Abstract
The bacterial diversity associated with citrus leaf midribs was characterized for citrus groves that contained the Huanglongbing (HLB) pathogen, which has yet to be cultivated in vitro. We employed a combination of high-density phylogenetic 16S rRNA gene microarrays and 16S rRNA gene clone library sequencing to determine the microbial community composition for symptomatic and asymptomatic citrus midribs. Our results revealed that citrus leaf midribs can support a diversity of microbes. PhyloChip analysis indicated that 47 orders of bacteria in 15 phyla were present in the citrus leaf midribs, while 20 orders in 8 phyla were observed with the cloning and sequencing method. PhyloChip arrays indicated that nine taxa were significantly more abundant in symptomatic midribs than in asymptomatic midribs. “Candidatus Liberibacter asiaticus” was detected at a very low level in asymptomatic plants but was over 200 times more abundant in symptomatic plants. The PhyloChip analysis results were further verified by sequencing 16S rRNA gene clone libraries, which indicated the dominance of “Candidatus Liberibacter asiaticus” in symptomatic leaves. These data implicate “Candidatus Liberibacter asiaticus” as the pathogen responsible for HLB disease.
Citrus is the most important commercial fruit crop in Florida. In recent years, citrus Huanglongbing (HLB) disease, also called citrus greening, has severely affected Florida's citrus production and hence has drawn an enormous amount of attention. HLB disease is one of the most devastating diseases of citrus (6, 14) and is characterized by blotchy mottling with green islands on leaves, as well as stunting, fruit decline, and small, lopsided fruits with poor coloration. The disease tends to be associated with a phloem-limited fastidious alphaproteobacterium that has been given provisional Candidatus status (“Candidatus Liberobacter spp.,” later changed to “Candidatus Liberibacter spp.”) (19, 26, 35). Previous studies indicated that HLB infection causes disorder in the phloem and severely impairs the translocation of assimilates in host plants (5, 28, 41). Tatineni and colleagues discovered that the HLB bacteria were unevenly distributed in phloem of bark tissue, vascular tissue of the leaf midrib, roots, and different floral and fruit parts (44).
Unsuccessful attempts to culture this pathogen have notably hampered efforts to understand its biology and pathogenesis mechanism. Using a modified Koch's Postulates approach, Jagoueix and colleagues were able to reinfect periwinkle plants using a mixed microbial community harvested from plants with HLB disease (26). Emergence of the disease in otherwise healthy plants led to the conclusion that HLB disease was associated with “Candidatus Liberibacter sp.” based on its 16S rRNA gene sequence (19, 26). Currently, three species of the pathogen are recognized for trees with HLB disease based on the 16S rRNA gene sequence: “Candidatus Liberibacter asiaticus,” “Candidatus Liberibacter africanus,” and “Candidatus Liberibacter americanus”; “Ca. Liberibacter asiaticus” is the most prevalent species in trees with HLB disease (5, 13, 19, 26, 45). “Ca. Liberibacter asiaticus” is naturally transmitted to citrus by the psyllid Diaphorina citri Kuwayama and can be artificially transmitted by grafting from citrus to citrus and from dodder (Cuscuta campestris) to Madagascar periwinkle (Catharanthus roseus) or tobacco (Nicotiana tabacum cv. Xanthi) (5). Based on current research regarding the associations of “Ca. Liberibacter” in planta, there is not enough evidence to implicate “Ca. Liberibacter” as the definitive causal agent of HLB disease due to its resistance to cultivation in vitro. It is possible that HLB disease may be the result of complex etiology where “Ca. Liberibacter” interacts with other endophytic bacteria. However, there is not enough evidence regarding its association(s) in planta to reach this conclusion, nor is it known whether associated microbial communities play a role in the expression of pathogenic traits.
It has been noticed that certain trees (called escape plants) may survive in citrus groves heavily infected with the HLB pathogen. Because these escape plants have the same genotype as susceptible plants and have developed under similar edaphic and climatic conditions, a possible explanation for the lack of HLB disease symptoms may lie in the nature of the microbial community associated with these plants. In a study of the endophytic bacteria associated with Xylella fastidiosa-infected citrus branches, the endophyte Curtobacterium flaccumfaciens was found more frequently in asymptomatic citrus trees infected by X. fastidiosa (2). It was also reported that C. flaccumfaciens was able to reduce symptoms caused by X. fastidiosa when C. roseus was used as the host plant (30). Microbial community analysis may lead to isolation and identification of novel bacteria that are potential biocontrol agents for use against the HLB pathogen. Identification of biocontrol organisms obtained from a niche similar to that of the pathogen would be particularly promising for effective disease control.
Microbial community analysis may solve the puzzles regarding the causal agent of HLB disease and differences in symptoms among citrus trees in infected groves. Except for the results of some studies of the citrus phyllosphere and X. fastidiosa-infected citrus (2, 52), little is known about the composition of the bacterial community associated with citrus. The phloem microbiome can be characterized by either cultivation-based or cultivation-independent methods. However, the portion of microbial diversity estimated by conventional culture techniques is only 0.1 to 10% of the total diversity (47), indicating that techniques based on laboratory cultivation might be significantly biased. In fact, it has been observed that in many environmental samples the bacteria that are most dominant and abundant are not cultivable (29, 38, 42). Due to the limitations of cultivation-based methods, in recent years molecular methods have been widely used for community analysis. Multiple methods have been developed, and the 16S rRNA gene-based methods are the most popular due to remarkably high conservation of this gene in all bacteria, which enables a universal phylogeny to be determined (48). 16S rRNA gene-based phylogenetic analysis has been commonly employed to characterize the microbial diversity in a variety of ecological niches, such as plants (10, 43), soils (29), and subsurface sediments and rocks (8, 9, 40). A high-density 16S rRNA gene oligonucleotide microarray, the PhyloChip microarray, has recently been developed and effectively used to study bacterial population diversity, and it is more powerful and sensitive for identifying bacteria in the environment (7, 15).
The main objective of the study was to test the hypothesis that bacteria other than “Ca. Liberibacter spp.” are associated with citrus greening disease. The differences between the relative abundance, species richness, and phylogenetic diversity of the microbial communities associated with the leaf midribs of symptomatic and asymptomatic citrus trees with the HLB pathogen were investigated using PhyloChip high-density 16S rRNA gene microarray and 16S rRNA gene clone library methods.
MATERIALS AND METHODS
Plant material collection and DNA extraction.
Leaf samples were collected from citrus groves in Dover (grove 1) and Lake Placid (grove 2) in Florida. Asymptomatic leaves (eight leaves from each of six trees in each of two groves) and leaves showing HLB blotchy mottling (eight leaves from each of six trees in each of two groves) were randomly collected and brought to the lab in a cooler with ice in January and February 10 days apart in 2008. The two citrus groves chosen for this study were confirmed to be HLB disease positive for more than the previous 2 years. The two groves are separated by 110 km, and both groves are planted with Valencia oranges (Citrus sinensis). The leaves were washed in tap water and surface sterilized in 35% bleach (2% active Cl−) and 70% (vol/vol) ethanol for 2 min each and then rinsed three times with sterile water. Later, midribs of leaves were separated, frozen in liquid nitrogen, and stored at −80°C. All of the midribs of the eight leaves from a single tree were pooled, and DNA was extracted using a Wizard genomic DNA purification kit (Promega Corp., Madison, WI) by following the protocol for isolating genomic DNA from plant tissue. The DNA was again purified once with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) and chloroform-isoamyl alcohol (24:1, vol/vol) by using the standard protocol (39). The DNA was precipitated, washed with 70% (vol/vol) ethanol, and resuspended in RNase- and DNase-free water. For cloning, bacterial plasmid DNA was isolated with the Wizard miniprep DNA purification system (Promega).
PCR detection of “Ca. Liberibacter asiaticus.”
PCR using primers A2 and J5 (24) was performed to confirm the presence of “Ca. Liberibacter asiaticus” in the samples. All PCRs in this study were performed in a DNAEngine Peltier thermal cycler (Bio-Rad Laboratories, Hercules, CA). Amplification of the DNA was conducted in a 20-μl (total volume) mixture using Speed Star HS polymerase (Takara Bio Inc., Otsu, Shiga, Japan). The PCR conditions for Speed Star HS polymerase were 2 min of predenaturation at 94°C, followed by 35 cycles of 10 s of denaturation at 94°C, 10 s of annealing at 65°C, 1 min of extension at 72°C and then a single final extension of 4 min at 72°C.
PCR amplification and sample preparation for 16S rRNA gene PhyloChip analysis.
For PhyloChip analysis, DNA from six asymptomatic and six symptomatic trees from each of the two groves sampled were amplified separately. Amplification of DNA was performed in a 25-μl PCR mixture using 1.5 U of Ex Taq polymerase (Takara Bio Inc., Otsu, Shiga, Japan). Gradient PCR was conducted with annealing at 48 to 58°C for 25 cycles (the gradient temperatures were 48.0, 48.3, 48.9, 49.7, 50.8, 52.3, 54.0, 55.4, 56.5, 57.3, 57.8, and 58.0°C). Primers 27f (5′-AGAGTTTGATCMTGGCTCAG) and 1492r (5′-GGYTACCTTGTTACGACTT) were used to amplify the 16S rRNA gene regions of bacteria (32). The PCR products of all 12 gradients from one DNA template sample were pooled before electrophoresis. Amplified PCR products were electrophoresed on a 1% agarose gel, and the desired ∼1.5-kb bands were gel purified and shipped on ice before PhyloChip analysis.
PhyloChip analysis.
Each PCR product was quantified using Egels (Invitrogen Corp. Carlsbad, CA), and 1,000 ng of bacterial PCR product was fragmented with DNase, labeled with biotin, and hybridized as previously described (7). The microbial community was resolved as a subset of 8,743 potentially detected taxa with corresponding hybridization scores expressed in arbitrary units. Each taxon consists of a set of 25 to 30 perfect match-mismatch probe pairs. For a taxon to be reported in this analysis, 92% of the probe pairs in its set (probe fraction, >0.92) must meet the following conditions: (i) the perfect match has an intensity that is at least 1.3 times higher than the intensity of the mismatch, and (ii) the intensities of both the perfect match and the mismatch are 500-fold greater than the background intensity. Hybridization scores for a taxon are reported for all samples if at least 1 of the 12 samples has a probe fraction of >0.92. Hybridization scores are averages of the differences between the perfect match and mismatch fluorescence intensities of all probe pairs except the highest and lowest values. Final hybridization scores were normalized to an average of 2,500 arbitrary units for each PhyloChip. For presentation of the relative abundances of reported taxa, hybridization scores were converted to 16S rRNA copy numbers based on the empirically determined log-linear relationship between the copy number of the applied 16S rRNA gene PCR product and the hybridization score; for analysis of the richness by group, presence or absence was determined based on a probe fraction cutoff of 0.9 for each taxon within the group (7).
Statistical analysis.
To estimate richness, we used a probe fraction value of 0.9 as a cutoff below which the taxon was deemed absent. Previously, the probe fraction was found to correlate well with richness patterns displayed by clone library analysis (15).
All statistical analyses were performed using JMP (SAS Institute, Inc., Cary, NC), PCOrd (McCune and Mefford), or R (R Team [37a]). The multiresponse permutation procedure was used to test the null hypothesis that the ordination contained distinct subgroups that were statistically separate from one another. All statistical significance was evaluated at a P value of 0.05, unless otherwise noted. Regression analysis of environmental variables against the ordination coordinates was performed as previously published (4). Student's t tests were performed as unpaired, two-tailed tests evaluated at a significance level of 0.05.
PCR amplification, cloning, and sequencing of bacterial 16S rRNA genes.
As the DNA extracted from citrus midribs contained a mixture of plant and bacterial DNA, it was necessary to use a PCR primer that is specific for the bacterial 16S rRNA gene sequence. We used universal primers 799f (5′-AACMGGATTAGATACCCKG) (10) and 1492r (32), which were shown to amplify the DNA of most bacterial species but not plastid DNA. The PCR products obtained from mitochondria using the 799f/1492r primer pair were approximately 1.5 times larger than the bacterial 16S rRNA gene product, which easily allowed separation of the PCR products of mitochondria from those of bacteria. DNA extracted from six asymptomatic and six symptomatic trees from each grove (six asymptomatic and six symptomatic samples from two groves resulted in 24 samples) were used as templates to amplify the bacterium-specific 16S rRNA gene region using the 799f and 1492r primers. Primer 1492r amplifies the 16S rRNA gene region of most eubacteria (32). The PCR conditions and number of cycles were exactly the same as those described above for preparation of samples for 16S rRNA gene PhyloChip analysis. The PCR products for all 12 gradients for one sample type (e.g., symptomatic leaves of tree 1 from grove 1) were pooled before electrophoresis. The PCR products were electrophoresed on a 1% agarose gel, and the bacterium-specific DNA band of the expected size (735 bp) was gel purified using the Wizard SV gel and PCR clean-up system (Promega) and ligated into the pGEM-T Easy cloning vector (Promega). The ligation mixture was transformed into chemically competent Escherichia coli DH5α, and transformants were selected on LB agar containing ampicillin (50 μg/ml). The positive clones with desired plasmids were screened by blue-white screening using 40 μl of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (2%, wt/vol) and 7 μl of isopropyl-β-d-thiogalactopyranoside (IPTG) (20% wt/vol) per plate. The white colonies were picked, and plasmids containing 16S rRNA gene inserts were sequenced using the T7 universal primer. Sequencing was performed at the sequencing facility of the Interdisciplinary Center for Biotechnology Research at University of Florida.
Phylogenetic analysis of 16S rRNA gene clone library.
The sequenced rRNA gene regions were compared to Ribosomal Database Project II (release 10, update 3) (http://rdp.cme.msu.edu/index.jsp) (11) using “Naive Bayesian rRNA Classifier,” version 2.0, to identify the nearest phylogenetic neighbor (confidence level, 95%). Homologies of the sequences were further verified using the Basic Local Alignment Search Tool (BLAST) algorithm (1). Sequences with more than 98% similarity were considered to be members of the same operational taxonomic unit.
Nucleotide sequence accession numbers.
A total of 1,276 sequences have been deposited in the GenBank database under accession numbers FJ387589 to FJ388209, FJ388211 to FJ388325, FJ388331, FJ388332, FJ388334, FJ388336 to FJ388340, FJ388342 to FJ388344, and FJ388346 to FJ388874.
RESULTS
PCR detection of “Ca. Liberibacter asiaticus” in test samples.
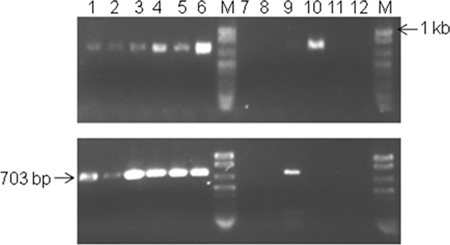
All plant midribs used to identify the bacterial populations associated with citrus leaf midribs were screened for the presence of “Ca. Liberibacter asiaticus” using PCR assays performed with primers A2 and J5 (24). Midribs were chosen since they are phloem rich and we intended to identify microbiomes in the same niches as “Ca. Liberibacter asiaticus” since “Ca. Liberibacter asiaticus” is known to be limited to the phloem. An expected 703-bp PCR product was amplified from all 12 symptomatic plants and 2 of 12 asymptomatic plants in grove 1 (Dover) and grove 2 (Lake Placid) (Fig. 1). Negative controls showed no amplification (data not shown).
FIG. 1.
Agarose gel electrophoresis of PCR products amplified using primers specific for “Ca. Liberibacter asiaticus.” Specific primers A2 and J5 target the 16S rRNA gene of “Ca. Liberibacter asiaticus,” resulting in a 703-bp amplicon (24). Total DNA extracted from midribs of symptomatic and asymptomatic leaves of sweet orange trees were used as templates for PCR amplification. Lanes 1 to 6, symptomatic leaf midribs; lanes 7 to 12, asymptomatic leaf midribs; lane M, DNA molecular weight markers. (Upper panel) Grove 1; (lower panel) grove 2.
PhyloChip bacterial community analysis.
The microbial communities detected in the vascular tissues of citrus leaves were comprised of 117 taxa in 15 different phyla of bacteria spanning the diversity of the bacterial phylogenetic tree (Table 1; see Table S1 in the supplemental material). There were 15 hits that were homologous to chloroplasts that were assumed to be plant derived and excluded from further analysis. Alphaproteobacteria were the most prevalent organisms, accounting for 26.5% of all of the taxa detected. Other phyla that were well represented included Acidobacteria, Deltaproteobacteria, and Gammaproteobacteria (Table 1; see also Table S1 in the supplemental material).
TABLE 1.
Richness of microbial communities in different groves or evidence of symptoms for citrus groves with HLB-disease based on PhyloChip analysisa
| Taxon | Richness of microbial communities
|
Analysis of variance (P)
|
|||||
|---|---|---|---|---|---|---|---|
| Asymptomatic
|
Symptomatic
|
||||||
| Grove 1 | Grove 2 | Grove 1 | Grove 2 | Grove | Symptomatic | Cross-product | |
| Domain Bacteria | 18.7 ± 2.2 | 22.17 ± 0.94 | 22.7 ± 2.5 | 51.2 ± 13.4 | <0.05 | <0.05 | |
| Phylum, class, or other taxon | |||||||
| Acidobacteria | 0.17 ± 0.17 | 0.17 ± 0.17 | 0.33 ± 0.21 | 3.33 ± 1.15 | <0.05 | ||
| Actinobacteria | 0.67 ± 0.42 | 0.83 ± 0.17 | 0.83 ± 0.17 | 1.83 ± 0.6 | |||
| AD3 | 0.67 ± 0.21 | 0 | 0.83 ± 0.17 | 0.5 ± 0.22 | |||
| Bacteroidetes | 0.83 ± 0.17 | 0 | 0.83 ± 0.17 | 2.33 ± 0.61 | <0.05 | ||
| BRC1 | 0.33 ± 0.21 | 0.83 ± 0.17 | 0.83 ± 0.17 | 0.83 ± 0.17 | |||
| Chlamydiae | 0.17 ± 0.17 | 0.17 ± 0.17 | 0.5 ± 0.22 | 0.67 ± 0.21 | <0.05 | ||
| Chlorobi | 0.17 ± 0.17 | 0.17 ± 0.17 | 0.33 ± 0.21 | 0.83 ± 0.17 | <0.05 | ||
| Chloroflexi | 0 | 0.33 ± 0.21 | 0.33 ± 0.21 | 1.0 ± 0.52 | |||
| Firmicutes | 0 | 0 | 0 | 0.67 ± 0.84 | |||
| Gemmatimonadetes | 0 | 0 | 0 | 0.5 ± 0.34 | ND | ||
| Marine group A | 0 | 0 | 0 | 0.5 ± 0.22 | ND | ||
| NC10 | 0 | 0 | 0 | 0.17 ± 0.17 | ND | ||
| Planctomycetes | 1.33 ± 0.21 | 1.67 ± 0.21 | 1.33 ± 0.21 | 2.0 ± 0.0 | <0.05 | ||
| Proteobacteria | 0.5 ± 0.34 | 0.5 ± 0.22 | 0.33 ± 0.33 | 14.0 ± 6.8 | |||
| Alphaproteobacteria | 0.17 ± 0.17 | 0 ± 0 | 0.17 ± 0.17 | 19.0 ± 9.1 | <0.05 | ||
| Betaproteobacteria | 0 | 0.33 ± 0.21 | 0.33 ± 0.21 | 1.83 ± 1.25 | |||
| Deltaproteobacteria | 0.17 ± 0.17 | 0 ± 0 | 0 ± 0 | 4.5 ± 2.16 | |||
| Epsilonproteobacteria | 0 | 0 | 0 | 0.33 ± 0.21 | |||
| Gammaproteobacteria | 0.17 ± 0.17 | 0 | 0.33 ± 0.21 | 3.17 ± 1.66 | <0.05 | <0.05 | |
| TM7 | 0.17 ± 0.17 | 0 | 0 | 0.5 ± 0.22 | ND | ||
| Unclassified | 0.83 ± 0.17 | 1.0 ± 0 | 1.0 ± 0.26 | 2.0 ± 0.52 | |||
| Verrucomicrobia | 0.5 ± 0.22 | 1.0 ± 0 | 1.17 ± 0.31 | 1.83 ± 0.6 | <0.05 | ||
The values are the mean ± one standard error of the mean for 5 df. A statistical analysis was performed using analysis of variance for each phylum, and statistical significance is indicated by a P value of <0.05. No statistical analysis was performed for phyla containing fewer than five taxa (ND). The factors used in the analyses of variance include grove, symptomatic plants, and cross-product.
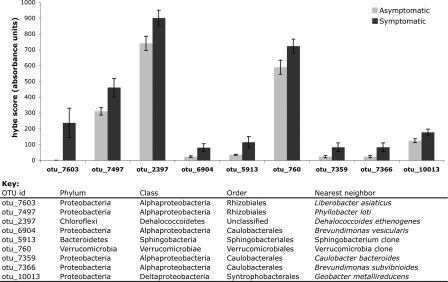
When the differences in richness between symptomatic and asymptomatic plants in grove 1 and grove 2 are examined, it is evident that there are some populations that covary with evidence of pathogenesis and grove location (Table 1). There was an overall increase in the bacterial richness in the symptomatic plants in grove 2. The increased richness seems to result mostly from bacteria in the phylum Alphaproteobacteria and the orders Caulobacterales, Sphingomonadales, and Rhizobiales. Measures of richness do not indicate changes in absolute abundance, but hybridization scores do have a linear relationship with absolute abundance when a single taxon is examined for different treatments (7). For the 117 taxa detected as members of the leaf midrib microbial community, we separately examined the individual taxa that were significantly different (P < 0.05) in the symptomatic and asymptomatic plants. Only nine taxa were significantly different, and all of them were more abundant in symptomatic plants than in asymptomatic plants (Fig. 2). In general, the differences were modest and ranged from a 50% increase to a doubling of relative abundance, with one notable exception. The otu_7603 taxon, representing “Ca. Liberibacter asiaticus,” was detected at a very low level in asymptomatic plants, but it was over 200 times more abundant in symptomatic plants.
FIG. 2.
Mean hybridization scores (hybe score) for 9 of 117 taxa detected in the leaf midrib microbial community. These nine taxa were significantly different (P < 0.05) for the symptomatic and asymptomatic leaves in each grove. The error bars indicate standard errors.
There were no discernible differences between the overall microbial communities in symptomatic and asymptomatic plants, as determined by ordination (data not shown). This community analysis was based on relative abundance of individual taxa detected by PhyloChip analysis. There were three symptomatic plants in grove 2 that were separated from the rest of the trees based on analysis of the microbial community; the bacterial richness of samples from these three trees (G2S3, G2S4, and G2S6) was consistently elevated compared to the richness of the rest of the samples, suggesting that either grove 2 in general or these plants specifically harbored a more complex microbial community.
16S rRNA gene clone library sequencing and phylogenetic analysis.
In order to verify the PhyloChip data and understand the relative abundance of different bacteria associated with HLB disease-affected citrus, the 16S rRNA gene amplicons used for the PhyloChip analysis were employed to construct a 16S rRNA gene clone library. However, sequencing of 192 clones indicated that they were all from chloroplasts. This was due to the dominance of citrus plant DNA and the fact that primers 27f and 1492r could not differentiate chloroplast 16S rRNA genes from bacterial 16S rRNA genes. Chelius and Triplett (10) designed primer 799f and used it in combination with 1492r (32), which successfully differentiated bacterial 16S rRNA genes from chloroplast DNA and mitochondrial products. Thus, clone libraries of 16S rRNA genes were constructed using the 16S rRNA gene PCR products amplified using primers 799f and 1492r.
A total of 2,062 clones were generated from cloning of the 16S rRNA gene regions amplified using the same set of genomic DNA samples that were used to amplify rRNA gene regions for the PhyloChip analysis. All sequences that were homologous to chloroplast or cyanobacterial sequences were assumed to be plant derived, likely from plastids, and excluded from the analysis. For the total population of sequenced clones, the database search placed clones into eight phyla, (i) Proteobacteria (47.1%), (ii) Bacteroidetes (14.1%), (iii) Dictyoglomi (0.01%), (iv) Actinobacteria (0.3%), (v) Chlamydiae (0.2%), (vi) Firmicutes (0.1%), (vii) TM7 (0.05%), and (viii) Verrucomicrobia (0.05%), and 37.6% of the clones originated from chloroplasts (Table 2).
TABLE 2.
Relative abundance of clones from asymptomatic or symptomatic citrus leaf midribsa
| Phylum | Class | Order | Family | Relative abundance
|
|||
|---|---|---|---|---|---|---|---|
| Grove 1
|
Grove 2
|
||||||
| Asymptomatic | Symptomatic | Asymptomatic | Symptomatic | ||||
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | 0.547 | 0.8845 | 0.1067 | 0.8443 |
| Bradyrhizobiaceae | 0 | 0.0024 | 0.0112 | 0.0019 | |||
| Rhizobiaceae | 0 | 0 | 0 | 0.0019 | |||
| Unclassified | 0 | 0 | 0.0056 | 0 | |||
| Caulobacterales | Caulobacteraceae | 0.0210 | 0.0049 | 0.0617 | 0.0019 | ||
| Sphingomonadales | Sphingomonadaceae | 0 | 0.0049 | 0.0112 | 0 | ||
| Unclassified | 0.0052 | 0.0073 | 0 | 0 | |||
| Betaproteobacteria | Burkholderiales | Comamonadaceae | 0.0052 | 0.0024 | 0.0112 | 0.0019 | |
| Incertae sedis 5 | 0.0105 | 0 | 0.0224 | 0 | |||
| Oxalobacteraceae | 0 | 0 | 0.0224 | 0 | |||
| Alcaligenaceae | 0 | 0 | 0.0056 | 0 | |||
| Unclassified | 0.0052 | 0 | 0 | 0 | |||
| Hydrogenophilales | Hydrogenophilaceae | 0.0052 | 0 | 0 | 0 | ||
| Rhodocyclales | Rhodocyclaceae | 0.0052 | 0 | 0 | 0 | ||
| Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.0157 | 0 | 0 | 0.0019 | |
| Pseudomonadaceae | 0 | 0 | 0.0056 | 0 | |||
| Oceanospirillales | Halomonadaceae | 0 | 0 | 0 | 0.0059 | ||
| Unclassified | 0 | 0.0024 | 0 | 0 | |||
| Enterobacteriales | Enterobacteriaceae | 0 | 0 | 0 | 0.0059 | ||
| Deltaproteobacteria | Desulfuromonadales | Geobacteraceae | 0 | 0 | 0 | 0.0019 | |
| Unclassified | 0 | 0.0024 | 0 | 0 | |||
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | 0 | 0.0049 | 0.0056 | 0.0019 |
| Nocardioidaceae | 0 | 0 | 0.0056 | 0 | |||
| Propionibacteriaceae | 0 | 0 | 0.0056 | 0 | |||
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 0 | 0 | 0.0112 | 0 |
| Lactobacillales | Carnobacteriaceae | 0 | 0 | 0 | 0 | ||
| Chlamydiae | Chlamydiae | Chlamydiales | Simkaniaceae | 0 | 0 | 0.0224 | 0 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Bacteroidaceae | 0 | 0.0024 | 0 | 0 |
| Flavobacteria | Flavobacteriales | Flavobacteriaceae | 0.3684 | 0.0614 | 0.6573 | 0.1157 | |
| Unclassified | 0 | 0.0024 | 0 | 0.0019 | |||
| Unclassified | 0 | 0 | 0 | 0.0099 | |||
| Sphingobacteria | Sphingobacteriales | Flexibacteraceae | 0 | 0.0122 | 0.0224 | 0.0019 | |
| Saprospiraceae | 0 | 0 | 0.0056 | 0 | |||
| Verrucomicrobia | Verrucomicrobiales | Verrucomicrobiaceae | 0.0052 | 0 | 0 | 0 | |
| Dictyoglomi | Dictyoglomi | Unclassified | 0.0052 | 0.0024 | 0 | 0 | |
| TM-7 | 0 | 0.0024 | 0 | 0 | |||
| Total | 1 | 1 | 1 | 1 | |||
The numbers of clones in the libraries and data set were as follows: grove 1 asymptomatic, 190; grove 1 symptomatic, 407; grove 2 asymptomatic, 178; and grove 2 symptomatic, 501. There were 786 clones matching chloroplast or mitochondrial products that were not included in this analysis.
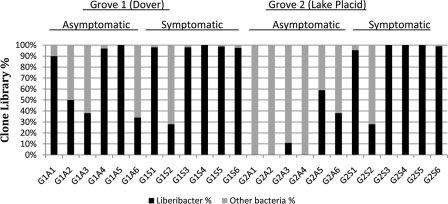
On average, more than 86% of the clones from symptomatic trees belonged to the phylum Proteobacteria, to which “Ca. Liberibacter asiaticus” belongs (Fig. 3). Further analysis of individual sequences of the clones described above revealed 99% identity with Asian strain “Sihui” (GenBank accession number EU644449) and a Florida strain (GenBank accession number EU982421). “Ca. Liberibacter asiaticus” was the only common bacterium found in all 12 symptomatic trees in two citrus groves. “Ca. Liberibacter asiaticus” was found in five of six asymptomatic trees in grove 1 and in three of six asymptomatic trees in grove 2 (Fig. 3). Significantly, midribs from symptomatic leaves contained higher percentages of clones whose genes matched “Ca. Liberibacter asiaticus” 16S rRNA genes (grove 1 symptomatic leaves, 88%; grove 2 symptomatic leaves, 84%), whereas the percentages for asymptomatic leaves were lower (grove 1 asymptomatic leaves, 55%; grove 2 asymptomatic leaves, 10%).
FIG. 3.
Prevalence of “Ca. Liberibacter asiaticus” in clone libraries for asymptomatic and symptomatic trees in each of the two groves sampled.
Comparison of cloning with PhyloChip analysis.
A comparison between clone library sequencing and PhyloChip analysis of the microbial community showed that PhyloChip analysis detected a broader richness of taxa than cloning: PhyloChip analysis detected 15 phyla in the citrus leaf midribs, whereas cloning detected 8 phyla (Table 3). Otherwise, the results of the two methods are largely in accordance. The PhyloChip analysis detected all of the phyla identified by cloning except Dictyoglomi. Both methods detected an overabundance of Alphaproteobacteria in general and “Ca. Liberibacter asiaticus” specifically.
TABLE 3.
Phyla detected in different samples by high-density PhyloChip analysis or by cloning and sequencing
| Plylum, class, or other taxon | Detection bya:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PhyloChip analysis
|
Cloning and sequencing
|
|||||||
| Asymptomatic
|
Symptomatic
|
Asymptomatic
|
Symptomatic
|
|||||
| Grove 1 (G1A) | Grove 2 (G2A) | Grove 1 (G1S) | Grove 2 (G2S) | Grove 1 (G1A) | Grove 2 (G2A) | Grove 1 (G1S) | Grove 2 (G2S) | |
| Proteobacteria | Y | Y | Y | Y | Y | Y | Y | Y |
| Alphaproteobacteria | Y | Y | Y | Y | Y | Y | Y | Y |
| Betaproteobacteria | N | Y | Y | Y | Y | Y | Y | Y |
| Deltaproteobacteria | Y | N | N | Y | N | N | Y | Y |
| Episilonproteobacteria | N | N | N | Y | N | N | N | N |
| Gammaproteobacteria | Y | N | Y | Y | Y | Y | N | Y |
| Acidobacteria | Y | Y | Y | Y | N | N | N | N |
| Actinobacteria | Y | Y | Y | Y | N | Y | Y | Y |
| AD3 | Y | N | Y | Y | N | N | N | N |
| Bacteroidetes | Y | N | Y | Y | Y | Y | Y | Y |
| BRC1 | Y | Y | Y | Y | N | N | N | N |
| Chlamydiae | Y | Y | Y | Y | N | Y | N | N |
| Chlorobi | Y | Y | Y | Y | N | N | N | N |
| Chloroflexi | N | Y | Y | Y | N | N | N | N |
| Dictyoglomi | N | N | N | N | Y | Y | Y | N |
| Firmicutes | N | N | N | Y | N | Y | N | N |
| Gemmatimonadetes | N | N | N | Y | N | N | N | N |
| NC10 | N | N | N | Y | N | N | N | N |
| Planctomycetes | Y | Y | Y | Y | N | N | N | N |
| TM7 | Y | N | N | Y | N | Y | N | N |
| Unclassified bacteria | Y | N | Y | Y | Y | Y | Y | Y |
| Verrucomicrobia | Y | Y | Y | Y | Y | N | N | N |
Y, positive; N, negative.
DISCUSSION
Molecular techniques based on PCR have made it possible to study the diversity of microorganisms in natural environments without culturing (51). These techniques are valuable for increasing our understanding of microbial communities despite some demonstrated amplification biases due to primer selection, the number of amplification cycles, and template concentration (37, 49). A diverse assemblage of microorganisms was observed in citrus leaf midribs from HLB disease-positive citrus groves with both PhyloChip analysis and 16S rRNA gene clone library sequencing. The fact that so many more orders of bacteria were detected by PhyloChip analysis (47 orders in 15 phyla) than by cloning and sequencing (20 orders in 8 phyla) indicates that PhyloChip analysis is more comprehensive for identification of microorganisms in environmental samples than 16S rRNA gene clone library sequencing. This is consistent with previous reports that compared clone library data with PhyloChip community analysis data for environmental samples (8, 15). The PhyloChip array used in this study contains 8,741 taxa representing all 121 demarcated bacterial and archaeal orders (7). The size of the clone library might also contribute to the difference in the data. It has been suggested that 40,000 sequencing reactions are required to document 50% of the richness of certain environmental samples, which is laborious, costly, and time-consuming (8). Typical 16S rRNA gene clone libraries include fewer than 1,000 sequences (16, 18, 27, 34). The numbers of clones obtained for our asymptomatic and symptomatic samples are 957 and 1,105, respectively. The different primers selected for PCR amplification for PhyloChip analysis (primers 27f and 1492r) and construction of a clone library (primers 799f and 1492r) might have contributed to the difference even though both sets of primers include universal primers for bacteria (10, 50). In addition, it is also possible that the PhyloChip array approach causes nonspecific hybridization, leading to false positives, although this is most likely to hamper discrimination of taxa at the genus or family level (8, 15). While this may inflate the number of species-level taxa detected per family, it does not affect either the phylum-level richness of the community or the change in relative abundance of “Ca. Liberibacter asiaticus,” which are the two main points that we meant to address with PhyloChip analysis.
Our study indicated that “Ca. Liberibacter sp.” is the dominant bacterium that is always detected in citrus showing HLB disease symptoms. 16S rRNA gene cloning and sequencing showed that “Ca. Liberibacter asiaticus” was the only common bacterium found in all 12 symptomatic trees in two citrus groves. The PhyloChip study indicated that nine taxa were significantly different, and all of them were more abundant in symptomatic plants than in asymptomatic plants. However, “Ca. Liberibacter asiaticus” was the dominant organism in the symptomatic leaves but not in the asymptomatic leaves, and the observation that “Ca. Liberibacter asiaticus” was the dominant organism in the symptomatic leaves supports the association between HLB disease and “Ca. Liberibacter asiaticus” in Florida (5). By using PhyloChip analysis the otu_7603 taxon, representing “Ca. Liberibacter asiaticus,” was detected at a very low level in asymptomatic plants, but it was over 200 times more abundant in symptomatic plants. Other than “Ca. Liberibacter asiaticus,” the taxa which were more abundant in symptomatic plants than in asymptomatic plants included representatives of the taxa Phyllobacter, Dehalicoccoides, Brevundimonas sp. strains 6904 and 7359, Sphingobacterium, Verrucomicrobia, Caulobacter, and Syntrophobacter (Fig. 2), and these bacteria have not been reported to cause plant diseases so far. Their roles in HLB disease symptom development remain to be investigated.
The abundance of some of the bacteria that were detected was greater for asymptomatic samples with HLB disease than for symptomatic samples. For example, incertae sedis 5, Oxalobacteraceae, Alcaligenaceae, Hydrogenophilaceae, Rhodocylclaceae, Pseudomonadaceae, Nocardioidaceae, Propionibacteriaceae, Bacillaceae, Simkaniaceae, Verrucomicrobiaceae, and Saprospiraceae, some of which have biocontrol and plant growth-promoting potential (3, 12, 22, 36), were found only in asymptomatic samples based on cloning. It is not known whether these bacteria play significant roles in suppressing HLB disease symptoms. The lack of symptoms might in some cases be due to the low titer of “Ca. Liberibacter asiaticus” in the phloem, considering that previous results indicated that a minimal “Ca. Liberibacter asiaticus” population is required for symptom development (N. Wang, unpublished data). Interestingly, clone library analysis and “Ca. Liberibacter asiaticus”-specific PCR suggest that there might be a few escape trees (asymptomatic trees with heavy loads of the putative pathogen “Ca. Liberibacter asiaticus”). Both methods found “Ca. Liberibacter asiaticus” in asymptomatic tree G1A4, while clone library analysis also indicated the presence of “Ca. Liberibacter asiaticus” at high titers in asymptomatic trees G1A1 and G2A5 and PCR showed that “Ca. Liberibacter asiaticus” was present in asymptomatic trees G1A4 and G2A3. Whether such trees can survive with large populations of “Ca. Liberibacter asiaticus” without showing any disease symptoms and whether the endophytic microbial community plays a role in symptom suppression remain to be determined.
Citrus leaves can support a diversity of microbes either epiphytically or endophytically. PhyloChip analysis revealed the presence of 47 orders of bacteria in 15 phyla, while 20 orders in 8 phyla were observed with the cloning and sequencing method for the citrus leaf midribs. Actinobacteria, Proteobacteria, and Firmicutes have previously been reported to be associated with plant leaves (23). The majority of these bacteria are insect transmitted or endosymbionts of insects. “Ca. Liberibacter asiaticus” has been shown to be psyllid transmitted. Most of the clones in the 16S rRNA gene library were closely related to bacteria reported to be endosymbionts of various insects (17, 20, 21, 46). Lacava et al. (31) have reported similarity between the endophytes of host plants and bacteria inhabiting the head region of the glassy-winged sharpshooter, Homalodisca vitripennis, an important vector of various strains of X. fastidiosa. Our study also indicates that there may be multipartite interactions between the host plant, the insect vector, and the associated microbial diversity. However, some bacteria, such as Chlamydiae, AD3, Bacteroidetes, and mgA-2, have never been reported to be associated with plant leaves (3, 7, 25, 33, 53). This indicates that our understanding of the extent of microbial diversity associated with plant leaves is still incomplete. It is not surprising that the bacterial population associated with citrus midribs seems to be quite different from and more diverse than the citrus phyllosphere population (52). The majority of the bacteria in our study are likely endophytes since surface sterilization was used. Surface sterilization has been shown to eliminate most, but not all, microbes on the leaf surface (10). The microbiome associated with citrus leaves from HLB pathogen-infected groves in Florida is very different from that of X. fastidiosa-infected citrus groves in Brazil (2). Curtobacterium flaccumfaciens, Enterobacter cloacae, Methylobacterium spp. Nocardia sp., and Pantoea agglomerans were reported for X. fastidiosa-infected citrus branches in Brazil, while they were not found in our study (2). This might have been due to differences in the environmental conditions in the two geographic locations where the plants were grown (e.g., geographic areas and weather conditions), dominant pathogens associated with the plants, or the tissues sampled (leaf midrib or branch).
This study included an extensive molecular analysis of the bacteria in citrus leaf midribs from HLB pathogen-positive citrus groves. We demonstrated that both symptomatic and asymptomatic leaves contain a diverse assemblage of bacteria. Some bacteria other than “Ca. Liberibacter” have been identified from citrus with HLB-disease. “Ca. Liberibacter asiaticus” is the dominant organism in the symptomatic leaves compared to the asymptomatic leaves, implicating this organism as the causal agent of HLB disease.
Supplementary Material
Acknowledgments
This work was supported by the Florida Citrus Production Research Advisory Council.
Footnotes
Published ahead of print on 16 January 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araújo, W. L., J. Marcon, W. Maccheroni, Jr., J. D. Van Elsas, J. W. L. Van Vuurde, and J. L. Azevedo. 2002. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl. Environ. Microbiol. 68:4906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacon, C. W., and D. M. Hinton. 2002. Endophytic and biological control potential of Bacillus mojavensis and related species. Biol. Control 23:274-284. [Google Scholar]
- 4.Balser, T. C., and M. K. Firestone. 2005. Linking microbial community composition and soil processes in two California ecosystems. Biogeochemistry 73:395-415. [Google Scholar]
- 5.Bové, J. M. 2006. Huanglongbing: a destructive, newly emerging, century-old disease of citrus. J. Plant Pathol. 88:7-37. [Google Scholar]
- 6.Bové, J. M., E. C. Calavan, S. P. Capoor, R. E. Cortez, and R. E. Schwarz. 1974. Influence of temperature on symptoms of California stubborn, South African greening, Indian citrus decline and Philippines leaf mottling diseases, p. 12-15. In Proceedings of the 6th Conference of the International. Organization of Citrus Virologists. International Organization of Citrus Virologists, Riverside, CA.
- 7.Brodie, E. L., T. Z. DeSantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodie, E. L., T. Z. DeSantis, J. P. Parker, I. X. Zubietta, Y. M. Piceno, and G. L. Andersen. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. USA 104:299-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler, D. P., J. K. Fredrickson, and F. J. Brockman. 1997. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol. Ecol. 6:475-482. [DOI] [PubMed] [Google Scholar]
- 10.Chelius, M. K., and E. W. Triplett. 2001. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41:252-263. [DOI] [PubMed] [Google Scholar]
- 11.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:169-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compant, S., B. Reiter, A. Sessitsch, J. Nowak, C. Clement, and E. A. Barkia. 2005. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71:1685-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Graça, J. V. 1991. Citrus greening disease. Annu. Rev. Phytopathol. 29:109-136. [Google Scholar]
- 14.da Graça, J. V., and L. Korsten. 2004. Citrus huanglongbing: review, present status and future strategies, p. 229-245. In N. Samh (ed.), Diseases of fruits and vegetables, vol. I. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 15.DeSantis, T. Z., E. L. Brodie, J. P. Moberg, I. X. Zubieta, Y. M. Piceno, and G. L. Andersen. 2007. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 53:371-383. [DOI] [PubMed] [Google Scholar]
- 16.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn, A. K., and E. V. Stabb. 2005. Culture-independent characterization of the microbiota of the ant lion Myrmeleon mobilis (Neuroptera: Myrmeleontidae). Appl. Environ. Microbiol. 71:8784-8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elshahed, M. S., J. M. Senko, F. Z. Najar, S. M. Kenton, B. A. Roe, T. A. Dewers, et al. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnier, M., S. Jagoueix-Eveillard, P. Cronje, H. Le Roux, and J. M. Bové. 2000. Genomic characterisation of a liberibacter present in an ornamental rutaceous tree, Calodendrum capense, in the Western Cape province of South Africa. Proposal for “Candidatus Liberibacter africanus subsp. capensis.” Int. J. Syst. Evol. Microbiol. 50:2119-2125. [DOI] [PubMed] [Google Scholar]
- 20.Gruwell, M. E., G. E. Morse, and B. B. Noemark. 2007. Phylogenetic congruence of armored scale insects (Hemiptera: Diaspididae) and their primary endosymbionts from the phylum Bacteroidetes. Mol. Phylogenet. Evol. 44:267-280. [DOI] [PubMed] [Google Scholar]
- 21.Gruwell, M. E., C. D. von Dohlen, K. Patch, and B. B. Normark. 2005. Preliminary PCR survey of bacteria associated with scale insects (Hemiptera: Coccoidea), p. 101-116. In L. B. Erkılıç and M. B. Kaydan (ed.), Proceedings of the Tenth International Symposium on Scale Insect Studies, Adana, Turkey, 19 to 23 April 2004. Scientific and Technical Research Council of Turkey, Ankara.
- 22.Haas, D., and G. Defago. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307-319. [DOI] [PubMed] [Google Scholar]
- 23.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hocquellet, A., J. M. Bové, and M. Garnier. 1999. Isolation of DNA from the uncultured “Candidatus Liberobacter” species associated with citrus huanglongbing by RAPD. Curr. Microbiol. 38:176-182. [DOI] [PubMed] [Google Scholar]
- 25.Horn, M. 2008. Chlamydiae as symbionts in eukaryotes. Annu. Rev. Microbiol. 62:113-131. [DOI] [PubMed] [Google Scholar]
- 26.Jagoueix, S., J. M. Bové, and M. Garnier. 1994. The phloem-limited bacterium of greening disease of citrus is a member of alpha subdivision of the Proteobacteria. Intl. J. Sys. Bacteriol. 44:379-386. [DOI] [PubMed] [Google Scholar]
- 27.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J., U. S. Sagaram, J. K. Burns, and N. Wang. 2009. Response of sweet orange (Citrus sinensis) to Candidatus Liberibacter asiaticus infection: microscopy and microarray analyses. Phytopathology 99:50-57. [DOI] [PubMed] [Google Scholar]
- 29.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacava, P. T., W. L. Araújo, and J. L. Azevedo. 2007. Evaluation of endophytic colonization of Citrus sinensis and Catharanthus roseus seedlings by endophytic bacteria. J. Microbiol. 45:11-14. [PubMed] [Google Scholar]
- 31.Lacava, P. T., J. Parker, F. D. Andreote, F. Dini-Andreote, J. L. Ramirez, and T. A. Miller. 2007. Analysis of the bacterial community in glassy-winged sharpshooter heads. Entomol. Res. 37:261-266. [Google Scholar]
- 32.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 33.Lodewyckx, C., M. Mergeay, J. Vangronsveld, H. Clijsters, and D. Van der Lelie. 2002. Isolation, characterization, and identification of bacteria associated with the zinc hyperaccumulator Thlaspi caerulescens subsp. calaminaria. Int. J. Phytoremediat. 4:101-115. [DOI] [PubMed] [Google Scholar]
- 34.McCaig, A. E., C. J. Phillips, J. R. Stephen, G. A. Kowalchuk, S. M. Harvey, R. A. Herbert, T. M. Embley, and J. I. Prosser. 1999. Nitrogen cycling and community structure of proteobacterial beta-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl. Environ. Microbiol. 65:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, R. G. E., and K.-H. Schleifer. 1994. Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes. Int. J. Syst. Bacteriol. 44:174-176. [DOI] [PubMed] [Google Scholar]
- 36.Ongena, M., and P. Jacques. 2008. Bacillus subtilis lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16:115-125. [DOI] [PubMed] [Google Scholar]
- 37.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 38.Romero, J., M. García-Varela, J. P. Laclette, and R. T. Espejo. 2002. Bacterial 16S rRNA gene analysis revealed that bacterial related to Arcobacter spp. constitute an abundant and common component of the oyster microflora (Tiostrea chilensis). Microb. Ecol. 44:365-371. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Santelli, C. M., B. N. Orcutt, E. Banning, W. Bach, C. L. Moyer, M. L. Sogin, H. Staudigel, and K. J. Edwards. 2008. Abundance and diversity of microbial life in ocean crust. Nature 453:653-656. [DOI] [PubMed] [Google Scholar]
- 41.Schneider, H. 1968. Anatomy of greening diseased sweet orange shoots. Phytopathology 58:1155-1160. [Google Scholar]
- 42.Singh, B. K., P. Millard, A. S. Whiteley, and J. C. Murrell. 2004. Unravelling rhizosphere microbial interactions: opportunities and limitations. Trends Microbiol. 12:386-393. [DOI] [PubMed] [Google Scholar]
- 43.Sun, L., F. Qiu, X. Zhang, X. Dai, X. Dong, and W. Song. 2008. Endophytic bacteria diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb. Ecol. 55:415-424. [DOI] [PubMed] [Google Scholar]
- 44.Tatineni, S., U. S. Sagaram, S. Gowda, C. J. Robertson, W. O. Dawson, T. Iwanami, and N. Wang. 2008. In plant distribution of ‘Candidatus Liberibacter asiaticus’ as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology. 98:592-599. [DOI] [PubMed] [Google Scholar]
- 45.Teixeira, D. C., J. Ayres, J. L. Danet, S. Jagoueix-Eveillard, C. Saillard, and J. M. Bové. 2005. First report of a huanglongbing-like disease of citrus in São Paulo, Brazil, and association of a new Liberibacter species, “Candidatus Liberibacter americanus,” with the disease. Plant Dis. 89:107. [DOI] [PubMed] [Google Scholar]
- 46.Thao, M. L., and P. Baumann. 2004. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 70:3401-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 48.Urakawa, H., S. El Fantroussi, H. Smidt, J. C. Smoot, E. Tribou, J. J. Kelly, et al. 2003. Optimization of single-base-pair mismatch discrimination in oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webster, G., C. J. Newberry, J. C. Fry, and A. J. Weightman. 2003. Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA based techniques: a cautionary tale. J. Microbiol. Methods 55:155-164. [DOI] [PubMed] [Google Scholar]
- 50.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, K. H., R. B. Blitchington, and R. C. Greene. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, C.-H., D. E. Crowley, J. Borneman, and N. T. Keen. 2001. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 98:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, J.-Z., X. Beicheng, H. Huang, D. S. Treves, L. J. Hauser, R. J. Mural, A. V. Palumbo, and J. M. Tiedje. 2003. Bacterial phylogenetic diversity and a novel candidate division of two humid region, sandy surface soils. Soil Biol. Biochem. 35:915-924. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.