Abstract
Although the health-promoting roles of bifidobacteria are widely accepted, the diversity of bifidobacteria among the human intestinal microbiota is still poorly understood. We performed a census of bifidobacterial populations from human intestinal mucosal and fecal samples by plating them on selective medium, coupled with molecular analysis of selected rRNA gene sequences (16S rRNA gene and internally transcribed spacer [ITS] 16S-23S spacer sequences) of isolated colonies. A total of 900 isolates were collected, of which 704 were shown to belong to bifidobacteria. Analyses showed that the culturable bifidobacterial population from intestinal and fecal samples include six main phylogenetic taxa, i.e., Bifidobacterium longum, Bifidobacterium pseudocatenulatum, Bifidobacterium adolescentis, Bifidobacterium pseudolongum, Bifidobacterium breve, and Bifidobacterium bifidum, and two species mostly detected in fecal samples, i.e., Bifidobacterium dentium and Bifidobacterium animalis subp. lactis. Analysis of bifidobacterial distribution based on age of the subject revealed that certain identified bifidobacterial species were exclusively present in the adult human gut microbiota whereas others were found to be widely distributed. We encountered significant intersubject variability and composition differences between fecal and mucosa-adherent bifidobacterial communities. In contrast, a modest diversification of bifidobacterial populations was noticed between different intestinal regions within the same individual (intrasubject variability). Notably, a small number of bifidobacterial isolates were shown to display a wide ecological distribution, thus suggesting that they possess a broad colonization capacity.
The human gastrointestinal tract (GIT) is a complex ecosystem which contains representative species of the three different domains of life: the Eukarya, the Bacteria, and the Archaea. This very complex population, also known as the gut microbiota, represents one of the highest cellular densities in natural ecosystems, reaching 1011 to 1012 cells/ml of luminal content (31). However, in contrast to other ecological niches, the biodiversity found in the human gut microbiota is relatively low (46). In fact, this ecosystem appears to be dominated by a relatively small number of bacterial taxa, in particular by representatives of Bacteroidetes and Firmicutes (4, 6, 8, 37, 60). The structure and composition of the human GIT microbiota reflect natural selection at both the microbial and host levels through complex competitive and symbiotic interactions. Bacterial colonization of the GIT occurs immediately after delivery and is influenced by numerous factors, including infant diet (e.g., breast-feeding or formula feeding) and hygiene conditions (7). One of the first groups of bacterial colonizers is represented by the bifidobacteria, whose numbers decline with the age of the host, although it has been claimed that the native bifidobacterial microbiota remains stable during the entire life span of a human being (38). The genus Bifidobacterium includes gram-positive bacteria belonging to the Actinobacteria phylum, which are characterized by nonmotile, nonsporulating, non-gas-producing, anaerobic microorganisms. Most bifidobacterial species described so far have been isolated from the GIT of mammals, insects, or birds (for a review, see reference 56). Some bifidobacterial species (e.g., Bifidobacterium bifidum, Bifidobacterium breve, and Bifidobacterium longum subsp. longum) are thought to be strictly of human origin, while others, such as Bifidobacterium gallinarum, Bifidobacterium angulatum, and Bifidobacterium cuniculi, appear to be exclusively associated with animal feces (21). The use of fecal material as a representation of the entire gut microbiota should be considered with caution. In fact, the fecal microbiota consists of not only the mucosa-adherent members of the human GIT microbiota but also bacteria that are considered to be transiently present, being derived from diet or other environmental contaminations (6). Certain members of the genus Bifidobacterium have been added for decades as viable bacteria to various functional foods, conferring health-promoting or probiotic effects (2-3). These probiotic activities include reduction of infectious diseases (e.g., bacterial and viral diarrhea), alleviation of chronic inflammatory diseases (e.g., pouchitis and ulcerative colitis), improvement of various physiological conditions (e.g., lowering the rate of cholesterol or lactose intolerance), and finally, reduction of certain risks that impact health (e.g., dental caries, allergy, and even cancer) (25). Due to these health-promoting effects, significant efforts have been made to discover and exploit novel Bifidobacterium strains with scientifically proven probiotic properties and isolated from their natural ecological niche, i.e., the human intestine. Within this context, it is obvious that understanding the contribution of bifidobacteria to the total human gut microbiota represents a crucial step in order to elucidate their role in such a complex ecosystem.
In recent years, molecular tools based on 16S rRNA genes, such as species-specific PCR primers (27-28, 52), amplified restriction polymorphism (51), or alternative molecular markers, such as atpD (50), tuf (49), recA (20, 49), groEL (18, 54), dnaJ (53), purF (53), rpoC (53), dnaB (53), xfp (53, 63), and clpC (53, 58), have been applied for specific identification/tracing of bifidobacteria retrieved from different ecological environments (e.g., human gut or foods) (8, 44). However, most of these studies were based on the analysis of fecal samples, with only few attempting to analyze the mucosa-adherent bifidobacterial component of the intestinal microbiota using colonoscopic samples. Furthermore, little is known about the diversity of bifidobacterial populations occurring between individuals and between different compartments of the GIT within the same individual.
The aim of this study was to perform a large-scale comparative analysis of 16S rRNA gene-internally transcribed spacer (ITS) sequences to better characterize the culturable adherent mucosal and fecal bifidobacterial communities, and to examine how these bifidobacterial communities differ between subjects and between sampling sites.
MATERIALS AND METHODS
Sampling and processing of samples.
Fifty-nine healthy volunteers were selected for this study. These individuals had not undergone any recent surgery, were not taking any medication, and had no gastrointestinal complaints. Furthermore, they were not on any special diet and had not taken probiotics or antibiotics for 2 months prior to sampling. The samples taken or collected from these volunteers included 30 colonic mucosa samples from adolescents or adults and 29 fecal samples from infants. For six subjects, two different mucosa samples (scrapings) were collected for the sigmoid and rectum tracts. The fresh samples were collected in sterile containers, transported to the laboratory in anaerobic jars containing Anaerocult A (Oxoid), and processed immediately upon receipt. Biopsies were taken during colonoscopy with sterilized biopsy forceps (PCF 160 AL colonoscope and FB2441 biopsy forceps; Olympus, GmbH, Germany). The time lapse between biopsy taking and retrieval of the sample was between 20 and 30 s. The biopsy channel was flushed with sterile saline after each sample was taken. Two biopsy samples were collected for each location and were vigorously washed in 900 μl reduced saline solution (0.25% cysteine [Sigma Chemical Co., St. Louis, MO], 10 μg líter−1 vitamin K1 [Sigma], and 0.02 g liter−1 hemin [Sigma]). The study was approved by the relevant human ethics committees (University of Parma and National University of Ireland).
Recovery of bifidobacteria on selective medium, purification, DNA isolation, and MIC analysis.
Serial dilutions of the supernatants derived from mucosa samples or fecal samples were pour plated onto bifidobacterium selective agar (BSM) for selective outgrowth of bifidobacteria. The BSM selective medium was prepared by the addition of 0.05% (wt/vol) l-cysteine hydrochloride and 50 μg mupirocin (Delchimica, Italy) per liter of MRS as described previously (41). Agar plates were incubated anaerobically (anaerobic jars with Anaerocult A gas packs; Oxoid) at 37°C for 72 h. To analyze the dominant Bifidobacterium population of each subject, about 100 to 200 colonies (20 to 30 colonies from each plate), chosen based on different morphologies and sizes, were randomly selected from each sample, and subcultured in Eugon agar (Difco, Italy) and MRS broth for 24 to 48 h. DNA was extracted from each isolate through rapid mechanical cell lysis as described previously (52). Ten colonies are believed to provide adequate representation of the major bacterial strains cultured on a selective medium (16, 61).
MICs for mupirocin for all bacterial isolates were determined by the broth microdilution procedure (34). The inoculum was derived from a broth culture which was incubated for 18 h at 37°C, from which 10 μl was used to inoculate each tube. The trays were covered, placed in plastic bags, and anaerobically incubated at 37°C for 18 to 20 h. The MIC was defined as the lowest concentration of antibiotic giving a complete inhibition of visible growth in comparison to an antibiotic-free control tube. The experiments were replicated at least three times to verify the methodology and reproducibility.
Development of genus-specific Bifidobacterium primers.
All available bifidobacterial and other actinobacterial 16S rRNA genes and 16S-23S rRNA gene spacer region sequences were retrieved from the EMBL and GenBank databases and aligned using the ClustalW software program (17). Two potentially genus-specific primers for Bifidobacterium were designed on the basis of this multiple sequence alignment. These primers were synthesized (MWG Oligo Synthesis) and used in the PCR procedure, as outlined below.
PCR amplification.
PCR was used to amplify the 16S and ITS sequences of all investigated (and suspected) Bifidobacterium isolates. A DNA fragment corresponding to the 16S and ITS region was amplified using the oligonucleotides BIF-specific (5′-GGTGTGAAAGTCCATCGCT-3′) and 23S_bif (5′-GTCTGCCAAGGCATCCACCA-3′).
Each PCR mixture (25 μl) contained 1.5 mM of MgCl2, 20 mM of Tris-HCl, 50 mM of KCl, 200 μM of each deoxynucleoside triphosphate, 25 pmol of each of the two primers, 1 U of Taq DNA polymerase (Taq PCR master mix kit; Qiagen, United Kingdom), and 50 ng of DNA template. Each PCR cycling program consisted of an initial denaturation step of 3 min at 94°C, followed by amplification for 35 cycles as follows: denaturation (30 s at 94°C), annealing (30 s at 56.5°C), and extension (1 min at 72°C). The PCR was completed with a single elongation step (10 min at 72°C). The resulting amplicons were separated on 0.8% agarose gels, followed by ethidium bromide staining. PCR fragments were purified using the PCR purification kit (Qiagen, United Kingdom) and subsequently sequenced.
Molecular typing.
Molecular typing was performed by a PCR-enterobacterial repetitive intergenic consensus sequences (ERIC) approach using the PCR primers ERIC-1 (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC-2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) (55). Each 25-μl reaction mixture contained 10 mM Tris-HCl, 50 mM KCl, 3 mM MgCl2, 200 μM of each deoxynucleoside triphosphate (GBRL, United Kingdom), 1 μM of each primer, 2.5 U of Taq DNA polymerase (GBRL, United Kingdom), and 25 ng of template DNA (extracted as described in reference 48). Amplifications were performed using an Applied Biosystem Verity thermal cycler (Applied Biosystems, Foster City, CA) with the following temperature profiles: 1 cycle at 94°C for 3 min; 35 cycles at 94°C for 30 s, at 48°C for 30 s, and at 72°C for 4 min; and 1 cycle at 72°C for 6 min. Aliquots of each amplification reaction mixture (15 μl each) were separated by electrophoresis in 2% (wt/vol) agarose gels at a voltage of 7 V/cm. Gels were ethidium bromide stained (0.5 μg/ml) and photographed under 260-nm UV light.
DNA sequencing and phylogenetic analysis.
Nucleotide sequencing of both strands from PCR amplicons was performed by Agencourt Bioscience Corporation using the primers bif-sec (5′-CATGCCCCTACGTCCAG-3′) and 23S-bif (5′-CAAGGCATCCACCATACGC-3′). Sequence data assembly and analysis was performed using the DNASTAR software program (version 5.05; DNAstar, Madison, WI). Assembled sequences were then compared to those present in public databases using BLASTn (1). Sequences were grouped into phylotypes using Olsen corrected similarity matrices, such as the least-similar pair within the phylotype exhibiting at least 98% identity (23). Sequence alignments were performed using the MultiAlign program and the ClustalW package. Phylogenetic analysis and trees were calculated using the PHYLIP software package, version 3.5c (9), and NJplot. Trees were calculated using the neighbor-joining method with Kimura's two-parameter model as the substitution model (19). Phylogenetic trees were also calculated by the maximum-likelihood method using the PHYML software program (13). Bootstrap values were computed by resampling 1,000. Dendrograms from gene sequences were also drawn using the ClustalX program (National Center for Biotechnology Information) and were visualized with the TreeView program.
Statistical analysis.
Double principal coordinate analysis (DPCoA) was chosen to characterize the bifidobacterial diversity of the mucosal and fecal samples based on the Rao diversity index (5). These analyses were performed using the R statistical software package (www.r-project.org). Redundancy analysis (1,000 permutations) applied to the results of DPCoA was employed to evaluate the statistical significance of categorical explanatory variables. Cluster analysis methods were used to analyze the relationships between bifidobacterial ITS as detected among each subject and between different intestinal regions within the same individual.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S-ITS sequences obtained in this study are as follows: Bifidobacterium magnum LMG11591, FJ231741; Bifidobacterium pseudolongum subsp. pseudolongum LMG11571, FJ231745; Bifidobacterium minimum LMG11592. FJ231743; Bifidobacterium pseudocatenulatum 249B, FJ231740; Bifidobacterium pseudocatenulatum LMG10505, FJ231744; and Bifidobacterium catenulatum LMG11043, FJ231742.
RESULTS
Isolation of bifidobacteria from human samples.
The bifidobacterial population of the collected intestinal mucosa samples and that from fecal samples were isolated using mupirocin-based medium (BSM), which has been described previously to be selective for bifidobacteria (36, 41). In order to assess the sensitivity of the different bifidobacterial species to mupirocin, we performed MIC tests involving two representative strains, including the type strain, of each of the 30 bifidobacterial species so far described (51). All strains analyzed were shown to be resistant to mupirocin levels of at least 50 μg/ml medium (data not shown). Generally, the total viable counts retrieved from fecal samples were 4 logs higher than those from colonoscopic samples (108/g versus 104/g of material), indicating that culturable mucosal adherent bifidobacteria are significantly less abundant than bifidobacteria in the fecal environment. To analyze the biodiversity of the culturable bifidobacterial fraction, 20 to 30 colonies assumed to represent the dominant flora of a given sample were randomly selected from each plate and further purified on another bifidobacterium-selective medium (Eugon Agar). A total of 900 bacterial isolates were thus collected and analyzed by bifidobacterium-specific PCR primers, followed by sequencing of rRNA gene sequences.
Phylogenetic analysis based on 16S-ITS region.
A phylogenetic analysis based on the 16S rRNA gene sequences of the type strains plus other strains (where available) of each recognized species and subspecies/biotype of the genus Bifidobacterium was performed. This analysis resulted in a phylogenetic tree (Fig. 1a), which was shown to be consistent with previously described bifidobacterial taxonomic analysis (53). The 16S rRNA gene allows the discrimination of most species of the genus Bifidobacterium. However, closely related taxa show 16S rRNA gene sequence homologies higher than 99% (Table 1). Moreover, the variation in the 16S rRNA gene sequences of closely related bifidobacterial strains is not sufficient to allow a clear determination of evolutionary distances (Fig. 1a and Table 1). The evolutionary rate of the 16S rRNA sequence in bifidobacteria is too low to provide useful information to delineate discrimination between closely related taxa or to measure the intraspecies relationship. Thus, to assess intraspecific phylogenetic relationships in bifidobacteria, we decided to investigate a more variable rRNA sequence, such as the 16S-23S ITS sequence. The 16S-23S rRNA gene spacer regions from 44 bifidobacterial taxa, including the same strains used to study phylogenetic relationships based on 16S rRNA gene sequences, were used to investigate phylogenetic relationships among the members of this genus. The sizes of the ITS sequences in all species analyzed varied from 300 bp to 500 bp, and generally only very limited variations (less than 5%) in ITS size were noticed within members of the same species. The variations in the ITS length may be attributable to the fact that such bacterial 16S-23S spacer regions carry tRNA sequences (5, 22). However, analysis of the available bifidobacterial ITS sequences for the presence of such sequences did not reveal any tRNA structures. These findings are consistent with previous data showing that no tRNA genes occur in the 16S-23S spacer regions of other members of the Actinobacteria phylum (e.g., Frankia and Streptomyces) (56). The phylogenetic tree based on the bifidobacterial ITS sequences was topologically similar to that based on the 16S rRNA gene sequences (Fig. 1b). In fact, the ITS tree clearly distinguished six supported phylogenetic groups, which corroborated the evolutionary development previously observed in the Bifidobacterium genus using other molecular markers, such as the groEL gene (54), the tuf gene (49), or the dnaK gene (59) or a multigene approach (53). However, comparative analysis of ITS sequences suggests that multiple substitutions may have occurred at the same nucleotide position as a consequence of the high evolutionary rate of this sequence. This suggests that ITS sequences suffer from mutational saturation, which will cause difficulties when inferring phylogeny among distantly related bacterial taxa (33). Furthermore, the level of sequence identity between any two strains of the same species was shown to be higher than that between two given members of different species. Thus, the high levels of the ITS sequence variation observed in bifidobacteria between members of the same species (Table 1) make these sequences a suitable molecular marker to infer phylogeny at the intraspecific level and between closely related taxa. Overall, the phylogenetic relationships observed in both trees, i.e., 16S rRNA gene and ITS sequences, were similar, which supported the hypothesis that ITS sequences could be used in phylogenetic analysis of bifidobacteria. Only a small number of discrepancies were observed for the B. adolescentis group (e.g., B. cuniculi ATCC 27916, Bifidobacterium mericycum 49931, and B. angulatum ATCC 27533) between these two trees. One way to explain the differences between these two trees is to assume the occurrence of multiple changes that accumulated in the ITS sequence, simulated false identities, and masked the actual number of evolutionary events.
FIG. 1.
Phylogenetic tree of the genus Bifidobacterium, computed on the basis of 16S rRNA gene sequences (a), ITS sequences (b), or concatenated sequences of the 16S rRNA gene and ITS (c). Bar scales indicate phylogenetic distances. Bootstrap values are reported for a total of 1,000 replicates. Trees were calculated by the neighbor-joining method as implemented in the neighbor module of PHYLIP. The trees were rooted using Nocardia farcinica. Shaded blocks indicate the different bifidobacterial phylogenetic clusters (53).
TABLE 1.
16S rRNA gene and ITS featuresa
| Species | Genetic distance
|
|||||
|---|---|---|---|---|---|---|
| 16S rRNA sequences
|
ITS sequences
|
Concatenated sequences
|
||||
| p-distance | K-distance | p-distance | K-distance | p-distance | K-distance | |
| B. adolescentis | 0.0295 | 0.0315 | 0.2158 | 0.2712 | 0.1288 | 0.1392 |
| B. bifidum | 0.0207 | 0.0214 | 0.1726 | 0.2302 | 0.0963 | 0.01107 |
| B. breve | 0.0142 | 0.0153 | 0.1613 | 0.1718 | 0.1421 | 0.1474 |
| B. lactis | 0.0082 | 0.0085 | 0.0739 | 0.1027 | 0.0491 | 0.0598 |
| B. longum | 0.0049 | 0.0051 | 0.1372 | 0.1464 | 0.1185 | 0.1205 |
| B. pseudocatenulatum | 0.0125 | 0.0128 | 0.1039 | 0.1212 | 0.0686 | 0.0754 |
| B. pseudolongum | 0.0039 | 0.004 | 0.0873 | 0.1183 | 0.0553 | 0.0652 |
Simple mean pairwise distances (p-distance) and mean pairwise distances calculated using Kimura's two-parameter model substitution (K-distance) are provided.
Recently the procedure of gene concatenation for the purpose of inferring bacterial phylogeny was shown to be a valid way to increase the robustness and efficacy of bacterial phylogenetic investigations (45, 53). The tree resulting from the concatenation of 16S rRNA gene and ITS sequences of bifidobacteria is presented in Fig. 1c. Interestingly, the concatenated tree offers the possibility of simultaneously inferring phylogeny at either the inter- or intraspecific level. Notably, the discriminatory power of the concatenated tree is much more significant than that observed with the single trees based on the 16S rRNA gene or ITS sequences. Moreover, concatenation allowed an increase in deep-node bootstrap values and thus led to a considerable increase in tree robustness (e.g., the 16S rRNA has an average bootstrap value around 831, versus a bootstrap value of 900 for the ITS sequences and a value of 996 for the concatenated sequences). This strong increase in the obtained bootstrap values demonstrates that the phylogenetic tree calculated from the concatenation of multiple gene sequences, such as those that represent alternative molecular markers relative to the 16S rRNA gene, may considerably improve the phylogenetic relevance.
Selection of Bifidobacterium genus-specific primer pair for PCR.
The analysis of the 16S rRNA gene sequences of the so far described 30 Bifidobacterium species (53) revealed a highly conserved region located at the 5′ end of the 16S rRNA gene (at nucleotide position 578 of the B. animalis subsp. lactis rRNA locus, accession number no. X89513), which is different from that of other closely related actinobacterial taxa (e.g., the genera Scardovia, Parascardovia, and Aeroscardovia). Based on the comparative analysis of these nucleotide sequences, one PCR primer (Bif-specific) was designed for the specific detection of members of the Bifidobacterium genus. This primer was coupled with another oligonucleotide (23S-specific) targeting a highly conserved DNA sequence located at the 3′ end of the ITS region (at nucleotide position 2040 of the rRNA locus of B. animalis subsp. lactis, accession no. X89513). The application of the oligonucleotide pair Bif-specific and 23S-specific resulted in a PCR amplicon of 1,440 bp. No amplicons were detected with these primers using template DNA derived from bacteria belonging to genera that are closely related to the genus Bifidobacterium or from other bacteria commonly residing in the human gut (data not shown).
Efficacies of primers for identification of bifidobacteria from human gut samples.
In order to evaluate the ability of the primer combination Bif-specific/23S-specific to monitor the distribution of bifidobacterial taxa in human gut samples, i.e., biopsy and fecal samples, all 900 isolates were assayed by PCR using these primers. In 704 cases, this resulted in a PCR amplicon specific for the genus Bifidobacterium, whereas for 196 isolates, the PCR did not produce a Bifidobacterium genus-specific amplification product. Furthermore, DNA sequencing of the 16S rRNA genes of some of these last isolates demonstrated that they belong to other bacterial taxa, such as Lactobacillus and Clostridium (data not shown). This suggests that the mupirocin-based selective medium, in contrast to what was described previously (41), is not completely selective for bifidobacteria, since it allows growth of other gram-positive bacteria. In contrast, sequence analysis of all 704 amplicons that had been generated by the Bif-specific/23S-specific primers showed them to be derived from members belonging to the genus Bifidobacterium.
Identification of bifidobacterial species from feces and colonoscopic biopsies.
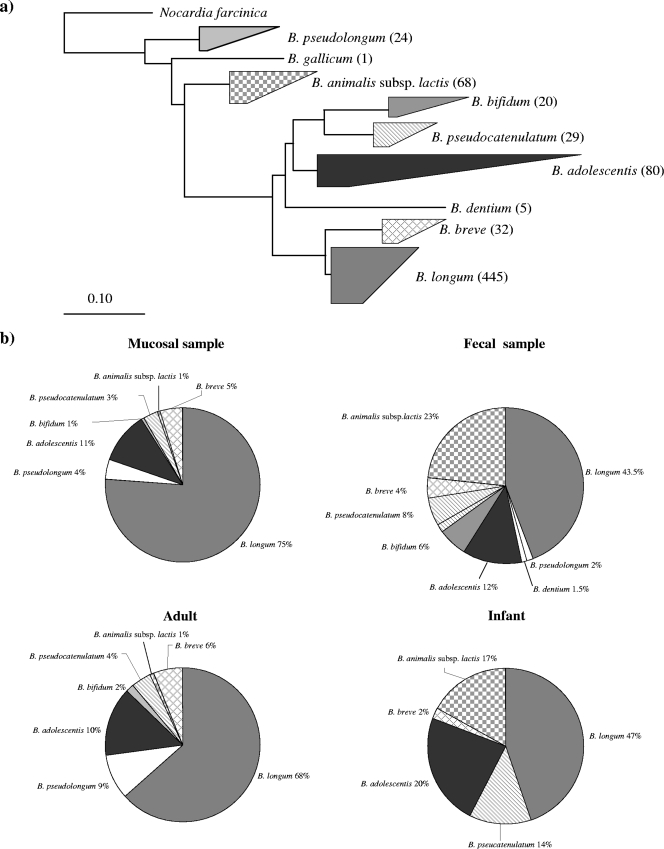
Each 16S rRNA gene-ITS sequence generated from individual colonies originating from fecal samples or mucosal biopsies was subjected to a BLAST search against GenBank. All 704 sequences thus obtained showed more than 98% sequence identity to their nearest database entries, and thus, no new bifidobacterial species were identified. Based on the BLAST results, all sequences were assigned to eight phylogenetic taxa, representing B. bifidum, B. adolescentis, B. pseudocatenulatum, B. breve, B. longum subsp. longum, B. dentium, Bifidobacterium gallicum, and B. animalis subsp. lactis. The phylogenetic relationship between the 704 isolated bifidobacterial strains was analyzed using the 16S-ITS sequences concatenation approach. The resulting concatenated tree clearly demonstrated the separation into nine distinct phylogenetic clusters, which correspond to the bifidobacterial species identified by BLAST analysis (Fig. 2a). Phylogenetic analysis showed that the majority of the investigated strains (63% of the total isolates) belonged to B. longum subsp. longum. The relative abundances of different bifidobacterial taxa presented in colonoscopic biopsies and fecal samples are depicted in Fig. 2b. Notably, most of the dominant bifidobacterial taxa detected in this study, i.e., B. longum subsp. longum, B. adolescentis, B. breve, B. pseudocatenulatum, and B. pseudolongum, were found both in mucosal and in fecal samples, thus suggesting that there is no major difference in terms of the variety of dominant bifidobacteria between these two environments. However, some bifidobacterial species, such as B. animalis subsp. lactis and B. dentium, appear to be more abundant or present only in stool samples, thus suggesting that these taxa do not represent a dominant component of the mucosa-associated bifidobacteria.
FIG. 2.
Relative abundances of bifidobacteria isolated from intestinal mucosa and fecal samples. Panel a shows the phylogenetic distribution of the bifidobacterial isolates among the different bifidobacterial species based on the combined human intestinal and fecal 16S rRNA gene-ITS sequence data set. The angle where each triangle joins the tree represents the relative abundance of sequences, and the lengths of the two adjacent sides indicate the range of branching depths with that clade. The numbers indicated in parentheses indicate the number of isolates obtained for each species. Panel b indicates the percentage and the total number of bifidobacterial isolates identified in this study from different environments (intestine versus feces) and from different subjects (adult versus infant).
Analysis of variability of bifidobacterial populations.
The isolation of a relatively large number of bifidobacterial strains derived from different subjects, anatomical sites (e.g., sigmoid or rectum regions), and sampling material (e.g., mucosa or stool) allowed us to investigate possible environment-based differences in bifidobacterial species distribution. We used techniques that are based on the relative abundance of the 16S rRNA gene-ITS sequences within the bifidobacterial communities and the extent of genetic divergence between sequences. The analysis of the inter- and intrasubject variabilities was performed using DPCoA (32). The Rao diversity coefficient (32), which accounts for bacterial dissimilarity, was higher between all the individuals, thus indicating the existence of a large variability at the intersubject level (Fig. 3a). In fact, the first two axes captured only 16.6% of the total variability, thus suggesting that each bifidobacterial population present in a given subject is specific for only that individual (Fig. 3a). Furthermore, we performed an analysis of bifidobacterial variability between different anatomical sampling sites within the same subject (intrasubject diversity), using a subset of eight individuals for which two different mucosal samples, corresponding to the sigmoid and rectum tracts, were available. Similar to what has been described previously about the intestinal microbiota (6), we found that the mucosa-associated bifidobacterial community sampled from the rectum was similar to that of the sigmoid site from the same subject, with two exceptions (Fig. 3b). Such analyses suggest a pattern of patchiness in the distribution of mucosal bifidobacteria rather than a homogeneous gradient along the longitudinal axis of the colon. The analyses also suggest that fecal bifidobacterial flora is a representation of shed mucosal bifidobacteria (here designated autochthonous bifidobacterial flora) combined with a nonadherent luminal bifidobacterial population (here designated allochthonous bifidobacterial flora). However, this suggestion must be handled with caution, given the fact that mucosal and stool samples were derived from different individuals. When DPCoA analyses were performed using both colonoscopic samples and fecal samples, a considerable variability in the bifidobacterial population was revealed (Fig. 3c). This would indicate that a clearly distinct species architecture exists between the autochthonous (i.e., mucosa-adherent) and allochthonous bifidobacterial populations.
FIG. 3.
DPCoA analysis for colonic mucosa (a), colonic mucosal sites alone (b), or for mucosa and stool (c). In panel a, the Rao dissimilarity values captured by each axis are represented by the histogram, where each histogram represents a Rao dissimilarity value. In panels b and c, percentages shown along the axes indicate the proportion of total Rao dissimilarity captured by that axis. Ellipses indicate the distribution of bifidobacterial strain per sample, except in panel c, where all mucosal samples and fecal samples are represented by one ellipse.
Common species and strains in the mucosa-adherent bifidobacterial flora.
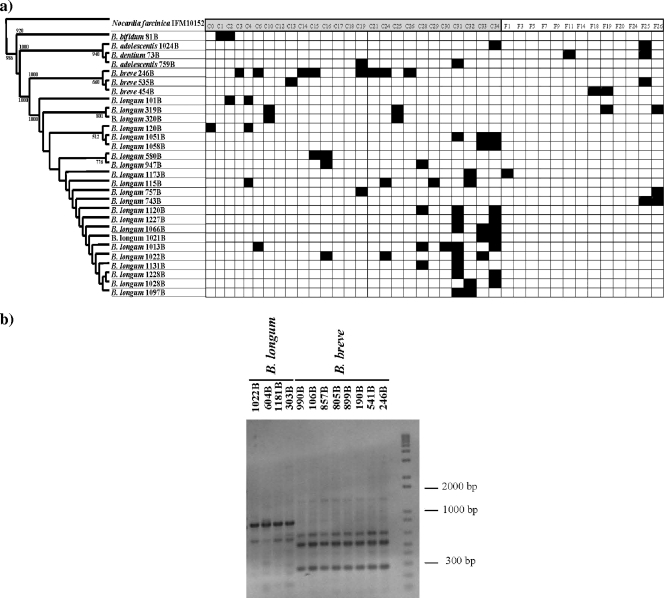
The analysis of bifidobacterial species distribution based on host age revealed that specific bifidobacterial taxa, such as B. pseudolongum and B. bifidum, are exclusively dominant in the adult human bifidobacterial population (e.g., specialist commensal) whereas certain other bifidobacterial species, i.e., B. longum, B. breve, B. pseudocatenulatum, and B. adolescentis, were found to be widely distributed irrespective of host age (generalist commensal), even if the relative abundance is different in adult-derived versus infant-derived samples (Fig. 2b). These results indicate that in contrast to what was observed for the mucosal versus fecal environments, a clear ecological specialization exists for a small number of bifidobacterial taxa when species distribution according to host age is considered. In contrast, other species, such as B. dentium, were isolated mostly from fecal samples, which suggest that these species do not belong to the mucosa-adherent members of the intestinal bifidobacteria microbiota but rather represent transient bifidobacteria, which are derived from food ingestion or from bacterial shedding from the proximal region of the human GIT (e.g., oral cavity). As described above, the high degree of variability of the ITS sequences at the intraspecific level in bifidobacteria renders this molecular marker suitable for the specific molecular characterization of bifidobacterial strains. When we compared the ITS sequences from all bifidobacteria isolated from mucosal samples and infant stools, we identified only a small number of strains which showed 100% identity at their ITS sequences and which had been isolated from different subjects. Bifidobacterial isolates with identical ITS sequences may be clonal representatives of the same strain despite the fact that they are present in different individuals (Fig. 4a). Since ERIC-PCR has been demonstrated to be an informative method suitable for strain identification in bifidobacteria (60), we used this technique to confirm or complement our ITS sequence-based typing results. The obtained ERIC-PCR patterns were indistinguishable within the same set of strains that also possessed identical ITS sequences (Fig. 4b; also data not shown), indicating that little or no genomic variability exists within such strains. Among these strains, a small number of bifidobacterial isolates, such as B. breve 246B, were shown to be widely distributed within the intestinal tracts of the individuals (Fig. 4a) enrolled in this study, suggesting that this strain possesses a high capacity to adapt to different hosts. The wide ecological distribution of B. breve 246B isolates may at first glance suggest that this strain is derived from previous probiotic treatment (more than 2 months before the sampling date) of the individuals involved in this study. However, the comparison of the ITS sequences of B. breve 246B strain with those of other probiotic bifidobacterial strains currently on the market clearly indicates the unique strain identity of B. breve 246B (data not shown).
FIG. 4.
(a) Distribution of bifidobacterial isolates that were found to be widely distributed among different subjects. The y axis of this panel is a neighbor-joining phylogenetic tree containing all the 32 bifidobacterial isolates that were found to be present more than one time in different individuals from this study. Each column is labeled by subject and sample (C, colonoscopic mucosa; F, fecal sample). The presence of the strain is indicated by a black box. (b) ERIC-PCR patterns of different bifidobacterial strains. The sets of strains displaying the same ITS sequences are clustered. Strains used are indicated above each lane. Lane M, 1-kb DNA ladder (Gibco BRL).
DISCUSSION
The human GIT microbiota is composed of mucosa-adherent bacteria as well as nonadherent or luminal microbial components that are derived from the diet (for a review, see reference 41). In the last decade, significant efforts have been made to elucidate the microbial composition of the GIT microbiota using culture-based methods (7, 14, 30, 35) and culture-independent approaches (e.g., metagenomic analyses) (6, 11, 15, 24, 29, 40, 47, 62-65). However, in all of these studies, only fragmentary information has become available regarding the contribution of bifidobacteria to the indigenous component of the human microbiota. Moreover, most of the so-far-published ecological studies that aimed to explore the biodiversity of bifidobacteria occurring in human gut were based on the analyses of fecal samples. However, it has become clear that the microbial community of the human intestine includes a luminal fraction as well as an adherent mucosal community, which is very different from that identified in stool samples (31). Thus, the human fecal microbiota cannot simply be considered a direct reflection of the intestinal microbiota (6, 31). This is also corroborated by the fact that a large number of bacteria present in fecal samples are derived from diet or from environmental contaminations and only partially from the mucosa-adherent bacterial components of the human GIT microbiota (6, 31, 62). In this study, the involvement of a polyphasic approach, where we combined a culture-based method with the determination of 16S-ITS rRNA gene sequences, allowed the assessment of the biodiversity of bifidobacterial populations present in human intestinal biopsy samples. Notably, only a restricted number of bifidobacterial species, i.e., B. longum, B. adolescentis, B. breve, B. pseudocatenulatum, and B. pseudolongum, appear to be dominant in the investigated biopsies and may therefore be considered widespread bifidobacterial species, whereas certain other bifidobacterial species seem to be restricted to a particular ecological niche (e.g., B. bifidum and B. pseudolongum). A large proportion of the currently used bifidobacterial probiotic strains belong to the taxon B. animalis subsp. lactis (e.g., Bb12, DN-173, and HN019), which are considered to confer beneficial effects on the host through interactions with this host and with other components of the intestinal microbiota (39). However, in this ecological study, we found that this bifidobacterial species is only rarely found in intestinal biopsy samples whereas it is frequently detected in fecal samples, suggesting that this taxon may not be abundant among the intestine-adherent component of the human intestinal bifidobacteria. This hypothesis is consistent with a recent report showing that the molecular impact of the probiotic B. animalis subsp. lactis strains on a resident member of the intestinal microbiota (i.e., B. thetaiotaomicron) was negligible compared to the effects induced by indigenous bifidobacterial components (42). However, surface-adherent and luminal bifidobacterial populations may fulfill different roles within the ecosystem. In fact, it has been recognized that the biofilm-like architecture of the mucosal microbiota, being in close contact with the underlying epithelium, promotes beneficial functions, including nutrient exchange and induction of host innate immunity (42). Thus, the mucosa-adherent properties, the human intestinal ecological origin, and consequently the capacity to interact with the human host may be important prerequisites for the design of novel probiotic bifidobacteria.
Culture-based methods are known to provide an image of bacterial diversity that is limited to only those bacteria that can be cultivated on synthetic media. In order to overcome this limitation, culture-independent approaches, such as metagenomic analyses, have been developed. However, the results of these methods may be affected by the experimental procedures used to extract DNA or by the conditions of DNA amplification (46). This might provide a reason as to why the so-far-published metagenomic reports on the human intestinal microbiota have assigned a rather modest contribution of bifidobacteria (about 4% of the obtained rRNA gene sequences was attributed to bifidobacteria) toward the abundance and diversity of this microbial ecosystem, (6, 12, 62). Recently Palmer et al. described the development of the intestinal microbiota in infants by the use of culture-independent approaches (31). In this study, a surprisingly low frequency and abundance of bifidobacteria in the fecal infant microbiota were described, which is in striking conflict with findings reported here and elsewhere (40). This suggests that the exclusive use of a culture-independent approach may not provide a complete image of the architecture of the human intestinal microbiota. In contrast, a polyphasic approach, such as that described here, constitutes a valuable method for overcoming these limitations and for providing an in-depth analysis of the bifidobacterial components of the human intestinal microbiota. The use of polyphasic approaches also overcomes the limitations of selective media. This has also been noticed in the current study, where the use of a mupirocin-based medium, in contrast to what was described previously (36, 41), was shown not to be completely selective for bifidobacteria since it allowed growth of other gram positive bacteria as well. Moreover, our findings may allow the isolation of ecologically competitive strains, which could be submitted to further biochemical, physiological, and genetic investigations. In spite of these positive aspects, this polyphasic method does suffer from certain disadvantages, such as the fact that it is time-consuming and rather laborious.
Comparisons of 16S rRNA have been demonstrated to be highly useful for inferring phylogenetic relationships among bifidobacteria from the level of domains to the level of species (26, 43, 55, 57). However, the evolutionary distances exhibited by the 16S rRNA gene sequences among closely related taxa (e.g., between subspecies) or at the intraspecific level are very modest, and thus, this molecular marker cannot be used in order to perform phylogenetic analysis below the species level. In contrast, we have shown here that the evolutionary rate of the ITS sequences is significantly higher than that of the 16S rRNA gene sequences, suggesting that these two molecules provide different but complementary phylogenetic information. In fact, the analysis of 16S rRNA gene sequences represents a valid tool for inferring inter- and intrageneric relationships (10), whereas we show here that the comparison of the ITS sequences provides data about intraspecific evolutionary development. By applying a multisequence tree approach based on the concatenation of the 16S rRNA gene and ITS sequences, we were able to simultaneously analyze isolated bifidobacteria at both interspecific and intraspecific levels. The ITS sequences provide insights about the precise identity and distribution of the bifidobacterial isolates present in different human subjects, intersubject variability, and differences between two intestinal regions of the same individual, i.e., intrasubject variability. We discovered significant intersubject variability and differences between stool and mucosa-adherent community composition. Moreover, we found that fecal bifidobacterial populations from infants are dominated by a relatively small number of bifidobacterial species and strains, exhibiting rather simple structures, although they revealed a high interindividual variation in taxonomic composition. In contrast, the bifidobacterial populations obtained from adults and children appeared much more complex but showed a remarkable conservation of bifidobacterial species and strains. The use of ITS sequences also revealed the likely presence of identical strains in different individuals. We identified a set of 32 strains which appeared to be commonly distributed among different individuals. Among these, the B. breve 246B isolate was shown to be one of the most commonly isolated mucosa-associated bifidobacterial strains, which suggests that this strain possesses a superior ecological fitness to adapt to different hosts and possibly various ecological niches. We can argue that bifidobacteria, such as B. breve 246B, which frequently contribute to the microbiota must possess very efficient nutrient acquisition strategies, by which these microorganisms can efficiently compete and adapt to the adult-type intestinal environment and establish strong symbiotic relationships with their host.
A future avenue of research will be directed at highlighting how the adult-type bifidobacterial microbiota develops and if it will be possible to identify a common set of bifidobacterial species and strains in the human gut, representing a core set of bifidobacteria. Such an analysis will expand our knowledge of the contribution of commensal bifidobacteria to gastrointestinal well-being.
Acknowledgments
This work was financially supported by Parmalat SpA, Parma, Italy, by the Italian Award for Outstanding Young Researcher, program “Incentivazione alla mobilità di studiosi stranieri e italiani residente all'estero,” 2005 to 2009, and a Marie Curie Reintegration Grant (MERG-CT-2005-03080) to M.V., and by the Science Foundation Ireland CSET award to the Alimentary Pharmabiotic Centre, located at University College Cork, and a DAF/HRB FHRI award to the ELDERNET project to D.V.S.
We thank Carlos Canchaya for bioinformatics support and helpful discussions. Further, we thank Angelo Pavesi and Matteo Manfredini for their statistical support. We thank all students and coworkers for contributing data and for their enthusiasm.
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitoyannis, I. S., and M. Van Houwelingen-Koukaliaroglou. 2005. Functional foods: a survey of health claims, pros and cons, and current legislation. Crit. Rev. Food Sci. Nutr. 45:385-404. [DOI] [PubMed] [Google Scholar]
- 3.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 4.Booijink, C. C., E. G. Zoetendal, M. Kleerebezem, and W. M. de Vos. 2007. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future Microbiol. 2:285-295. [DOI] [PubMed] [Google Scholar]
- 5.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 6.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanaro, S., V. Vigi, R. Chierici, and G. Boehm. 2003. Fecal flora measurements of breastfed infants using an integrated transport and culturing system. Acta Paediatr. 92:634-635. [DOI] [PubMed] [Google Scholar]
- 8.Favier, C. F., F. Vaughan, W. M. de Vos, and D. L. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 10.Fox, G. E., J. D. Wisotzkey, and P. Jurtshunk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 11.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variation of bifidobacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 14.Harmsen, H. J. M., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 15.Harmsen, H. J. M., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 66:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartley, C. L., H. M. Clements, and K. B. Linton. 1977. Escherichia coli in the faecal flora of man. J. Appl. Bacteriol. 43:261-269. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, D. G., A. J. Bleasby, and R. Fuchs. 1992. CLUSTALV: improved software for multiple alignment. Comput. Appl. Biosci. 8:189-191. [DOI] [PubMed] [Google Scholar]
- 18.Jian, W., L. Zhu, and X. Dong. 2001. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 51:1633-1638. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 20.Kullen, M. J., L. J. Brady, and D. J. O'Sullivan. 1997. Evaluation of using a short region of the recA gene for rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol. Lett. 154:377-383. [DOI] [PubMed] [Google Scholar]
- 21.Lamendella, R., J. W. Santo Domingo, C. Kelty, and D. B. Oerther. 2008. Bifidobacteria in feces and environmental waters. Appl. Environ. Microbiol. 74:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loughney, K., E. Lund, and J. E. Dahlberg. 1982. tRNA genes are found between the 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 10:1607-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangin, I., A. Suau, F. Magne, D. Garrido, M. Gotteland, C. Neut, and P. Pochart. 2006. Characterization of human intestinal bifidobacteria using competitive PCR and PCR-TTGE. FEMS Microbiol. Ecol. 55:28-37. [DOI] [PubMed] [Google Scholar]
- 25.Marco, M. L., S. Pavan, and M. Kleerebezem. 2006. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 17:204-210. [DOI] [PubMed] [Google Scholar]
- 26.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuki, T., K. Watanabe, R. Tanaka, and H. Oyaizu. 1998. Rapid identification of human intestinal bifidobacteria by 16S rRNA targeted species and group specific primers. FEMS Microbiol. Lett. 167:113-121. [DOI] [PubMed] [Google Scholar]
- 28.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen, D. S., P. L. Moller, V. Rosenfeldt, A. Paerregaard, K. F. Michaelsen, and M. Jakobsen. 2003. Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon. Appl. Environ. Microbiol. 69:7545-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palframan, R. J., G. R. Gibson, and R. A. Rastall. 2003. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr. Issues Intest. Microbiol. 4:71-75. [PubMed] [Google Scholar]
- 31.Palmer, C., E. M. Bik, D. B. Digiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavoine, S., A. B. Dufour, and D. Chessel. 2004. From dissimilarities among species to dissimilarities among communities: a double principal coordinate analysis. J. Theor. Biol. 228:523-537. [DOI] [PubMed] [Google Scholar]
- 33.Philippe, H., and A. Adoutte. 1996. What can phylogenetic patterns tell us about the evolutionary process generating biodiversity?, p. 41-59. In M. Hochberg, J. Clobert, and R. Barbault (ed.), Aspects of the genesis and maintenance of biological diversity. Oxford University Press, Oxford, United Kingdom.
- 34.Phillips, I., J. M. Andrews, E. Bridson, E. M. Cooke, R. C. Spencer, H. A. Holt, R. Wise, A. J. Bint, D. F. J. Brown, D. Greenwood, A. King, and R. J. Williams. 1991. A guide to sensitivity testing, report of the Working Party on Antibiotic Sensitivity Testing of the British Society for Antimicrobial Chemotherapy. J. Antimicrob. Chemother. 27(Suppl. D):1-50. [PubMed] [Google Scholar]
- 35.Poxton, I. R., R. Brown, A. Sawyerr, and A. Ferguson. 1997. Mucosa-associated bacterial flora of the human colon. J. Med. Microbiol. 46:85-91. [DOI] [PubMed] [Google Scholar]
- 36.Rada, V., and J. Koc. 2000. The use of mupirocin for selective enumeration of bifidobacteria in fermented milk products. Milchwiessenchaft 55:65-67. [Google Scholar]
- 37.Rajilić-Stojanović, M., H. Smidt, and W. M. de Vos. 2007. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9:2125-2136. [DOI] [PubMed] [Google Scholar]
- 38.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 39.Sanders, M. E. 2006. Summary of probiotic activities of Bifidobacterium lactis HN019. J. Clin. Gastroenterol. 40:776-783. [DOI] [PubMed] [Google Scholar]
- 40.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson, P. J., G. F. Fitzgerald, C. Stanton, and R. P. Ross. 2004. The evaluation of a mupirocin-based selective medium for the enumeration of bifidobacteria from probiotic animal feed. J. Microbiol. Methods 57:9-16. [DOI] [PubMed] [Google Scholar]
- 42.Sonnenburg, J. L., C. T. Chen, and J. I. Gordon. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4:e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 44.Sul, S. Y., H. J. Kim, T. W. Kim, and H. Y. Kim. 2007. Rapid identification of Lactobacillus and Bifidobacterium in probiotic products using multiplex PCR. J. Microbiol. Biotechnol. 17:490-495. [PubMed] [Google Scholar]
- 45.Teichmann, S. A., and G. Mitchison. 1999. Is there a phylogenetic signal in prokaryote proteins? J. Mol. Evol. 49:98-107. [DOI] [PubMed] [Google Scholar]
- 46.Turroni, F., A. Ribbera, E. Foroni, D. van Sinderen, and M. Ventura. 2008. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie van Leeuwenhoek 94:35-50. [DOI] [PubMed] [Google Scholar]
- 47.van Tongeren, S. P., J. P. J. Slaets, H. J. M. Harmsen, and G. W. Welling. 2005. Fecal microbiota composition and frailty. Appl. Environ. Microbiol. 71:6438-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 68:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ventura, M., and R. Zink. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69:7517-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ventura, M., C. Canchaya, D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70:3110-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ventura, M., M. Elli, R. Reniero, and R. Zink. 2001. Molecular microbial analysis of human intestinal Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 52.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ventura, M., C. Canchaya, A. Del Casale, F. Dellaglio, E. Neviani, G. F. Fitzgerald, and D. van Sinderen. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 30:734-759. [DOI] [PubMed] [Google Scholar]
- 54.Ventura, M., C. Canchaya, R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analysis. Appl. Environ. Microbiol. 70:6197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ventura, M., C. Canchaya, G. F. Fitzgerald, R. S. Gupta, and D. van Sinderen. 2007. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie van Leeuwenhoek 91:351-372. [DOI] [PubMed] [Google Scholar]
- 56.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, K. Chater, G. F. Fitzgerald, and D. van Sinderen. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie van Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 58.Ventura, M., G. F. Fitzgerald, and D. van Sinderen. 2005. Genetic and transcriptional organization of the clpC locus in Bifidobacterium breve UCC 2003. Appl. Environ. Microbiol. 71:6282-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ventura, M., R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and its application in bifidobacterial tracing. Appl. Environ. Microbiol. 71:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ventura, M., V. Meylan, and R. Zink. 2003. Identification and tracing of Bifidobacterium species by use of enterobacterial repetitive intergenic consensus sequences. Appl. Environ. Microbiol. 69:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wall, R., G. Fitzgerald, S. Hussey, T. Ryan, B. Murphy, P. Ross, and C. Stanton. 2007. Genomic diversity of cultivable Lactobacillus populations residing in the neonatal and adult gastrointestinal tract. FEMS Microb. Ecol. 59:127-137. [DOI] [PubMed] [Google Scholar]
- 62.Wang, M., S. Ahrnè, B. Jeppson, and G. Molin. 2005. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 54:219-231. [DOI] [PubMed] [Google Scholar]
- 63.Yin, X., J. R. Chambers, K. Barlow, A. S. Park, and R. Wheatcrof. 2005. The gene encoding xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (xfp) is conserved among Bifidobacterium species within a more variable region of the genome and both are useful for strain identification. FEMS Microbiol. Lett. 246:251-257. [DOI] [PubMed] [Google Scholar]
- 64.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zoetendal, E. G., K. Ben-Amor, H. J. M. Harmsen, F. Schut, A. D. L. Akkermans, and W. M. De Vos. 2002. Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl. Environ. Microbiol. 68:4225-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]