of only two amino acid residues. This peptide is similar to those reported for circularin A and uberolysin, which have three and six N-terminal amino acids in their leader peptides, respectively. However, the cleavage reaction for maturation and the functions of the short leader peptides have not been unraveled yet. Lactocyclicin Q was expected to undergo a dehydration reaction between the N-terminal leucine residue and the C-terminal tryptophan residue during maturation. In general, cyclic bacteriocins produced by LAB have aromatic amino acids, such as tyrosine or tryptophan, in the N- or C-terminal region. For example, in gassericin A and other highly homologous cyclic bacteriocins tyrosine and tryptophan are the second and third N-terminal amino acid residues, respectively. On the other hand, in enterocin AS-48 and uberolysin tryptophan is at the end of the C terminus. Furthermore, lactocyclicin Q, uberolysin, and enterocin AS-48 have a K-X-X-X-X-X-W sequence at the end of the C terminus, in contrast to gassericin A. Additionally, in most cyclic bacteriocins leucine or isoleucine is at the end of the N terminus. These amino acids might be one of the factors affecting secretion and cyclization. A possible example would be a consensus sequence for a recognition site for a cyclization enzyme and/or a transporter.
Cyclization mechanisms of cyclic peptides, including those from LAB, have not been well characterized. Recently, two enzymes, McjB and McjC, encoded near mcjA (microcin J25 structural gene), were proven to be necessary for the conversion of McjA to mature microcin J25 (9), which is a cyclic peptide bacteriocin produced by E. coli AY 25 (4, 29). However, microcin J25, a non-LAB bacteriocin, does not have aromatic amino acids in its N- or C-terminal region, indicating that cyclic bacteriocins produced by LAB have another cyclization mechanism. For enterocin AS-48, the genes as-48BCC1DD1 located near the structural gene as-48A have been confirmed to be necessary for production and immunity, and all of their products are predicted to be located in the membrane (25). The cyclization mechanism of lactocyclicin Q is also thought to involve cyclization enzymes and transmembrane proteins.
The genus Lactococcus, which produces lactocyclicin Q, is one of the safest and most important microorganisms in human life. This means that lactocyclicin Q may be used not only for food preservation but also in the creation of medicinal drugs. In addition, the great stability of lactocyclicin Q due to its cyclic structure means that maintaining strong antimicrobial activity after various treatments and environmental stresses is more likely with this bacteriocin than with the other LAB bacteriocins, like nisin. However, the biosynthesis mechanism for cyclic bacteriocins, including lactocyclicin Q, has not been proven yet. If this mechanism is demonstrated, it may enable us to synthesize various kinds of cyclic peptides efficiently in vivo and in vitro. In addition, intentionally created cyclic peptides may have new characteristics and novel functions, such as a unique antimicrobial spectrum. In the future, cyclic peptides, including lactocyclicin Q, and the derived cyclization technique will likely be used in a variety of fields, including drug development.
Acknowledgments
We are grateful to K. Ogawa (Asahi Kasei Pharma Corporation, Ohito, Japan) for amino acid composition analysis, K. Furukawa of Kyushu University, Japan, for kindly giving us access to an amino acid sequencer, and F. Yoneyama of Kyushu University for experimental assistance.
This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (JSPS) and by a research project for utilizing advanced technologies in agriculture, forestry, and fisheries of the Ministry of Agriculture, Forestry and Fisheries of Japan.
Footnotes
Published ahead of print on 9 January 2009.
REFERENCES
- 1.Abriouel, H., E. Valdivia, A. Gálvez, and M. Maqueda. 1998. Response of Salmonella choleraesuis LT2 spheroplasts and permeabilized cells to the bacteriocin AS-48. Appl. Environ. Microbiol. 64:4623-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abriouel, H., E. Valdivia, A. Gálvez, and M. Maqueda. 2001. Influence of physico-chemical factors on the oligomerization and biological activity of bacteriocin AS-48. Curr. Microbiol. 42:89-95. [DOI] [PubMed] [Google Scholar]
- 3.Abriouel, H., R. Lucas, N. Ben Omar, E. Valdivia, M. Maqueda, M. Martinez-Canamero, and A. Galvez. 2005. Enterocin AS-48RJ: a variant of enterocin AS-48 chromosomally encoded by Enterococcus faecium RJ16 isolated from food. Syst. Appl. Microbiol. 28:383-397. [DOI] [PubMed] [Google Scholar]
- 4.Bayro, M. J., J. Mukhopadhyay, G. V. Swapna, J. Y. Huang, L. C. Ma, E. Sineva, P. E. Dawson, G. T. Montelione, and R. H. Ebright. 2003. Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J. Am. Chem. Soc. 125:12382-12383. [DOI] [PubMed] [Google Scholar]
- 5.Cobos, E. S., V. V. Filimonov, A. Galvez, M. Maqueda, E. Valdivia, J. C. Martinez, and P. L. Mateo. 2001. AS-48: a circular protein with an extremely stable globular structure. FEBS Lett. 505:379-382. [DOI] [PubMed] [Google Scholar]
- 6.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 7.Craik, D. J., N. L. Daly, I. Saska, M. Trabi, and K. J. Rosengren. 2003. Structures of naturally occurring circular proteins from bacteria. J. Bacteriol. 185:4011-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drider, D., G. Fimland, Y. Héchard, L. M. McMullen, and H. Prévost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duquesne, S., D. Destoumieux-Garzon, S. Zirah, C. Goulard, J. Peduzzi, and S. Rebuffat. 2007. Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem. Biol. 14:793-803. [DOI] [PubMed] [Google Scholar]
- 10.Fujita, K., S. Ichimasa, T. Zendo, S. Koga, F. Yoneyama, J. Nakayama, and K. Sonomoto. 2007. Structural analysis and characterization of lacticin Q, a novel bacteriocin belonging to a new family of unmodified bacteriocins of gram-positive bacteria. Appl. Environ. Microbiol. 73:2871-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galvez, A., M. Maqueda, E. Valdivia, A. Quesada, and E. Montoya. 1986. Characterization and partial purification of a broad spectrum antibiotic AS-48 produced by Streptococcus faecalis. Can. J. Microbiol. 32:765-771. [DOI] [PubMed] [Google Scholar]
- 12.Garneau, S., N. I. Martin, and J. C. Vederas. 2002. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84:577-592. [DOI] [PubMed] [Google Scholar]
- 13.Gross, E., and J. L. Morell. 1971. The structure of nisin. J. Am. Chem. Soc. 93:4634-4635. [DOI] [PubMed] [Google Scholar]
- 14.Joosten, H. M., M. Nunez, B. Devreese, J. Van Beeumen, and J. D. Marugg. 1996. Purification and characterization of enterocin 4, a bacteriocin produced by Enterococcus faecalis INIA 4. Appl. Environ. Microbiol. 62:4220-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai, Y., R. Kemperman, J. Kok, and T. Saito. 2004. The circular bacteriocins gassericin A and circularin A. Curr. Protein Pept. Sci. 5:393-398. [DOI] [PubMed] [Google Scholar]
- 16.Kawai, Y., T. Saito, H. Kitazawa, and T. Itoh. 1998. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci. Biotechnol. Biochem. 62:2438-2440. [DOI] [PubMed] [Google Scholar]
- 17.Kawai, Y., Y. Ishii, K. Arakawa, K. Uemura, B. Saitoh, J. Nishimura, H. Kitazawa, Y. Yamazaki, Y. Tateno, T. Itoh, and T. Saito. 2004. Structural and functional differences in two cyclic bacteriocins with the same sequences produced by lactobacilli. Appl. Environ. Microbiol. 70:2906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemperman, R., A. Kuipers, H. Karsens, A. Nauta, O. Kuipers, and J. Kok. 2003. Identification and characterization of two novel clostridial bacteriocins, circularin A and closticin 574. Appl. Environ. Microbiol. 69:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 20.Leer, R. J., J. M. van der Vossen, M. van Giezen, J. M. van Noort, and P. H. Pouwels. 1995. Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141:1629-1635. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Lara, I., A. Galvez, M. Martinez-Bueno, M. Maqueda, and E. Valdivia. 1991. Purification, characterization, and biological effects of a second bacteriocin from Enterococcus faecalis ssp. liquefaciens S-48 and its mutant strain B-48-28. Can. J. Microbiol. 37:769-774. [DOI] [PubMed] [Google Scholar]
- 22.Maisnier-Patin, S., E. Forni, and J. Richard. 1996. Purification, partial characterisation and mode of action of enterococcin EFS2, an antilisterial bacteriocin produced by a strain of Enterococcus faecalis isolated from a cheese. Int. J. Food Microbiol. 30:255-270. [DOI] [PubMed] [Google Scholar]
- 23.Maqueda, M., A. Galvez, M. M. Bueno, M. J. Sanchez-Barrena, C. Gonzalez, A. Albert, M. Rico, and E. Valdivia. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr. Protein Pept. Sci. 5:399-416. [DOI] [PubMed] [Google Scholar]
- 24.Maqueda, M., M. Sanchez-Hidalgo, M. Fernandez, M. Montalban-Lopez, E. Valdivia, and M. Martinez-Bueno. 2008. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol. Rev. 32:2-22. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Bueno, M., M. Maqueda, A. Galvez, B. Samyn, J. Van Beeumen, J. Coyette, and E. Valdivia. 1994. Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J. Bacteriol. 176:6334-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulders, J. W., I. J. Boerrigter, H. S. Rollema, R. J. Siezen, and W. M. de Vos. 1991. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 201:581-584. [DOI] [PubMed] [Google Scholar]
- 27.Nes, I. F., D. B. Diep, L. S. Håvarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 28.Noonpakdee, W., C. Santivarangkna, P. Jumriangrit, K. Sonomoto, and S. Panyim. 2003. Isolation of nisin-producing Lactococcus lactis WNC 20 strain from nham, a traditional Thai fermented sausage. Int. J. Food Microbiol. 81:137-145. [DOI] [PubMed] [Google Scholar]
- 29.Rosengren, K. J., R. J. Clark, N. L. Daly, U. Goransson, A. Jones, and D. J. Craik. 2003. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 125:12464-12474. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez-Barrena, M. J., M. Martinez-Ripoll, A. Galvez, E. Valdivia, M. Maqueda, V. Cruz, and A. Albert. 2003. Structure of bacteriocin AS-48: from soluble state to membrane bound state. J. Mol. Biol. 334:541-549. [DOI] [PubMed] [Google Scholar]
- 31.Toba, T., E. Yoshioka, and T. Itoh. 1991. Potential of Lactobacillus gasseri isolated from infant faeces to produce bacteriocin. Lett. Appl. Microbiol. 12:228-231. [Google Scholar]
- 32.Toba, T., S. K. Samant, E. Yoshioka, and T. Itoh. 1991. Reutericin 6, a new bacteriocin produced by Lactobacillus reuteri LA6. Lett. Appl. Microbiol. 13:281-286. [Google Scholar]
- 33.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1997. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol. 179:7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Troutt, A. B., M. G. McHeyzer-Williams, B. Pulendran, and G. J. Nossal. 1992. Ligation-anchored PCR: a simple amplification technique with single-sided specificity. Proc. Natl. Acad. Sci. USA 89:9823-9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughan, A., V. G. Eijsink, and D. Van Sinderen. 2003. Functional characterization of a composite bacteriocin locus from malt isolate Lactobacillus sakei 5. Appl. Environ. Microbiol. 69:7194-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirawan, R. E., K. M. Swanson, T. Kleffmann, R. W. Jack, and J. R. Tagg. 2007. Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 153:1619-1630. [DOI] [PubMed] [Google Scholar]
- 37.Zendo, T., M. Fukao, K. Ueda, T. Higuchi, J. Nakayama, and K. Sonomoto. 2003. Identification of the lantibiotic nisin Q, a new natural nisin variant produced by Lactococcus lactis 61-14 isolated from a river in Japan. Biosci. Biotechnol. Biochem. 67:1616-1619. [DOI] [PubMed] [Google Scholar]
- 38.Zendo, T., N. Eungruttanagorn, S. Fujioka, Y. Tashiro, K. Nomura, Y. Sera, G. Kobayashi, J. Nakayama, A. Ishizaki, and K. Sonomoto. 2005. Identification and production of a bacteriocin from Enterococcus mundtii QU 2 isolated from soybean. J. Appl. Microbiol. 99:1181-1190. [DOI] [PubMed] [Google Scholar]
- 39.Zendo, T., S. Koga, Y. Shigeri, J. Nakayama, and K. Sonomoto. 2006. Lactococcin Q, a novel two-peptide bacteriocin produced by Lactococcus lactis QU 4. Appl. Environ. Microbiol. 72:3383-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]