Abstract
We investigated the efficacy of a loop-mediated isothermal amplification (LAMP) assay for detection of chicken meat samples naturally contaminated with Campylobacter jejuni and Campylobacter coli. A total of 144 Preston enrichment broth cultures from chicken meat samples were assessed by using the LAMP assay and conventional culture methods, which consist of a combination of Preston enrichment culturing and plating onto Butzler and modified charcoal cefoperazone deoxycholate agars. Compared with C. jejuni-C. coli isolation using the conventional culture test, the LAMP results showed 98.5% (67/68) and 97.4% (74/76) sensitivity and specificity, respectively, and the positive and negative predictive values were 97.1% (67/69) and 98.7% (74/75), respectively. The conventional culture test required more than 3 to 4 days to isolate and identify C. jejuni and C. coli in the Preston enrichment cultures. In contrast, the LAMP assay was markedly faster, requiring less than 90 min from the beginning of DNA extraction to final detection and differentiation of C. jejuni and C. coli. In total, the LAMP assay required 23.5 to 25.5 h from the beginning of the enrichment culture to final determination. These results suggest that our LAMP assay is a powerful tool for rapid, sensitive, and practical detection of C. jejuni and C. coli which may facilitate surveillance and control of C. jejuni-C. coli contamination in chicken, as well as investigations of food poisoning incidents caused by these organisms. This is the first report of a highly sensitive and specific LAMP assay to detect and differentiate C. jejuni and C. coli in chicken meat samples.
Campylobacter is widely acknowledged as one of the most frequent causes of acute bacterial gastroenteritis in humans worldwide (20, 26), and Campylobacter jejuni and Campylobacter coli are the predominant causes of Campylobacter gastroenteritis (20, 26). Poultry is often implicated as a main source of human infection due to the high prevalence of C. jejuni-C. coli contamination. Outbreaks and sporadic cases associated with the consumption of raw or undercooked chicken meat have frequently been reported in Japan (14) and worldwide (4, 7, 27), which indicates that ingestion of raw or undercooked chicken meat contaminated with C. jejuni-C. coli is a risk factor in humans (29, 31).
Since stool samples from patients often contain large numbers of viable campylobacters, it is not difficult to isolate the organisms by direct plating on selective media without enrichment. Food products, however, may harbor only small numbers of campylobacters, and they may be injured. Therefore, enrichment is essential to recover Campylobacter cells. To determine the causative food in incidents caused by C. jejuni-C. coli, microbiological examinations have been performed. The rate of isolation of C. jejuni-C. coli from food samples associated with food poisoning incidents is, however, approximately 10% in Japan (14). The remaining 90% of the causative foods are unknown due to the difficulty of recovering C. jejuni-C. coli from food samples (14, 17).
The test procedure includes plating a sample onto selective agar medium and subsequent identification of suspected colonies on the agar medium after a pure culture is obtained. The bacterial culture tests for campylobacters from samples require 2 to 3 days and are time-consuming and laborious. Further, an enrichment procedure is required for food samples to increase the small numbers of campylobacters and reduce the growth of the background flora before selective plating of cultures. Therefore, it takes at least 3 to 4 days for identification of the isolates. Given these limitations, a rapid, simple, and practical assay to detect C. jejuni and C. coli in chicken meat is required.
Several investigators have developed loop-mediated isothermal amplification (LAMP) assays (19, 21, 25) to detect pathogenic microorganisms, such as C. jejuni and C. coli, verotoxin-producing, enterotoxigenic, and enteroinvasive Escherichia coli, Shigella, Salmonella, Helicobacter pylori, Vibrio cholerae, Mycobacterium tuberculosis, and norovirus (10, 11, 16, 18, 30, 32, 33, 35, 36). A LAMP assay is faster and easier to perform than conventional PCR assays, as well as more specific (10, 33). Furthermore, because the LAMP assay synthesizes a large amount of DNA and its by-product, an insoluble white precipitate of magnesium pyrophosphate, the by-product can be detected by a simple turbidity analysis. Also, the increase in the turbidity of the reaction mixture due to the production of the white precipitate correlates with the amount of DNA synthesized (10, 32, 33). Thus, compared to PCR assays, expensive equipment is not necessary to obtain a high level of precision (10, 11, 33). In addition, there are fewer preparation steps for the LAMP assay than for conventional PCR and real-time PCR assays, and LAMP assays require less time than the latter assays (12). These features allow simple, rapid, and cost-effective detection (10, 16).
We previously developed a LAMP assay to identify and directly detect C. jejuni and C. coli contaminants in pure cultures and human fecal specimens (32). In the present study, we established a simple and practical DNA extraction protocol to detect small numbers of C. jejuni and C. coli cells in spiked chicken samples. Subsequently, we evaluated the efficacy of a combination of the new DNA extraction protocol with the LAMP assay for rapid and sensitive detection of chicken samples naturally contaminated by C. jejuni and C. coli.
MATERIALS AND METHODS
Isolation of C. jejuni and C. coli from naturally contaminated chicken meat.
A total of 144 chicken samples, purchased in 2007 and 2008 at supermarkets in Osaka and Shiga prefectures, Japan, were used; these samples included 39 thighs, 25 breasts, 25 ground meat samples, 21 white meat samples, 14 wings, 4 livers, 3 skin samples, 2 hearts, 2 gizzards, and 9 chicken meat samples (unknown portions). Approximately 2-g chicken meat samples were added to 18 ml of Preston enrichment broth (Oxoid Ltd., Hampshire, United Kingdom) supplemented with 5% (vol/vol) lysed horse blood, and this was followed by thorough mixing using a Vortex-Genie 2 (Scientific Industries Inc., New York). Samples were then incubated at 42°C for 22 to 24 h under microaerobic atmosphere conditions, generated using an AnaeroPack MicroAero (Mitsubishi Gas Chemical Co. Ltd., Tokyo, Japan), which maintained an atmosphere containing approximately 8% O2, 7% CO2, and 85% N2. For C. jejuni and C. coli isolation, one loopful (approximately 10 μl) of a Preston broth culture sample was streaked onto Butzler (Oxoid) and modified charcoal cefoperazone deoxycholate (mCCDA) (Oxoid) agars using a disposable loop (Inoculating Loop; Nunc Ltd., Roskide, Denmark), which was followed by incubation at 42°C for 44 to 48 h. A maximum of three typical Campylobacter-like colonies were selected from a single sample to avoid false-negative culture results due to selection of non-Campylobacter colonies, which on Butzler agar were slightly pink, round, convex, and smooth, as well as pinhole-like, small (diameter, ca. 1 mm), smooth, and semitransparent and on mCCDA agar either were greyish, flat, and moistened or had a metallic sheen. Selected samples were then processed by subculturing them at 42°C for 24 to 48 h. Isolates were confirmed based on their Gram stain appearance (Campylobacter isolates are gram negative and appear to be spiral rods), as well as by latex agglutination in a Campylobacter confirmation test (Microscreen Campylobacter; Microgen Bioproducts Ltd., Camberley, United Kingdom). The majority of isolates were positive for C. jejuni using a hippurate hydrolysis test (13), whereas hippurate hydrolysis-negative C. jejuni and C. coli isolates were identified using a species-specific PCR (34) or a LAMP (32) assay. The sensitivities of Butzler and mCCDA agars were compared.
DNA extraction from enrichment broth with three-step centrifugation procedures.
To improve the sensitivity of the LAMP assay, a DNA extraction protocol was modified with three-step centrifugation procedures. The purposes of the three-step centrifugation procedures are to remove larger debris from chicken samples and components of the Preston enrichment broth containing inhibitors of DNA amplification and to concentrate the small number of Campylobacter cells. Concurrent with strain isolation, C. jejuni and C. coli detection was performed by LAMP testing using 1 ml of each Preston enrichment culture sample, which was used for preparation of DNA templates. To remove larger debris in the chicken sample and components of the Preston enrichment broth, including horse blood, a Preston enrichment culture was thoroughly mixed and centrifuged at 900 × g for 1 min. The resulting supernatant was transferred to a new 1.5-ml microcentrifuge tube and centrifuged at 10,000 × g for 5 min. Fatty components visually confirmed to be on the microcentrifuge tube wall, which were derived from chicken meat, were removed using a sterile cotton swab. After removal of the supernatant, pellets were resuspended in 50 μl of NaOH (25 mmol liter−1), thoroughly mixed using a Vortex-Genie 2, and heated at 95°C for 5 min. After neutralization with 4 μl of Tris-HCl buffer (1 mol liter−1, pH 7.5), debris was centrifuged at 4°C and 20,000 × g for 5 min. Supernatants were stored at −20°C until use, at which time 2-μl samples were used as template DNA for the LAMP assay.
LAMP assay.
The LAMP assay was performed using a Loopamp DNA amplification kit (Eiken Chemical Co. Ltd., Tokyo, Japan) as previously described (32). LAMP assays, which target two sequences presumed to contain an oxidoreductase gene and a gufA gene in C. jejuni and C. coli, respectively (6, 28), were performed using single 25-μl 1× reaction mixtures (Eiken Chemical) containing 2 μl of template DNA from a Preston enrichment culture, 1 μl of Bst DNA polymerase (Eiken Chemical), 1.6 μmol liter−1 each of inner primers FIP and BIP, 0.2 μmol liter−1 each of outer primers F3 and B3, and 0.8 μmol liter−1 each of loop primers LoopF and LoopB. Primer sequences are shown in Table 1. All primers were produced by Hokkaido System Science Co. Ltd. (Sapporo, Japan). In a Loopamp real-time turbidimeter, the reaction mixtures were incubated at 65°C for 60 min, followed by 80°C for 2 min to complete the reaction (LA-320; Teramecs Co. Ltd., Kyoto, Japan). Amplification in the LAMP assay was detected in real time as turbidity at 650 nm using an LA-320 turbidimeter. A reaction was considered positive when the turbidity reached 0.1 within 60 min. Turbidity visible with the naked eye was also considered to indicate a successful LAMP procedure (19). C. jejuni-C. coli-positive samples were further differentiated by the LAMP assay using the primer sets used to identify C. jejuni and C. coli and LAMP conditions identical to those described above. The LAMP assay was performed in a blinded fashion.
TABLE 1.
LAMP primers used for assays
| Target gene | GenBank accession no. | Primer | Sequence (5′ to 3′) |
|---|---|---|---|
| Cj0414a | AL111168 | CJ-FIP | ACAGCACCGCCACCTATAGT-AGAAGCTTTTTTAAACTAGGGC (F1c-F2) |
| CJ-BIP | AGGCAGCAGAACTTACGCATT-GAGTTTGAAAAAACATTCTACCTCT (B1-B2c) | ||
| CJ-F3 | GCAAGACAATATTATTGATCGC (F3) | ||
| CJ-B3 | CTTTCACAGGCTGCACTT (B3c) | ||
| CJ-LF | CTAGCTGCTACTACAGAACCAC (LFc) | ||
| CJ-LB | CATCAAGCTTCACAAGGAAA (LB) | ||
| CCO0367b | AAFL01000003 | CC-FIP | AAGAGATAAACACCATGATCCCAG-TCATGAATGAGCTTACTTTAGC (F1c-F2) |
| CC-BIP | GCGGCAAAGACTTATGATAAAGC-TACCGCCATTCCTAAAACAAG (B1-B2c) | ||
| CC-F3 | TGGGAGCGTTTTTGATCT (F3) | ||
| CC-B3 | AATCAAACTCACCGCCAT (B3c) | ||
| CC-LF | CCACTACAGCAAAGGTGATG (LFc) | ||
| CC-LB | CCACGATAGCCTTTATGGA (LB) |
Determination of sensitivity of the LAMP assay with spiked chicken samples.
The sensitivity of the LAMP assay for detection of C. jejuni and C. coli in spiked chicken samples was determined using bacterial cells of C. jejuni subsp. jejuni LMG 8841T and C. coli JCM 2529T. For preparation of the template DNA, 10 g of a chicken thigh sample was added to 90 ml of Preston enrichment broth in a plastic stomacher bag and incubated at 42°C for 22 h under microaerobic atmosphere conditions. After light hand massaging for 30 s, a sample was confirmed to be Campylobacter negative by using the culture and LAMP assays mentioned above. Campylobacter-negative template DNA was used as a negative control for the sensitivity test. Colonies of the background flora in the Preston enrichment culture were counted on blood agar, and the number of CFU was calculated. In parallel, bacterial cultures were incubated at 37°C for 16 h in a microaerobic atmosphere. Serial 10-fold dilutions were prepared in phosphate-buffered saline, and 100 μl of each dilution was spiked into 1 ml of a Preston enrichment broth culture in a 1.5-ml microcentrifuge tube and then mixed well using a Vortex-Genie 2. Template DNA was concentrated in 54 μl of supernatant as mentioned above, and 2 μl of each sample was used in triplicate for determination of the sensitivity of the LAMP assay. In parallel, to determine the inoculum size, 100 μl of each dilution was plated onto blood agar (Oxoid) in duplicate and incubated at 37°C for 48 h in a microaerobic atmosphere. Further, the sensitivity of the LAMP assay for spiked Preston enrichment cultures with dilutions of C. jejuni and C. coli in mixtures containing different ratios of the bacteria (C. jejuni/C. coli ratios of 100:1, 10:1, 1:1, 1:10, and 1:100, which correspond to CFU ratios of 790:3.8, 79:3.8, 7.9:3.8, 7.9:38, and 7.9:380, respectively, in LAMP reaction tubes) was determined.
Preliminary study for evaluation of three-step centrifugation procedures.
A preliminary study was performed using 64 chicken samples and the method of the International Organization for Standardization (ISO) (15). Portions (25 g) of chicken samples were added to 225 ml Bolton enrichment broth (Oxoid) supplemented with 5% (vol/vol) lysed horse blood, homogenized with a stomacher (Pro-media SH-001; ELMEX Co. Ltd., Tokyo, Japan) for 1 min, and incubated at 37°C for 4 h and then at 42°C for 20 to 44 h under microaerobic atmosphere conditions. The protocol used for isolation of C. jejuni and C. coli was that mentioned above. In parallel, to evaluate the influence of inhibitors of DNA amplification from chicken samples and components of the enrichment broth and the effectiveness of centrifugation for concentrating the small number of C. jejuni-C. coli cells in enrichment broth, two DNA extraction protocols were compared. Following the manufacturer's instructions for a commercially available kit, template DNA was prepared from 42 chicken samples using a mixture containing 50 μl of a Bolton enrichment broth culture and 50 μl of Extraction Solution for Foods (Eiken Chemical), thoroughly mixed using a Vortex-Genie 2, and heated at 95°C for 5 min. After neutralization with 10 μl of Tris-HCl buffer (1 mol liter−1, pH 7.0), debris was centrifuged with a desktop centrifugation apparatus for 1 min. The other template DNA from 22 chicken samples from 1-ml Bolton enrichment broth cultures were prepared with three-step centrifugation procedures as mentioned above. All 64 template DNA were assayed by the LAMP assay.
RESULTS
To evaluate the efficacy of the LAMP assay for detecting C. jejuni and C. coli in Preston enrichment cultures, 144 chicken meat samples were assayed by using the LAMP assay, and the results were compared with those obtained with a conventional testing method consisting of Preston enrichment culture preparation followed by plating onto Butzler and mCCDA agars. The results are summarized in Tables 2 and 3.
TABLE 2.
Comparison of LAMP and C. jejuni-C. coli isolation results for 144 chicken meat samples
| C. jejuni-C. coli LAMP results | Culture results
|
|
|---|---|---|
| No. positive | No. negative | |
| Positive (n = 69) | 67 | 2 |
| Negative (n = 75) | 1 | 74 |
| Total (n = 144) | 68 | 76 |
TABLE 3.
Comparison of C. jejuni and C. coli detection results for 144 chicken meat samples obtained with the LAMP assay and by culture on Butzler and mCCDA agars
| LAMP results | No. culture positive on:
|
No. culture negative on Butzler and mCCDA agars | ||
|---|---|---|---|---|
| Butzler and mCCDA agars | Butzler agar | mCCDA agar | ||
| C. jejuni positive (n = 59) | 57 | 56 | 45 | 2 |
| C. coli positive (n = 5) | 5 | 2 | 5 | 0 |
| C. jejuni and C. coli positive (n = 5)a | 5 | 5 | 4 | 0 |
| Negative (n = 75) | 1 | 1 | 0 | 74 |
| Total (n = 144) | 68 | 64 | 54 | 76 |
After random selection of three isolates from five chicken samples, two and one samples showed three isolates of C. jejuni and C. coli, respectively. The remaining two samples showed one and two and two and one isolates of C. coli and non-Campylobacter, respectively.
Tables 2 and 3 show that C. jejuni or C. coli was isolated from 68 of the 144 chicken samples (60 C. jejuni-positive samples and 8 C. coli-positive samples) by plating; 67 of the these 68 samples were positive and 1 was negative with the LAMP assay. Of the remaining 76 C. jejuni and C. coli culture-negative chicken samples, 74 and 2 yielded C. jejuni-C. coli-negative and C. jejuni-positive results, respectively, when the LAMP assay was used. Compared to isolation of C. jejuni-C. coli by conventional culture testing, the LAMP assay showed 98.5% sensitivity (67 of 68 samples) and 97.4% specificity (74 of 76 samples), and the positive and negative predictive values were 97.1% (67 of 69 samples) and 98.7% (74 of 75 samples), respectively. As shown in Table 3, Butzler and mCCDA agars showed 94.1% (64/68) and 79.4% (54/68) sensitivity, respectively.
Of the 68 culture-positive samples, 67 were LAMP and culture positive, whereas 1 chicken thigh sample was LAMP negative and culture positive. For the 68 culture-positive samples, C. jejuni and C. coli were differentiated using the respective LAMP assays (Table 3). Among the 67 LAMP- and culture-positive samples, the LAMP assay identified 57 C. jejuni-positive, 5 C. coli-positive, and 5 C. jejuni- and C. coli-positive samples. The one remaining chicken thigh sample, which was LAMP negative and culture positive, was identified as C. jejuni positive by culture. Further, 74 of the 76 culture-negative samples were LAMP and culture negative, whereas 1 chicken wing sample and 1 thigh sample were C. jejuni LAMP positive and culture negative. An increase in turbidity was confirmed for all LAMP products using an LA-320 turbidimeter and by visual confirmation of turbidity. The results determined by the two methods consistently matched each other.
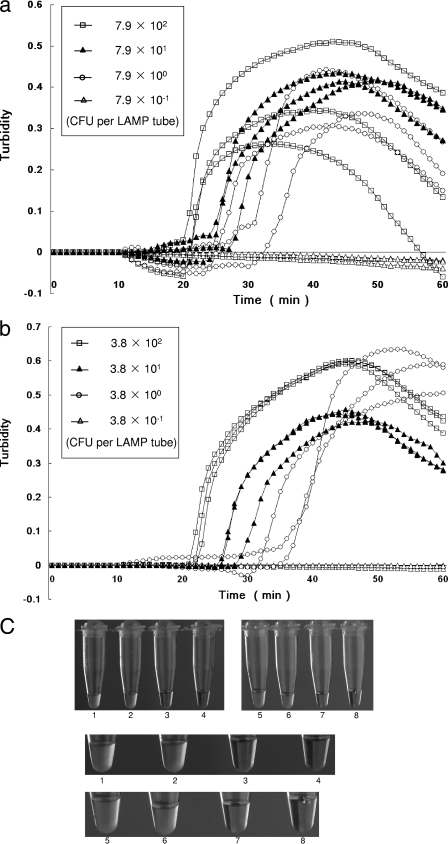
As shown in Fig. 1, the sensitivity of the LAMP assay for detection of C. jejuni and C. coli in spiked Preston enrichment cultures was determined in triplicate by using a real-time turbidimeter. The sensitivities for C. jejuni and C. coli were found to be 7.9 and 3.8 CFU per LAMP reaction tube. The dilutions yielding 790 to 7.9 CFU and 380 to 3.8 CFU showed an increase in turbidity, but the dilutions yielding 0.8 CFU and 0.4 CFU did not. Amplification in the LAMP assay could also be judged based on turbidity by visual assessment using the naked eye. The sensitivities determined by the two methods consistently matched. When the LAMP assay was performed in triplicate using 2-μl mixtures consisting of 1 μl template DNA containing 3.9 CFU C. jejuni and 1 μl template from a negative control sample, two of three samples showed positive results. Correspondingly, when 2-μl mixtures consisting of 1 μl template DNA containing 1.9 CFU C. coli and 1 μl template from a negative control sample were used, one of three samples showed a positive result. Further, all sensitivity tests with mixtures containing a known amount of C. jejuni and C. coli at different ratios showed positive results with the two methods, as expected (data not shown). The amount of background flora in the Preston enrichment cultures used for the sensitivity tests was approximately 106 CFU/ml.
FIG. 1.
Sensitivity test to detect C. jejuni subsp. jejuni LMG8841T and C. coli JCM2529T by using a real-time turbidimeter and visual assessment. The curves from left to right in panels a and b indicate decreasing concentrations of bacterial DNA (790 to 0.79 CFU per test tube in panel a and 380 to 0.38 CFU per test tube in panel b). (a) Detection of C. jejuni; (b) detection of C. coli; (c) visual detection of C. jejuni and C. coli by observation of turbidity. Tube 1, third sample containing 790 CFU for C. jejuni; tube 2, third sample containing 7.9 CFU for C. jejuni; tube 3, third sample containing 0.8 CFU for C. jejuni; tube 4, negative control; tube 5, third sample containing 380 CFU for C. coli; tube 6, third sample containing 3.8 CFU for C. coli; tube 7, third sample containing 0.4 CFU for C. coli; tube 8, negative control.
In the preliminary study using the template DNA prepared by the manufacturer's instructions for the commercially available Loopamp Campylobacter detection kit (Eiken Chemical), despite the 95.2% specificity (20/21), the results showed that there was insufficient sensitivity (76.2%, 16/21). In contrast, using the template DNA preparation obtained using 1 ml Bolton enrichment culture, followed by a three-step centrifugation, as described in Materials and Methods, yielded 100% specificity (15/15) and 100% sensitivity (7/7).
Isolation and identification of C. jejuni-C. coli from Preston enrichment samples using the bacterial culture test required more than 4 days. In contrast, the LAMP assay was markedly faster, requiring less than 90 min from the beginning of DNA extraction to final detection and differentiation of C. jejuni and C. coli. In total, the LAMP assay required 23.5 to 25.5 h from the beginning of the enrichment culture to final determination.
DISCUSSION
Here, we compared LAMP assay and conventional culture methods used for the detection and differentiation of C. jejuni and C. coli in chicken meat samples after an enrichment procedure. The results show that the combination of modified DNA extraction procedures and the LAMP assay is markedly simpler and more rapid than conventional culture methods, while the accuracy of the results is maintained. As shown in Tables 2 and 3, a high correlation was observed between LAMP and conventional culture results. These results suggest that the LAMP assay is useful as an adjunct to facilitate early detection of C. jejuni and C. coli in chicken meat samples.
ISO has described a horizontal method for the detection of thermotolerant Campylobacter in food and animal feedstuffs (15). This method uses Bolton enrichment broth, starting with incubation at 37°C for 4 to 6 h and followed by incubation at 41.5°C for another 40 to 48 h. Plating is on mCCDA agar and on blood-based selective medium as a second choice (15). A Bolton enrichment culture requires a long incubation period (1 day), and therefore, in the present study, we used Preston enrichment cultures to obtain the LAMP results within 1 day. In addition, to decrease the cost of broths and to enable rapid and simultaneous testing of a large number of samples, we used 18 ml Preston enrichment broth instead of 225 ml Bolton enrichment broth.
As shown in Table 3, the sensitivities of both Butzler and mCCDA agars suggest that use of a single selective medium is not sufficient to obtain a real positive value for C. jejuni-C. coli in Preston enrichment culture, and ISO recommends the use of two selective media. Further, it was reported previously that the growth of certain C. jejuni and C. coli strains was inhibited on these selective media supplemented with selective ingredients and that there was overgrowth of the background flora of chickens (3, 5, 22, 23); therefore, studies to compare C. jejuni-C. coli isolation using combinations of various enrichment broths and selective media have been performed to obtain the best detection value (1, 2).
Butzler agar is known for its remarkable inhibition of the growth of background flora in chicken meat, as well as of certain C. coli strains (5, 9). As shown in Table 3, five C. coli-positive chicken samples were detected using mCCDA agar, whereas only two samples were detected using Butzler agar, which suggests that Butzler agar inhibits specific C. coli strains. Although mCCDA agar appears to be suitable for C. coli detection, the results showed that the detection value was lower than that with Butzler agar due to the observed overgrowth of the background flora. This result is due to the difference in the selective ingredients of mCCDA and Butzler agars, which include amphotericin B and cefoperazone, whereas the selective ingredients of Butzler agar also include bacitracin, cephazolin, colistin, cycloheximide, and novobiocin (5). These findings suggest that compared to mCCDA agar, Butzler agar has a stronger inhibitory effect on a wide range of background organisms in chicken meat samples and also yields a higher detection value. Further, supplemental use of mCCDA agar appears to facilitate accurate detection of C. jejuni and C. coli in samples.
The amplified LAMP products of the two LAMP-positive, culture-negative samples analyzed by agarose gel electrophoresis showed ladder patterns unique to the LAMP assay (25) (data not shown). Therefore, instead of being caused by a false positive in the LAMP assay, this discrepancy may have been due to a false negative in the culture assay caused by overgrowth on Butzler and mCCDA agars of the normal flora that was present in the chicken samples (5, 9, 29) or was stressed enough that it resisted conventional culture in standard conditions (29). Studies have shown that a number of food components inhibit DNA amplification in LAMP and PCR assays (10, 24). One typical Campylobacter-like colony from the one LAMP-negative, culture-positive chicken thigh sample was confirmed to be C. jejuni by conventional culture and LAMP assays. Therefore, instead of being caused by a false positive in the culture assay, the discrepancy in this single sample may have been due to a false negative in the LAMP assay caused by inhibition of DNA amplification by components found in the chicken thigh sample. Further work is needed to confirm this hypothesis.
As shown in Table 3, the three C. coli and two C. jejuni culture-positive samples were both C. jejuni and C. coli LAMP positive. The sensitivity test with mixtures containing different ratios of C. jejuni and C. coli indicates that this result is due to the small number of subcultures found in the selective agar (n = 3). Further, the five chicken samples were suspected of mixed contamination with both C. jejuni and C. coli, which is potentially revealed using our assay without a requirement for tedious subculture.
Of the 68 culture-positive samples, 3 (2 C. jejuni and 1 C. coli) and 2 (2 C. coli) produced one and two typical Campylobacter-like colonies, respectively, on the two selective media. Two microliters of concentrated template DNA used for the LAMP assay corresponds to approximately 37 μl Preston enrichment culture, and approximately 10 μl of Preston enrichment culture streaked onto each medium corresponds to approximately 3.7 to 7.4 CFU DNA per LAMP reaction test tube for the five samples. The results show that the sensitivities of the LAMP assay for C. jejuni and C. coli were 3.9 to 7.9 CFU and 1.9 to 3.8 CFU per test tube, respectively, which is a sufficient detection level for the five samples.
In the preliminary study, the discrepancy in the results was possibly due to the small number of C. jejuni-C. coli cells in the enrichment culture in previous studies, which was not sufficient for LAMP detection. Given this finding, we used centrifugation to concentrate C. jejuni and C. coli in the enrichment broth. A study by Furuhata et al. (8), which compared a conventional culture method with a commercial LAMP assay kit for the detection of C. jejuni-C. coli, reported sensitivity, specificity, and positive and negative predictive values for the LAMP assay of 96.0% (24/25), 90.8% (99/109), 70.6% (24/34), and 99.0% (99/100), respectively. The correlation between these values and those obtained with conventional culture methods, however, appeared to be insufficient. Further, the LAMP assay kit is unable to differentiate C. jejuni and C. coli, and the primer set sequences are not publicly accessible. Therefore, in the present study, we used published LAMP primer sets for C. jejuni and C. coli, which were developed in our previous study (32).
C. jejuni-C. coli contamination in chicken meat is one of the most important public health hazards (20, 29, 31). The frequent worldwide outbreaks caused by C. jejuni-C. coli highlight the need to control these contaminants in chicken meat. Here, we successfully established a rapid and sensitive assay for detection of C. jejuni and C. coli in naturally contaminated chicken meat samples using a combination of modified DNA extraction procedures and a LAMP assay, while the accuracy of the results was maintained. The LAMP assay is a rapid, sensitive, and practical method that is able to differentiate between these contaminants and may potentially facilitate surveillance for C. jejuni-C. coli contamination in chicken meat, screening of contaminated chicken meat samples before they are consumed, and investigations seeking to determine the causative agents of food poisoning.
Acknowledgments
This work was supported in part by a grant-in-aid from the Daido Life Welfare Foundation.
Footnotes
Published ahead of print on 9 January 2009.
REFERENCES
- 1.Baylis, C. L., S. MacPhee, K. W. Martin, T. J. Humphrey, and R. P. Betts. 2000. Comparison of three enrichment media for the isolation of Campylobacter spp. from foods. J. Appl. Microbiol. 89:884-891. [DOI] [PubMed] [Google Scholar]
- 2.Bolton, F. J., D. Coates, P. M. Hinchliffe, and L. Robertson. 1983. Comparison of selective media for isolation of Campylobacter jejuni/coli. J. Clin. Pathol. 36:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks, B. W., M. M. Garcia, D. E. Fraser, H. Lior, R. B. Stewart, and A. M. Lammerding. 1986. Isolation and characterization of cephalothin-susceptible Campylobacter coli from slaughter cattle. J. Clin. Microbiol. 24:591-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, J. T., J. B. Ochieng, L. Kumar, G. Okoth, R. L. Shapiro, J. G. Wells, M. Bird, C. Bopp, W. Chege, M. E. Beatty, T. Chiller, J. M. Vulule, E. Mintz, and L. Slutsker. 2006. Surveillance for bacterial diarrhea and antimicrobial resistance in rural western Kenya, 1997-2003. Clin. Infect. Dis. 43:393-401. [DOI] [PubMed] [Google Scholar]
- 5.Corry, J. E., D. E. Post, P. Colin, and M. J. Laisney. 1995. Culture media for the isolation of campylobacters. Int. J. Food Microbiol. 26:43-76. [DOI] [PubMed] [Google Scholar]
- 6.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman, C. R., J. Niemann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 8.Furuhata, K., S. Kakimoto, T. Momoda, T. Kozima, M. Ikedo, and M. Fukuyama. 2006. Comparison of the loop-mediated isothermal amplification (LAMP) method and conventional culture method for the detection of Campylobacter species from retail chickens. Jpn. J. Food Microbiol. 23:237-241. (In Japanese.) [Google Scholar]
- 9.Gun-Munro, J., R. P. Rennie, J. H. Thornley, H. L. Richardson, D. Hodge, and J. Lynch. 1987. Laboratory and clinical evaluation of isolation media for Campylobacter jejuni. J. Clin. Microbiol. 25:2274-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara-Kudo, Y., M. Yoshino, T. Kojima, and M. Ikedo. 2005. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253:155-161. [DOI] [PubMed] [Google Scholar]
- 11.Hara-Kudo, Y., J. Nemoto, K. Ohtsuka, Y. Segawa, K. Takatori, T. Kojima, and M. Ikedo. 2007. Sensitive and rapid detection of vero toxin-producing Escherichia coli using loop-mediated isothermal amplification. J. Med. Microbiol. 56:398-406. [DOI] [PubMed] [Google Scholar]
- 12.Hara-Kudo, Y., N. Konishi, K. Ohtsuka, R. Hiramatsu, H. Tanaka, H. Konuma, and K. Takatori. 2008. Detection of verotoxigenic Escherichia coli O157 and O26 in food by plating methods and LAMP method: a collaborative study. Int. J. Food Microbiol. 122:156-161. [DOI] [PubMed] [Google Scholar]
- 13.Harvey, S. M. 1980. Hippurate hydrolysis by Campylobacter fetus. J. Clin. Microbiol. 11:435-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Infectious Agents Surveillance Report. 2006. Campylobacter enteritis in Japan, 1999-2005. IASR 27:167-168. http://idsc.nih.go.jp/iasr/. [Google Scholar]
- 15.International Organization for Standardization. 2006. Microbiology of food and animal feeding stuff—horizontal method for detection and enumeration of Campylobacter spp. Part 1: detection method. ISO 10272-1:2006. International Organization for Standardization, Geneva, Switzerland.
- 16.Iwamoto, T., T. Sonobe, and K. Hayashi. 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs-Reitsma, W., U. Lyhs, and J. Wagenaar. 2008. Campylobacter in the food supply, p. 627-644. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 18.Minami, M., M. Ohta, T. Ohkura, T. Ando, K. Torii, T. Hasegawa, and H. Goto. 2006. Use of a combination of brushing technique and the loop-mediated isothermal amplification method as a novel, rapid, and safe system for detection of Helicobacter pylori. J. Clin. Microbiol. 44:4032-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 20.Nachamkin, I. 2003. Campylobacter and Arcobacter, p. 902-914. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 21.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 22.Ng, L. K., M. E. Stiles, and D. E. Taylor. 1985. Inhibition of Campylobacter coli and Campylobacter jejuni by antibiotics used in selective growth media. J. Clin. Microbiol. 22:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng, L. K., D. E. Taylor, and M. E. Stiles. 1988. Characterization of freshly isolated Campylobacter coli strains and suitability of selective media for their growth. J. Clin. Microbiol. 26:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordstrom, J. L., M. C. Vickery, G. M. Blackstone, S. L. Murray, and A. DePaola. 2007. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl. Environ. Microbiol. 73:5840-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.On, S. L. W. 2005. Taxonomy, phylogeny, and methods for the identification of Campylobacter species, p. 13-42. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 27.Padungtod, P., and J. B. Kaneene. 2005. Campylobacter in food animals and humans in northern Thailand. J. Food Prot. 68:2519-2526. [DOI] [PubMed] [Google Scholar]
- 28.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 29.Sails, A. D., A. J. Fox, F. J. Bolton, D. R. Wareing, and D. L. Greenway. 2003. A real-time PCR assay for the detection of Campylobacter jejuni in foods after enrichment culture. Appl. Environ. Microbiol. 69:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song, T., C. Toma, N. Nakasone, and M. Iwanaga. 2005. Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243:259-263. [DOI] [PubMed] [Google Scholar]
- 31.Waage, A. S., T. Vardund, V. Lund, and G. Kapperud. 1999. Detection of small numbers of Campylobacter jejuni and Campylobacter coli cells in environmental water, sewage, and food samples by a seminested PCR assay. Appl. Environ. Microbiol. 65:1636-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamazaki, W., M. Taguchi, M. Ishibashi, M. Kitazato, M. Nukina, N. Misawa, and K. Inoue. 2008a. Development and evaluation of a loop-mediated isothermal amplification assay for rapid and simple detection of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 57:444-451. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki, W., K. Seto, M. Taguchi, M. Ishibashi, and K. Inoue. 2008b. Sensitive and rapid detection of cholera toxin-producing Vibrio cholerae using a loop-mediated isothermal amplification. BMC Microbiol. 8:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamazaki-Matsune, W., M. Taguchi, K. Seto, R. Kawahara, K. Kawatsu, Y. Kumeda, M. Kitazato, M. Nukina, N. Misawa, and T. Tsukamoto. 2007. Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subsp. hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J. Med. Microbiol. 56:1467-1473. [DOI] [PubMed] [Google Scholar]
- 35.Yano, A., R. Ishimaru, and R. Hujikata. 2007. Rapid and sensitive detection of heat-labile I and heat-stable I enterotoxin genes of enterotoxigenic Escherichia coli by loop-mediated isothermal amplification. J. Microbiol. Methods 68:414-420. [DOI] [PubMed] [Google Scholar]
- 36.Yoda, T., Y. Suzuki, K. Yamazaki, N. Sakon, M. Kanki, I. Aoyama, and T. Tsukamoto. 2007. Evaluation and application of reverse transcription loop-mediated isothermal amplification for detection of noroviruses. J. Med. Virol. 79:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]