Abstract
Enterococci are indicator bacteria used to assess the risk of acquiring enteric disease from swimming in marine waters. Previous work identified beach sands as reservoirs of enterococci which can be transported from the sand to the sea, where they may instigate beach advisories. The present study establishes that naturally occurring enterococci can replicate in beach sands under environmentally relevant conditions. In unseeded, nonsterile microcosm experiments, it was shown that intermittent wetting of sands by seawater, like that which would occur at the high tide line, stimulates the transient replication of enterococci at rates of 0.20 to 0.63 per day (equivalent to doubling times of 1.1 to 3.5 days). Replication was not observed in control microcosms that were not subjected to wetting. Enterococci were enumerated using both culture-dependent (membrane filtration and mEI media) and culture-independent (quantitative PCR [QPCR], 23S rRNA gene based) techniques, which allowed tracking of both culturable and total enterococcus populations. Inhibition of QPCR and DNA extraction efficiencies were accounted for in the interpretation of the QPCR results. The results provide evidence that enterococci may not be an appropriate indicator of enteric disease risk at recreational beaches subject to nonpoint sources of pollution.
Globally, exposure to coastal waters polluted with wastewater causes an estimated 120 million gastrointestinal and 50 million severe respiratory illnesses per year (39). In an effort to reduce recreational waterborne illnesses, beach-monitoring programs have been implemented around the world. These programs use densities of the fecal indicator bacteria Escherichia coli and enterococci to assess the suitability of water for human contact (48). Fecal indicator bacteria are used to evaluate beach water quality because their densities in coastal waters contaminated with wastewater and urban runoff have been linked quantitatively to swimmer illness in epidemiological studies (10, 19, 25, 43). Enterococci are preferred for monitoring marine waters as their densities correlate best to adverse health outcomes (43). Thus, reducing concentrations of enterococci is a goal of total-maximum-daily-load and remediation programs in marine settings.
Wet and dry beach sands in diverse climates and with diverse geographies are reservoirs of enterococci and E. coli (1a, 15, 34, 47, 49), which challenges total-maximum-daily-load and remediation programs, as well as the use of these organisms for assessing risk. The source of E. coli and enterococci in sand is often uncertain. In some cases, these organisms may originate from exogenous sources, such as animal feces, runoff, or spilled sewage. In other cases, where an exogenous source cannot be readily identified, it has been suggested that they may represent indigenous populations adapted to living and growing in beach sands (8, 34). The existence of indigenous populations of E. coli in beach sands has been investigated using multilocus enzyme electrophoresis, multilocus sequence typing, and ribotyping (9, 44), but similar experiments have not been undertaken for enterococci. The growth of E. coli in sands has been investigated using unseeded, unaltered sands (15, 41), unseeded sands with nutritive additives (9), and seeded sands (1, 12, 18, 20, 23, 27). However, investigations of enterococcus growth have been limited. Lee et al. (27) reported growth of seeded American Type Culture Collection (ATCC) strains of Enterococcus faecalis in autoclaved ocean water overlaying commercially available washed sand and autoclaved beach sands. Hartz et al. (20) investigated growth of seeded enterococcal strains in sterile beach sands. Desmarais et al. (15) showed that there was enterococcus growth in unseeded river sediments. Studies of enterococcus growth in beach sands have employed culture-dependent methods to document the change in density once sands are moistened or a nutritive substance is added. One drawback to this technique is that it is not possible to distinguish growth from recovery of unculturable cells once experimental treatments are applied.
Lovers Point in California is a sheltered, tide-dominated beach with elevated levels of enterococci in its beach sands. In a previous study, Yamahara et al. (49) found the highest concentrations of enterococci near the high tide line, in the section of the sand that is intermittently rewetted by the highest high tides (spring tides). In the present study, the hypothesis that enterococci in beach sands replicate during tidal wetting events that mimic the wetting that occurs during the spring tides was tested. This was accomplished using unseeded column microcosms with unaltered beach sands and both culture-dependent and culture-independent measurement techniques. There are limitations to using microcosms to study microbial processes, as natural field conditions (such as temperature, moisture, and concentration of predators) are difficult to maintain. However, microcosms are commonly used for studying microbial growth and transformations in environmental media (15, 24). Using this technique, the present study shows that the intermittent wetting of sands by seawater stimulates the transient replication of enterococci at rates of 0.20 to 0.63 per day (equivalent to doubling times of 1.1 to 3.5 days).
MATERIALS AND METHODS
Environmental sands and water.
Beach sand was collected from Lovers Point in California (36°37′29.88″N, 121°54′59″W) using presterilized 2-liter plastic bags. Sampling was conducted above the high tide line to a depth of 10 cm at 10 locations evenly spaced across the length of the beach. The sand samples were composited to obtain a final volume of approximately 20 liters. The 20-liter sample was then mechanically homogenized using a sterilized stainless steel mixing paddle for 20 min, followed by hand folding with a sterile stainless steel spoon for 10 min. This cycle was repeated four times. Sand characteristics were described previously by Yamahara et al. (49).
Seawater was collected from the surf zone at Lovers Point in acid-washed, triple-rinsed 20-liter low-density polyethylene containers on two occasions during the course of the experiment, just prior to watering simulations. Water samples were transported on ice to the laboratory, filter sterilized using a 0.45-μm polyethersulfone membrane (Nalgene, Rochester, NY), stored at 4°C, and used in the experiment within 5 days. Nutrient, dissolved organic carbon, and culturable and total enterococcus concentrations in filtered seawater were measured.
Column design.
To mimic a vertical section of sand at the beach, acrylic columns were constructed to contain sand microcosms. Each column consisted of an inner column and an outer column, which fit tightly together, and a small “catch basin” at the bottom to collect the leachate during watering simulations. The inner column consisted of three separable sections (“top,” “middle,” and “bottom”); each of the sections was 5.7 cm in diameter and 5.1 cm long. The outer column held the three inner column sections together and was 6.4 cm in diameter and 20.3 cm long. Stainless steel mesh (mesh size, 0.0625 mm) was used at both the top and the bottom of the column to retain the sand but allow water to flow into the catch basin. The total mass of sand in each column was 610 g (dry weight).
Experimental design.
Fifty columns were packed with homogenized sand. During the packing process, 50 ml of sand was set aside after every two or three columns were packed (to obtain a total of 13 samples) to determine the initial enterococcus concentration.
All columns were placed at 20°C in the dark for up to 45 days. Thirty of the 50 columns were designated treatment columns, and 20 columns were designated control columns. Treatment columns were subjected to intermittent wetting with 250 ml of filtered seawater and draining to simulate the natural wetting and draining process that occurs in the upper reaches of the beach during the highest spring tides. Here this process is referred to as “watering.” Watering was performed on days when the tides at Lovers Point exceeded 1.8 m (relative to mean sea level) in October and November 2007 (http://tbone.biol.sc.edu/tide). This threshold was chosen arbitrarily and is at the upper reaches of the tidal excursion. The tides exceeded this threshold on days 6, 7, and 8 and then on days 34, 35, 36, 37, and 38. These two sets of consecutive days are referred to below as the first and second watering simulations, respectively. Water drained from the columns under the influence of gravity, and the leachate was collected in column catch basins for analysis.
Control and treatment columns were sacrificed daily from day 1 through day 4. Thereafter, for the remaining 41 days of the experiment, treatment columns were sacrificed approximately every other day and control columns were sacrificed every third day. On days when watering was performed, treatment columns were sacrificed before watering. Column sacrificing consisted of removing the inner column from the outer column and separating the three inner column sections with a sterile stainless steel knife. Each column section was homogenized by hand mixing with a sterile spatula for 10 min. From each column section the following sand samples were collected: (i) triplicate 15- to 20-ml samples for elution in MilliQ water and enumeration on mEI solid media using membrane filtration, (ii) one 15-ml sample for direct extraction of DNA from sand, and (iii) one 15-ml sample for measuring the moisture content and total organic carbon. All elutions and filtrations were performed immediately after a column was sacrificed. Sand samples used for DNA extraction were stored at −80°C. Sand archived for moisture and total organic carbon content analyses was sealed with Parafilm in 15-ml polypropylene centrifuge tubes and stored at 20°C until analysis.
Nutrient, carbon, and moisture analyses.
Thirty milliliters of the filtered seawater used for wetting the columns was collected on the first 3 days of each watering simulation and stored at −20°C in clear glass scintillation vials (for dissolved organic carbon analyses) and brown high-density polyethylene plastic bottles (for nutrient analyses). The dissolved organic carbon content was determined using the nonpurgeable organic carbon method (4) and a total organic carbon analyzer (Shimadzu, Columbia, MD). The concentrations of orthophosphate, nitrate, nitrite, and ammonia were measured by standard methods (3) with a nutrient autoanalyzer (QuikChem 8000; Lachat, Loveland, CO).
The sand moisture contents for the top, middle, and bottom sections of each sacrificed column were determined by drying preweighed sand at 110°C for 24 h. It was assumed that the 10-min homogenization process and weighing had negligible effects on the moisture content. Moisture content was used to normalize all enterococcus concentrations to grams (dry weight) of sand. The total organic carbon of triplicate sand samples (representing composites of equal masses of dry sand samples from the bottom, middle, and top sections) from treatment columns on days 0, 16, and 45 was measured using the loss-on-ignition method (26).
Enumeration of cENT.
For day 0, each of 13 sand samples set aside during column packing was analyzed to determine the initial concentration of culturable enterococci (cENT). Thereafter, cENT were enumerated in triplicate sand samples from each section (top, middle, and bottom) of the sacrificed column. Enumeration was accomplished by elution in MilliQ water and membrane filtration onto mEI selective agar (EMD Chemicals, Gibbstown, NJ) (42) as described by Yamahara et al. (49). Colonies exhibiting typical enterococcus morphology were counted at 48 h. An extra 24 h was allowed for incubation because injured environmental bacteria may take longer to adapt and grow in high-nutrient conditions like those found in solid media (38). MilliQ water was membrane filtered and transferred onto mEI media each day to check the sterility of the filtration apparatus and the MilliQ rinse water. Colony counts were normalized to sand dry weights.
cENT in filtered seawater used to wet treatment columns and in treatment column leachates were enumerated using EPA method 1600 with 48 h of incubation. Duplicate 100-ml filtered seawater samples were membrane filtered before the onset of each watering simulation to enumerate the input of cENT into the columns. On day 6, the onset of the first watering simulation, 25 to 50 ml of leachate from each column (n = 46) was analyzed for cENT by membrane filtration. Thereafter, depending on the number of treatment columns remaining, leachates from three or four columns were combined and analyzed in duplicate for cENT.
Ten colonies isolated from mEI plates were picked for genus confirmation using Enterococcus-specific 16S rRNA gene primers and PCR cycling conditions described by Deasy et al. (13). The isolates were recovered from plates produced over the course of the experiment from both sands and leachates. Confirmation was accomplished by amplification of a 733-bp product that was visualized in a 1.5% agarose gel stained with ethidium bromide using a Gel Doc XR imager (Bio-Rad, Hercules, CA).
Extraction of DNA.
For day 0, 5-ml portions of 13 homogenized sand samples set aside during column packing were composited, and DNA was extracted from a ∼0.5-g subsample. Thereafter, equal wet weights of homogenized sand from the top, middle, and bottom sections of sacrificed columns were composited (∼0.5 g). DNA was extracted directly from sands using a FastDNA Spin kit for soil and a FastPrep instrument (MP Biomedicals, Santa Ana, CA) according to the manufacturer's protocols. One drawback to using small masses for DNA extraction is the potential for small-scale heterogeneities in the distribution of the target that affect the results. To quantify variation within columns, DNA was extracted from triplicate ∼0.5-g composite samples from six columns (treatment columns from days 19, 28, and 37 and control columns from days 20, 33, and 37).
DNA from filtered seawater used for wetting treatment columns and treatment column leachate was obtained by filtering water samples (200 ml for filtered seawater and 20 ml for leachate) onto 47-mm, 0.2-μm polycarbonate filters (GE Osmonics, Minnetonka, MN). DNA from water used for wetting was obtained on each watering day except days 7 and 38. For the leachates, the following sampling scheme was used. On day 6, the first watering day, leachates from 3 of the 46 treatment columns were processed for DNA extraction. On all other watering days, leachates from three or four columns were pooled to obtain composite samples, and two of these composite leachate samples were processed for DNA extraction. Filters were folded into FastDNA Spin kit for soil lysing matrix tubes and processed as described above.
Enumeration of tENT by QPCR.
Quantitative PCR (QPCR) analyses were performed using a Taqman assay targeting a portion of the 23S rRNA gene. PCR primer and hybridization probe sequences for total Enterococcus (tENT) are described by Ludwig and Schleifer (28) and Haugland et al. (21). Two microliters of DNA was used as the template in 25-μl reaction mixtures with the following composition: 1× Taqman universal master mix (Applied Biosystems, Foster City, CA), 1 μM forward primer ECST784F, 1 μM reverse primer ENC854R, 0.40 μM Taqman probe GPL813TQ (with 6-carboxytetramethylrhodamine and 6-carboxyfluorescein), and 0.2% bovine serum albumin fraction V (GIBCO, Carlsbad, CA). QPCR was carried out with a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA) in 96-well plates with the following thermal cycling conditions: 2 min at 50°C and 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. Fluorescence and cycle thresholds were automatically generated by the instrument.
Standards were generated using whole genomic DNA from Enterococcus faecium ATCC 19434 extracted using a QIAamp DNA minikit (Qiagen, Valencia, CA). DNA was quantified using a NanoDrop ND-1000 spectrophotometer (Wilmington, DE), and the mass of DNA per volume was converted to 23S rRNA copy number per volume by assuming that there were six rRNA gene operons per genome and that the genome size was 2,600 kb (32). The standards consisted of decimal dilutions ranging from 4.2 × 101 to 4.2 × 106 copies per reaction mixture. Standards were run in triplicate on each QPCR plate to create standard curves (see Fig. S1 in the supplemental material). On average, the efficiency of the QPCR was 93.5%. Enterococcus concentrations measured by QPCR are reported below in cell equivalents (CE). CE were calculated from copy numbers by assuming that there were six rRNA gene operons per cell (32).
Unknowns were run in triplicate. Two additional reaction mixtures for unknowns were spiked with a known copy of DNA target prior to QPCR to account for inhibition. The spiked reaction mixtures consisted of 1 μl of unknown sample DNA and 1 μl of a preparation containing ∼2.1 × 103 copies/μl target DNA. The exact copy number of the spike (S) was determined on each QPCR plate by running the spike in triplicate. Inhibition was determined by comparing the averages of the machine-reported copy numbers in the spiked reactions (Ya) and unspiked reactions (Xa). The true copy number (X) for each unknown reaction, accounting for inhibition, is given by X = XaS/(Ya− Xa).
The machine-reported standard deviations were scaled according to the same factor, S/(Ya − Xa). This formulation assumes that inhibition is independent of copy number. It should be noted that if Ya is orders of magnitudes larger than Xa, then it is hard to detect inhibition, so S must be chosen carefully. Here S was chosen so that Ya was close to the same order of magnitude as most of the Xa values. The inhibition factor (IF) was calculated as follows: IF = Xa/X. IF varies from 0 (complete inhibition) to 1 (no inhibition).
DNA extraction efficiencies were determined for each sample matrix and DNA extraction method. One milliliter of a 105 dilution of stationary-phase E. faecium ATCC 19434 was spiked (i) into 15 to 20 ml of sand for direct DNA extraction from sand and (ii) directly onto 0.2-μm polycarbonate filters. DNA extraction from sand and filters was performed as described above. QPCR was used to determine the concentration of tENT in DNA extracts from unspiked sand, spiked sand, spiked filters, and 1 ml of inoculum. The number of bacteria in the inoculum was also determined by direct cell counting using epifluorescence microscopy with SYBR green I dye (31). The efficiency of DNA extraction (η) was defined as the copy number detected (after correction for inhibition) divided by the copy number spiked into the matrix. Since the matrices from which DNA was extracted were relatively consistent throughout the experiments, it was assumed that η was constant throughout the experiment for a particular DNA extraction method. η was accounted for in all the reported tENT concentrations. The corrected copy number (Xcorrected) is given by Xcorrected = X/η.
Cloning and sequencing.
QPCR amplicons produced from treatment column sands on days 28 and 32 were chosen for cloning and sequencing to confirm that there was amplification of the intended target. Triplicate 25-μl reaction mixtures were pooled for each sample and purified using a MinElute PCR purification kit (Qiagen, Valencia, CA). A TOPO-TA cloning kit for sequencing using the pCR 4.0 vector and Mach competent cells was used according to the manufacturer's protocols (Invitrogen, Carlsbad, CA). Insert-bearing white clones were randomly selected and screened with conventional PCR for inserts using the M13 reverse and T7 forward vector primers (Invitrogen, Carlsbad, CA). Eighteen clones were randomly selected and sequenced with an ABI 3730xl sequencer (Elim Biopharmaceuticals, Inc., Hayward, CA). The resulting sequences were trimmed, edited, and aligned using Sequencher v4.8 (Gene Codes, Ann Arbor, MI) and then compared against the nonredundant GenBank sequence library.
Statistical analyses.
Statistical analyses were performed using Matlab 7.1.0.183 (R14) (Mathworks, Natwick, MA), SPSS v. 11 (SPSS, Chicago, IL), and Igor Pro (Wavemetrics, Lake Oswego, OR). Least-squares curve fits were used to estimate apparent growth rates (kg) and death rates (kd) with units of day−1 from slopes of natural log-transformed concentrations and time (11). A t test was used to test if the slope was significantly different from 0 (40). Slopes were compared using a t test as described by Neter et al. (30). The normality of data groups was determined using the Kolmogorov-Smirnov test. If variables were not normally distributed, they were log transformed to achieve normality prior to analyses. Comparisons between groups and samples were conducted using one-way analysis of variance and t tests; F and t statistics are reported below. It was assumed that statistical results were significant if the P value was <0.05. All errors reported below represent 95% confidence intervals. For log-normal variables, 95% confidence ranges are reported rather than errors for ease of interpretation. Sums of log-normal variables and their associated uncertainties were estimated using Monte Carlo simulations and are reported as medians and 95% reference ranges (defined as the 2.5th and 97.5th percentiles) (5).
Nucleotide sequence accession numbers.
Sequences obtained in this study have been deposited in the GenBank database under accession numbers FJ347143 to FJ347160.
RESULTS
Moisture, carbon, and nutrients.
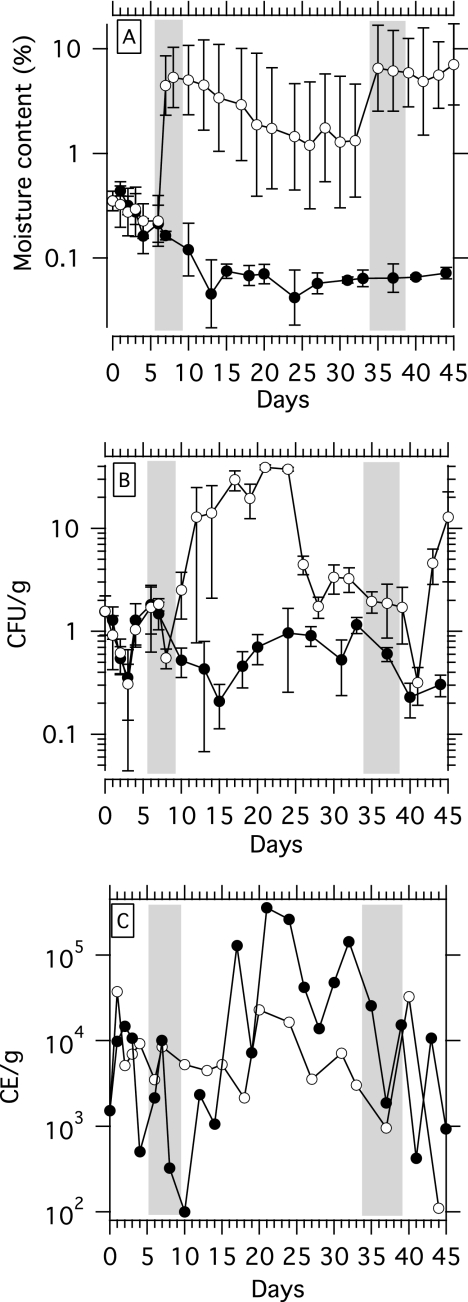
The control columns exhibited desiccation throughout the experiment, while the treatment columns did not. The initial moisture content of the sand was 0.35% (geometric mean for the top, middle, and bottom sections). In the control columns, which received no water addition, the moisture content decreased over the experiment to 0.07% on the final day (Fig. 1A). The treatment columns also exhibited desiccation during days 0 through 6, prior to the first watering simulation. After the first watering, the mean moisture content in treatment columns increased to 5.00% and remained above 1.98% for the remainder of the experiment.
FIG. 1.
Times series for moisture content (A), cENT concentration (B), and tENT concentration (C) in treatment (open circles) and control (filled circles) columns. Shading indicates days when treatment columns were wetted with filtered seawater. All enterococcal concentrations are normalized to the dry weight of sand. The error bars in panels A and B indicate 95% confidence intervals. In panel C, error bars are not shown; however, the error for each tENT data point is ±0.9 log unit.
The organic carbon content in treatment sands did not change significantly over the duration of the experiment (F = 0.18, df = 2, P = 0.84). The organic carbon contents in treatment columns on days 0, 16, and 45 were determined to be 0.40% ± 0.05%, 0.39% ± 0.03%, and 0.38% ± 0.06%, respectively (n = 3 for each measurement).
The filtered seawater used to wet the treatment columns contained 3.3 ± 0.9 ppm dissolved organic carbon (n = 6). As a total of 2,000 ml filtered seawater was applied to the treatment columns over the course of the experiment, approximately 7 mg dissolved organic carbon was added. The orthophosphate, ammonium, nitrate, and nitrite concentrations in the filtered seawater were 0.91 ± 0.05 μM, 2.75 ± 0.24 μM, 4.56 ± 0.21 μM, and 0.65 ± 0.17 μM (n = 6 for each nutrient), respectively. Over the course of the experiment 0.2 mg N and 0.05 mg P were applied to the treatment columns.
cENT.
The detection limits for cENT were 0.01 CFU/ml for filtered seawater, ∼0.03 CFU/ml for leachate, and ∼0.06 CFU/g for eluted sands. All method blanks indicated that sterility was maintained throughout the experiment, and no cENT were detected in the filtered seawater used to water the treatment columns.
Before the first watering simulation, the cENT concentrations in the treatment and control columns were the same. The cENT concentration on day 0 was 1.6 ± 0.6 CFU/g, which is on par with values previously reported for Lovers Point (49). The error reported for cENT represents both the analytical error and the variation within the column between the top, middle, and bottom sections. The latter is especially important after watering simulations were applied since they redistributed the target within the columns. From days 0 to 6, before the first treatment, the cENT concentrations in the control and treatment columns were not significantly different (t = 1.590, df = 5, P = 0.17) (Fig. 1B). The cENT concentration decreased on days 0 to 3 in both the treatment and control columns (kd = −0.53 day−1, t = −12.774, df = 6, P = 10−5) This was coincident with a decrease in the moisture content from 0.35% to 0.30% (Fig. 1A).
cENT exhibited significant growth in treatment columns between days 10 and 21 after the first watering simulation. The concentration increased from 2.53 CFU/g to 39.13 CFU/g at a rate (kg) of 0.20 day−1 (t = 3.466, df = 4, P = 0.026). After day 21, the cENT concentration in the treatment decreased through day 41. This period of time encompassed the second watering simulation, which occurred between days 34 and 38. The kd between days 21 and 32 was −0.28 day−1 (t = −2.93, df = 4, P = 0.04). After the second watering simulation, between days 39 and 41, there was a significant decrease in the cENT concentration (t = 2.821, df = 16, P = 0.012). The reason for this decrease is unknown, but it could have been caused by increased grazing, loss of culturability, or death. After day 41, there was a rebound in the cENT concentration in the treatment columns. The kg between days 41 and 45 was 0.92 day−1, but the increasing trend did not achieve statistical significance (t = 3.916, df = 1, P = 0.16).
The watering simulations removed a fraction of sand-associated cENT from the column via the wetting and draining process (Table 1). During the first watering simulation, a total of 3.3 ± 2.18 CFU/g was recovered in the leachate and thus removed from the column by draining seawater over the 3 days of watering. During the second watering simulation, a total of 2.3 ± 0.5 CFU/g was recovered in the leachate over the entire 5 days of watering.
TABLE 1.
Enterococcal concentrations in leachates on each watering day
| Watering day | Vol of leachate collected in catch basin (ml) | cENT concn in leachate (CFU/ml)a | Normalized cENT concn in leachate (CFU/g)a,b | No. of leachate samples analyzed for cENT | tENT concn in leachate (CE/ml)c | Normalized tENT concn in leachate (CE/g)b,c | No. of leachate samples analyzed for tENT |
|---|---|---|---|---|---|---|---|
| 6 | 204 | 0.73 ± 0.45 | 0.24 ± 0.15 | 46 | 240 (120-460) | 79 (41-150) | 4 |
| 7 | 236 | 7.17 ± 3.51 | 2.78 ± 1.36 | 8 | 27 (0.50-1,400) | 10 (0.20-540) | 2 |
| 8 | 239 | 0.92 ± 1.43 | 0.36 ± 0.56 | 8 | 48 (1.8-1,300) | 19 (0.71-500) | 2 |
| 34 | 223 | 0.17 ± 0.06 | 0.06 ± 0.02 | 8 | 120 (0.5-27,000) | 42 (0.2-9,800) | 2 |
| 35 | 237 | 4.72 ± 0.79 | 1.83 ± 0.31 | 6 | 250 (13-5,100) | 98 (4.9-2,000) | 2 |
| 36 | 242 | 0.91 ± 0.5 | 0.36 ± 0.2 | 6 | 18 (14-24) | 7.3 (5.5-9.5) | 2 |
| 37 | 238 | 0.12 ± 0.04 | 0.05 ± 0.02 | 6 | 5.4 (0.45-64) | 2.1 (0.17-25) | 2 |
| 38 | 243 | 0.03 ± 0.02 | 0.01 ± 0.01 | 6 | 5.3 (2.0-14) | 2.1 (0.81-5.6) | 2 |
Means with 95% confidence intervals.
The concentrations were normalized by the mass of sand in the column.
Means (95% confidence intervals).
cENT persisted in the control columns, but the concentrations did not increase as they did in the treatment columns. The average cENT concentration in the controls was 0.74 ± 0.22 CFU/g. There was a slight decreasing trend in densities over the course of the experiment (kd = −0.02 day−1), but the trend is not statistically significant (t = −1.928, df = 17, P = 0.071).
Ten isolates representing colonies of different sizes and shapes that were counted as cENT on mEI agar were tested using molecular methods to confirm that they were Enterococcus isolates. All of the tested isolates were confirmed to be Enterococcus isolates based on the presence of the intended PCR product.
tENT.
In order to report quantitative tENT concentrations, matrix η and IF were used to calculate corrected copy numbers. η for seeded sand was 89%, and η for filters spiked with enterococci was 83%. IF varied between samples and ranged from 0.081 to 1.0 (average, 0.53) for DNA extracted directly from sands and from 0.62 to 1.0 (average, 0.97) for DNA extracted from filters. The detection limits for tENT were ∼33 CE/g in sand, 0.83 CE/ml in leachate, and 0.083 CE/ml in filtered seawater. All method blanks, including no-template controls, indicated that there was no cross contamination during QPCR.
The filtered seawater used for wetting treatment columns had tENT concentrations ranging from 2.3 to 8.1 CE/ml (average, 3.8 ± 1.7 CE/ml; n = 6). This likely represented extracellular enterococcus DNA and unculturable or dead enterococci with diameters less than 0.45 μm that passed through the filter used to sterilize the water. Given that 250 ml of this solution was applied during each watering simulation, approximately 1.6 ± 0.7 CE/g was added to the column during each watering simulation. This quantity is insignificant given the concentrations of tENT observed in the columns, as described below.
The time series for tENT in the treatment and control columns are shown in Fig. 1C. Each data point represents the tENT concentration obtained using DNA extracted from a single 0.5-g sand sample, except for the treatment columns on days 19, 28, and 37 and the control columns on days 20, 33, and 37, for which triplicate sand samples were analyzed and the geometric mean values for tENT from the triplicate samples are reported. The 95% confidence intervals associated with these measurements, representing within-column variation, were assumed to be the same for each data point: ±0.9 log unit about the measured concentration. This value was calculated by comparing tENT concentrations for triplicate sand samples from six different columns. Because only ∼0.5 g (<0.1% of the column mass) was used for measuring tENT, this amount of within-column variation was not unexpected, as small-scale heterogeneities were likely to exist within the sand matrix. Similar variation in QPCR targets has been reported previously for ammonia-oxidizing bacteria and archaea in beach sands (36). The analytical errors associated with the QPCR measurements of the sand samples were insignificant compared to the within-column variation. On average, the analytical error, expressed as 95% confidence intervals, was 25% of the machine-derived tENT concentrations. Therefore, analytical errors are not discussed further here; all reported errors represent within-column variation.
The trends in tENT in the treatment and control columns match the trends described above for cENT, with the tENT concentration increasing after watering simulations in treatment columns and relatively constant in control columns (Fig. 1C). tENT concentrations were higher than cENT concentrations by 2 to 5 orders of magnitude. The initial tENT concentration was 1,500 CE/g (95% confidence interval, 190 to 1.2 × 104 CE/g). For days 0 through 6, prior to the first treatment, the control and treatment concentrations were not significantly different (t = 0.76, df = 5, P = 0.48).
The first watering simulation removed tENT from the treatment column via the wetting and draining process (Table 1). The sum of the tENT concentrations in leachate recovered on days 6, 7, and 8 was 150 CE/g (median; 95% reference range, 54 to 2,800 CE/g). After the first watering simulation, the tENT concentration increased in the treatment columns. Between days 10 and 21, kg was 0.63 day−1 (t = 3.50, df = 4, P = 0.02). After day 21, there was a declining trend in the tENT concentration (kd = −0.15), but unlike the trend during this period for cENT, the trend did not achieve statistical significance (t = −1.12, df = 4, P = 0.32).
During the second watering simulation (days 34 to 38), a total of 510 CE/g tENT (95% reference range, 28 to 1.2× 105 CE/g tENT) was recovered over the 5 days of watering. After the second watering simulation, the concentrations in the treatment columns were variable. When tENT measured on days 39 through 45 was used, an overall decrease was evident (kd = −0.26 day−1), but the trend is not statistically significant (t = −0.57, df = 2, P = 0.63).
In the control columns, no significant trends in the tENT concentration were observed over the course of study (kd = −0.031, t = −1.437, df = 17, P = 0.169), and the tENT concentrations throughout the 45 days did not deviate from ∼104 CE/g.
To ensure that the Taqman QPCR assay was specific for the target, PCR amplicons from days 28 and 32 were cloned. Eighteen clones were chosen randomly for sequencing. All of the sequences exhibited a 100% match to Enterococcus.
DISCUSSION
Growth of enterococci.
Growth of cENT and tENT was observed when there was an increase in the moisture content of unseeded, unaltered, nonsterile sands from Lovers Point in California (Fig. 1). Constant concentrations of cENT and tENT were maintained in the control columns over the course of the study, while the cENT and tENT concentrations increased in the treatment columns at rates of 0.20 day−1 (cENT) and 0.63 day−1 (tENT) after addition of seawater to the columns. The growth rates indicate that the doubling times were 3.5 days (cENT) and 1.1 days (tENT). While the growth rate of tENT was higher than that of cENT, the rates are not significantly different (t = 2.287, P = 0.052) (30).
Statistically significant increases in the cENT and tENT concentrations were not observed after the second watering simulation. One explanation for this is that the experiment was terminated too early to observe a second phase of growth. Only 3 days of data collection took place for the treatment columns after the second watering simulation. It would have been interesting to extend the experiment through another wetting event.
The growth rates measured for cENT and tENT are similar to those reported for marine bacterial communities and a single previously reported cENT growth rate. Moriarity (29) measured growth rates of bacterial communities in California marine waters that were between 0.2 and 2.3 day−1, and Karl and Novitsky (24) report growth rates for bacteria in surface marine sediments that were between 0.18 and 0.64 day−1. Results of several previous studies have suggested that enterococci can replicate in the environment, particularly in beach sands and sediments, as determined by culture-dependent methods (15, 20, 27). One of these studies (20) reported that the cENT growth rates were between 0.7 and 1.6 day−1 for sterile sands seeded with pure enterococcus cultures. The growth rates reported in the present study are close to these rates despite the different experimental conditions.
The organic carbon present in sand appears to be more than sufficient to support the number of tENT observed. Based on population size equations of Canfield et al. (11) and using molar growth yields of aerobically grown fecal streptococci determined by Brown and Collins (7), it is estimated that a 106-CE/g population of tENT requires less than 0.01% of the total organic carbon in the sands. This calculation assumes that the molar growth yield of streptococci is an appropriate approximation of the molar growth yield of environmental enterococci (for which a molar growth rate is not readily available). Considering that these genera have similar metabolisms, this assumption should yield a reasonable approximation.
Previous researchers have reported increases in cENT concentrations in beach sands subjected to tidal inundation and attributed the growth to an increase in moisture content (15). While growth was initiated in the experiments due to an increase in moisture content, growth ceased in response to a factor not related to moisture content. The moisture content in the treatment columns remained elevated throughout the experiment after the first watering simulation, yet growth ceased after day 21 (Fig. 1B and C). The organic carbon content in the sands did not change over the course of the experiment (it remained 0.39%), suggesting that this nutrient was not limiting. It is possible that dissolved organic carbon or another trace nutrient introduced with the seawater was necessary for the growth of enterococci and that growth ceased when this nutrient was depleted. The seawater at Lovers Point is rich in organic compounds and nutrients due to a large nearby kelp forest. Indeed, enterococci lack many biosynthetic capabilities and thus have multiple trace vitamin requirements (16). Another possibility is that a buildup of waste products from enterococci or other microorganisms inhibited growth.
The cENT and tENT trends differed after day 21, when growth ceased. The cENT concentrations exhibited a significant decline (kd = −0.28 day−1) from day 21 to day 32. However, the tENT concentrations remained relatively constant and exhibited no statistically significant decline. The difference in behavior of these two populations is consistent with a loss of culturability or death of readily eluted, loosely bound enterococci and persistence of viable but nonculturable enterococci, tightly bound enterococci (not removed by elution in MilliQ water), and extracellular enterococcus DNA in the columns.
Comparison of cENT and tENT.
The ratio of cENT to tENT ranged from 7 × 10−6 to 3 × 10−2 with a median of 2 × 10−4 in sands and from 1 × 10−3 to 3 × 10−1 with a median of 2 × 10−2 in leachate. The higher concentration of tENT than of cENT is not surprising because QPCR detects viable but nonculturable and dead cells, which cannot not be detected by culture-dependent methods.
The cENT/tENT ratios reported in the present study for sands and leachate are lower than and within in the range of, respectively, previously reported cENT/tENT ratios. Haugland et al. (21) reported cENT/tENT ratios of 0.06 and 0.17 for freshwater at West Beach and Huntington Beach on the Great Lakes, respectively. He and Jiang (22) reported cENT/tENT ratios of 0.003, 0.081, and 0.20 for estuarine and creek waters at Middle Newport Bay, Lower Newport Bay, and San Diego Creek, respectively, in southern California. The relatively high numbers of tENT relative to cENT in sands could be explained in part by PCR targets that were present in the sand matrix as extracellular DNA and tightly bound enterococci. Because DNA was extracted directly from sands, these targets contributed to the tENT signal. In contrast, cENT from sand was enumerated by elution in MilliQ water, so only loosely bound enterococci contributed to the cENT signal.
Enterococci in leachate.
During both watering simulations, cENT and tENT were removed from the columns with draining water (Table 1). These targets could have been present in thin films on the sand grains or weakly attached to sand grains. Mobilization of bacteria in partially saturated sands can occur via thin-film expansion release, air-water interface scouring, and shear mobilization (14). The cENT/tENT ratio in leachate was generally higher than that in the sand. This could be explained by differential mobilization of cENT and tENT. It may also have been a result of the leachate enumeration method; leachate was filtered and DNA was extracted from the filter, which would have excluded extracellular DNA. Further work needs to be done to confirm the mechanisms by which loosely and tightly bound enterococci and extracellular enterococcus DNA are mobilized in sands.
The cENT and tENT concentrations were sufficiently elevated in leachates that they could represent nonpoint sources of enterococci for coastal waters. This is particularly interesting in light of the finding that the leachate removed only fractions of enterococci from the sand, leaving sufficient cells behind to grow. The cycle of wetting and drying at beaches may move enterococci from sand to coastal water or groundwater (49) and promote the growth of enterococci that are left behind. This is consistent with findings that certain reaches of a beach (those remaining dry for a sufficient time between wetting events) harbor elevated enterococcus concentrations (37, 49). To fully understand the impact of the transient replication and die-off in beach sands and leaching from sands on enterococcal densities in coastal waters, particularly in relation to other potential sources, a mass balance model of enterococci in sand and water needs to be developed. This is an interesting area for further research.
Limitations of the experiments.
Caution should be taken in extending results from microcosms to the field. In the experiments, natural populations of organisms may have been affected by mixing of sands and storage in the dark at a constant temperature. In the field, it is likely that drying of the sand column after wetting would occur more quickly than it did in the present experiments due to winds and heating from the sun. However, these experiments are more representative of field conditions than previous experiments investigating enterococcus growth in beach sands because the sands were not sterilized prior to the experiment and populations of enterococci naturally present in the sands were tracked.
The growth rates reported here are apparent growth rates because growth, grazing, and death were simultaneously occurring in the experiments. If true growth rates were desired, a culture-independent technique, such as bromodeoxyuridine DNA labeling and immunocapture followed by PCR, could be used (2, 45).
Implications.
Naturally present populations of enterococci can grow in marine beach sands subjected to intermittent tidal wetting. Enterococci are the preferred indicator for monitoring recreational marine beaches. However, the results suggest that enterococci in beach sands violate an important criterion of indicator organisms; namely, indicator organisms should not multiply in the environment. The search for alternate indicators that do not multiply in the environment should continue.
Our findings lead to the question of whether pathogenic bacteria are also capable of multiplying in beach sands. Isolation of human bacterial pathogens, including Campylobacter, Vibrio spp., Salmonella, and Pseudomonas aeruginosa (6, 17, 33, 35), from beach sand has been reported. Further research needs to be conducted to determine if pathogenic bacteria can grow in beach sands.
Sand leachate represents a nonpoint source of cENT and tENT for coastal waters and potentially contributes to beach advisories and closures. In California, the single-sample standard for cENT in swimming waters is 1.04 CFU/ml. cENT concentrations in some of the treatment column leachates exceeded the single-sample standard, suggesting that these organisms may contribute to impairment of bodies of water. There are no standards for tENT concentrations in marine waters; however, the tENT concentrations in leachates were quite elevated (Table 1). The total maximum daily loads and remediation plans developed for marine beaches need to consider contributions of enterococci from beach sands.
Supplementary Material
Acknowledgments
This work was supported by NSF CAREER award BES-0641406 to A.B.B.
Daniel Keymer, Alyson Santoro, Nicholas De Sieyes, Francisco Tomayo, and Tim Julian assisted with the work and/or provided suggestions for improving the manuscript. Comments from three anonymous reviewers improved the manuscript.
Footnotes
Published ahead of print on 16 January 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alm, E. W., J. Burke, and E. Hagan. 2006. Persistence and potential growth of the fecal indicator bacteria, Escherichia coli, in shoreline sand at Lake Huron. J. Great Lakes Res. 32:401-405. [Google Scholar]
- 1a.Alm, E. W., J. Burke, and A. Spain. 2003. Fecal indicator bacteria are abundant in wet sand at freshwater beaches. Water Res. 37:3978-3982. [DOI] [PubMed] [Google Scholar]
- 2.Artursson, V., and J. K. Jansson. 2003. Use of bromodeoxyuridine immunocapture to identify active bacteria associated with arbuscular mycorrhizal hyphae. Appl. Environ. Microbiol. 69:6208-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlas, E. L., S. W. Hager, L. I. Gordon, and P. K. Park. 1971. A practical manual for the use of the technicon autoanalyzer in seawater nutrient analysis (revised). Technical report 215. Department of Oceanography, Oregon State University, Corvallis.
- 4.Bird, S. M., M. S. Fram, and K. L. Crepeau. 2003. Determination of dissolved organic carbon in water by high temperature catalytic oxidation, method validation, and quality-control practices. Technical Report Open File Report 03-366. U.S. Geological Survey, Sacramento, CA.
- 5.Bland, M. 2000. An introduction to medical statistics, vol. 9. Oxford University Press, New York, NY.
- 6.Bolton, F., S. B. Surman, K. Martin, D. R. Wareing, and T. J. Humphrey. 1999. Presence of Campylobacter and Salmonella in sand from bathing beaches. Epidemiol. Infect. 122:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, W. V., and E. B. Collins. 1977. End products and fermentation balances for lactic streptococci grown aerobically on low concentrations of glucose. Appl. Environ. Microbiol. 33:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 9.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, W. T. E. Ting, C. C. Tsen, and M. B. Nevers. 2006. Seasonal persistence and population characteristics of Escherichia coli and enterococci in deep backshore sand of two freshwater beaches. J. Water Health 4:313-320. [DOI] [PubMed] [Google Scholar]
- 10.Cabelli, V. J., A. P. Dufour, L. J. McCabe, and M. A. Levin. 1982. Swimming-associated gastroenteritis and water quality. Am. J. Epidemiol. 115:606-616. [DOI] [PubMed] [Google Scholar]
- 11.Canfield, D. E., E. Kristensen, and B. Thamdrup. 2005. Aquatic geomicrobiology, vol. 48. Elsevier, Burlington, MA. [DOI] [PubMed]
- 12.Davies, C. M., J. A. H. Long, M. Donald, and N. J. Ashbolt. 1995. Survival of fecal microorganisms in marine and freshwater sediments. Appl. Environ. Microbiol. 61:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deasy, B. M., M. C. Rea, G. F. Fitzgerald, T. M. Cogan, and T. P. Beresford. 2000. A rapid PCR based method to distinguish between Lactococcus and Enterococcus. Syst. Appl. Microbiol. 23:510-522. [DOI] [PubMed] [Google Scholar]
- 14.DeNovio, N. M., J. E. Saiers, and J. N. Ryan. 2004. Colloid movement in unsaturated porous media: recent advances and future directions. Vadose Zone J. 3:338-351. [Google Scholar]
- 15.Desmarais, T. R., H. M. Solo-Gabriele, and C. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devriese, L. A., M. D. Collins, and R. Wirth. 1992. The genus Enterococcus, p. 1465-1481. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, NY.
- 17.Elmanama, A. A., M. I. Fahd, S. Afifi, S. Abdallah, and S. Bahr. 2005. Microbiological beach sand quality in Gaza Strip in comparison to seawater quality. Environ. Res. 99:1-10. [DOI] [PubMed] [Google Scholar]
- 18.Gerba, C. P., and J. S. McLeod. 1976. Effect of sediment on the survival of E. coli in marine waters. Appl. Environ. Microbiol. 32:114-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haile, R., J. Witte, M. Gold, R. Cressey, C. McGee, R. Millikan, A. Glasser, N. H. abd C. Ervin, P. Harmon, J. Harper, J. Dermand, J. Alamille, K. Barrett, M. Nides, and C. Wang. 1999. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology 10:355-363. [PubMed] [Google Scholar]
- 20.Hartz, A., M. Cuvelier, K. Nowosielski, T. D. Bonilla, M. Green, N. Esiobu, D. McCorquodale, and A. Rogerson. 2008. Survival potential of Escherichia coli and enterococci in subtropical beach sand: implications for water quality managers. J. Environ. Qual. 37:898-905. [DOI] [PubMed] [Google Scholar]
- 21.Haugland, R. A., S. C. Siefring, L. J. Wymer, K. P. Brenner, and A. P. Dufour. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559-568. [DOI] [PubMed] [Google Scholar]
- 22.He, J. W., and S. Jiang. 2005. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karl, D. M., and J. A. Novitsky. 1988. Dynamics of microbial-growth in surface-layers of a coastal marine sediment ecosystem. Mar. Ecol. Prog. Ser. 50:169-175. [Google Scholar]
- 25.Kay, D., J. M. Fleisher, R. L. Salmon, F. Jones, M. D. Wyer, A. F. Godfree, J. Zelenauch, and R. Shore. 1994. Predicting likelihood of gastroenteritis from sea bathing—results from randomised exposure. Lancet 344:905-909. [DOI] [PubMed] [Google Scholar]
- 26.Kettler, T. A., J. W. Doran, and T. L. Gilbert. 2001. Simplified method for soil and particle-size determination to accompany soil quality analyses. Soil Sci. Soc. Am. J. 65:849-852. [Google Scholar]
- 27.Lee, C. M., T. Lin, C.-C. Lin, G. A. Kohbodi, A. Bhatt, R. Lee, and J. Jay. 2006. Persistence of fecal indicator bacteria in Santa Monica Bay beach sediments. Water Res. 40:2593-2602. [DOI] [PubMed] [Google Scholar]
- 28.Ludwig, W., and K.-H. Schleifer. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556-562. [DOI] [PubMed] [Google Scholar]
- 29.Moriarity, D. J. W. 1986. Measurement of bacterial growth rates in aquatic systems from rates of nucleic acid synthesis. Adv. Microb. Ecol. 9:245-292. [Google Scholar]
- 30.Neter, J., M. H. Kutner, and W. Wasserman. 1990. Applied linear statistical models: regression, analysis of variance, and experimental designs, 3rd ed. Irwin, Boston, MA.
- 31.Noble, R. T. 2001. Enumeration of viruses, p. 43-52. In J. Paul (ed.), Marine microbiology, vol. 30. Academic Press, San Diego, CA. [Google Scholar]
- 32.Oana, K., Y. Okimura, Y. Kawakami, N. Hayashida, M. Shimosaka, M. Okazaki, T. Hayashi, and M. Ohnishi. 2002. Physical and genetic map of Enterococcus faecium ATCC 19434 and demonstration of intra- and interspecific genomic diversity in enterococci. FEMS Microbiol. Lett. 207:133-139. [DOI] [PubMed] [Google Scholar]
- 33.Obiri-Danso, K., and K. Jones. 2000. Intertidal sediments as reservoirs for hippurate negative campylobacters, salmonellae and faecal indicators in three EU recognized bathing waters in North West England. Water Res. 34:519-527. [Google Scholar]
- 34.Oshiro, R., and R. Fujioka. 1995. Sand, soil, and pigeon droppings—sources of indicator bacteria in the waters of Hanauma Bay, Oahu, Hawaii. Water Sci. Technol. 31:251-254. [Google Scholar]
- 35.Pianetti, A., F. Bruscolini, L. Sabatini, and P. Colantoni. 2004. Microbial characteristics of marine sediments in bathing area along Pesaro-Gabicce coast (Italy): a preliminary study. J. Appl. Microbiol. 97:682-689. [DOI] [PubMed] [Google Scholar]
- 36.Santoro, A. E., C. A. Francis, N. R. De Sieyes, and A. B. Boehm. 2008. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10:1068-1079. [DOI] [PubMed] [Google Scholar]
- 37.Shibata, T., H. M. Solo-Gabriele, L. E. Fleming, and S. Elmir. 2004. Monitoring marine water recreational water quality using multiple microbial indicators in an urban tropical watershed. Water Res. 38:3119-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shrestha, P. M., M. Noll, and W. Liesack. 2007. Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ. Microbiol. 9:2464-2474. [DOI] [PubMed] [Google Scholar]
- 39.Shuval, H. 2003. Estimating the global burden of thalassogenic diseases: human infectious diseases caused by wastewater pollution in the environment. J. Water Health 1:53-64. [PubMed] [Google Scholar]
- 40.Snedecor, G. W., and W. G. Cochran. 1989. Statistical methods, 8th ed. Blackwell, Ames, IA.
- 41.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. Environmental Protection Agency. 1997. Method 1600: membrane filter test method for enterococci in water. Technical report. Office of Water and Hazardous Materials, U.S. Environmental Protection Agency, Washington, DC.
- 43.Wade, T. J., N. Pai, J. N. Eisenberg, and J. M. Colford, Jr. 2003. Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 111:1102-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walk, S. T., E. W. Alm, L. M. Calhoun, J. M. Mladonicky, and T. S. Whittam. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274-2288. [DOI] [PubMed] [Google Scholar]
- 45.Walters, S. P., and K. G. Fields. 2006. Persistence and growth of fecal Bacteroidales assessed by bromodeoxyuridine immunocapture. Appl. Environ. Microbiol. 72:4532-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reference deleted.
- 47.Whitman, R. L., and M. B. Nevers. 2003. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl. Environ. Microbiol. 69:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.World Health Organization. 2003. Guidelines for safe recreational water environments. World Health Organization, Geneva, Switzerland. http://www.who.int.
- 49.Yamahara, K. M., B. A. Layton, A. E. Santoro, and A. B. Boehm. 2007. Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters. Environ. Sci. Technol. 41:4515-4521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.