Abstract
This study investigated the roles of various environmental sources, such as truck-washing systems, waste-processing lagoons, and other sources, as potential contributors to the exposure and dissemination of Salmonella in commercial swine production systems. Four cohorts of nursery age swine herds which originated from distinct farm flows were selected. In addition, cross-sectional sampling of four truck wash stations selected based on the types of disinfectants and sources of water used for sanitizing trucks were tested. Salmonella isolates were recovered from pigs (feces, cecal contents, and mesenteric lymph nodes) and environmental sources (barn floor, lagoon, barn flush, trucks, and holding pens). Antimicrobial susceptibility testing and genotyping were conducted using Kirby-Bauer disk diffusion and amplified fragment length polymorphism, respectively. Salmonella prevalence significantly increased with age from late nursery to slaughter for all of the cohorts (P = 0.007). In two of three instances, all three pig holding pens (lairage) sampled at processing were Salmonella positive. The predominant antibiotypes for all sources included ACSSuT (51.8%), SSuT (16.8%), T (6%), and pansusceptible (7.4%). For the isolates obtained at the farms, the ACSSuT phenotype was 5.6 times more likely to be found in the animals than in the environment (95% confidence interval, 4.4 to 7.2 times). Serogroup B was the most common serogroup (79%), followed by serogroup E (10.4%). Despite the fact that the four production flows were independent, 1 of the 11 genotypic clusters (cluster A1) was commonly detected in any type of sample regardless of its origin. Five of the genotypic clusters (clusters A3, A4, A5, A6, and A7) contained isolates that originated from trucks and lairage swabs and also from cecal contents and/or mesenteric lymph nodes. More interestingly, genotypic clusters A3, A4, and A6 (but not clusters A5 and A7) were not detected on the farms. They originated from the trucks and lairage swabs and then were identified from the cecal contents and/or mesenteric lymph nodes. These findings underscore the significance of various environmental factors, including inadequate truck-washing systems, and emphasize the role of lairage contamination by Salmonella that has food safety significance.
Salmonellae are among the most important bacterial food-borne pathogens, causing a large number of food-borne gastroenteritis in humans worldwide. In the United States, Salmonella is responsible for an estimated 1.2 million illnesses annually, only 40,000 of which are reported or clinically diagnosed, and an estimated 500 deaths are attributed to Salmonella infections each year (21; http://www.cdc.gov/nczved/dfbmd/disease_listing/salmonellosis_gi.html). Although all serotypes of Salmonella enterica are considered potential public health problems, certain Salmonella serovars are predominant and of particular public health concern. In the United States, Salmonella serotypes of public health importance, such as S. enterica serotype Typhimurium, are commonly isolated from commercial swine herds (2, 10). Identification of factors that may contribute to introduction and dissemination of this food-borne pathogen is a very important issue facing the pork industry today, especially for large integrated pork production systems. In general terms, the on-farm risk factors that are believed to be responsible for increased shedding include biosecurity and environmental factors (3). To date, only very limited studies have been conducted to delineate specific environmental factors that could play a major role in Salmonella dispersion in the farm-to-table continuum.
Previous studies reported that the most common serotype in humans, serotype Typhimurium, is also very common in pigs, suggesting that pork products are likely significant in food safety (8-10). Salmonella serotype Typhimurium has also been commonly isolated in various parts of the world from swine herds (4, 20). In addition, this serotype has been reported to often be multidrug resistant (9-11, 16). Although a majority of the Salmonella isolates from swine are commonly isolated from apparently health pigs, some serotypes could also have clinical significance because of their zoonotic potential. A recent study reported that serotype Typhimurium ranked first with 26% prevalence among the top 10 S. enterica serotypes isolated from different diseased food animal species, including swine, in the United States (37).
Antimicrobial resistance associated with food-borne pathogens is a major public health concern. Salmonella isolates from swine herds, particularly strains of serotypes commonly isolated in human illnesses, such as serotype Typhimurium, have often been reported to exhibit multidrug resistance (5, 9, 11). Previous studies that investigated isolates obtained at different time points have also shown that there has been an increased rate of resistance to most classes of antimicrobials (4). Furthermore, the occurrence of resistant strains of Salmonella in various environmental sources associated with swine production systems has also increased (15, 31). However, there is a paucity of information concerning the specific roles of various common practices in swine production systems and their impact on the occurrence and persistence of Salmonella, particularly multidrug-resistant strains, in the environment.
Phenotypic and genotypic similarities of strains identified from cohorts of pigs at slaughter have been investigated in previous reports, and overall the findings show that slaughterhouse sampling alone is not a good indicator of farm status both qualitatively (serotypes, genotypes, etc.) and quantitatively (prevalence) (5). The findings imply that other, thus far not fully investigated intermediary factors (transportation and other associated environmental factors) are potentially significant factors in the introduction and dispersion of Salmonella in market age swine herds. Prompted by these earlier findings and the paucity of specific data for environmental sources in the farm-to-harvest continuum, the present study was carried out to investigate whether trucks, truck-washing systems, the barn environment, waste lagoons, and holding pens at processing facilities are potential factors that contribute to exposure to and dissemination of Salmonella. The prevalence, antimicrobial resistance phenotypes, and genotypic clonality of Salmonella isolates from swine and the environment were investigated.
MATERIALS AND METHODS
Study sites and sample collection.
In this longitudinal study, four cohorts of nursery age swine herds, which originated from independent production farm flows, were selected from one large commercial conventional swine production system. A farm flow represents a specific breeding herd allocated to distinct farrowing houses, and weaned pigs were moved to specific nursery barns and then moved together to finishing barns. Each farm flow tested in this study had its own set of farrowing, nursery, and finishing barns, and there was no overlap among flows. In addition, the sources of feed and water for each of the farm flows were distinctly different. None of the farm flows shared service personnel or farm workers. Within each farm flow, cohorts of pigs were identified by age. Fecal samples from 60 pigs from each cohort were collected from animals based on convenience within the cohort at different stages from nursery to slaughter as described in detail below. To identify sources of Salmonella at different stages of swine production from nursery to slaughter, samples were obtained from pigs and environmental sources, including trucks, lagoon water, barn swabs, holding pens, and swabs at truck wash facilities.
Sixty fecal samples were collected from pigs from one barn at each farm location at three stages during production, late nursery (just prior to relocation to the finisher), early finisher (2 weeks after placement in the finisher), and late finisher (within 24 h prior to transport to market for slaughter), and samples were also collected at slaughter (60 cecal content samples and 60 mesenteric lymph node samples). Samples for the late finisher stage and slaughter were not obtained for one of the farm flows (farm 3) due to loss of follow-up (early shipment of pigs), and therefore, data from this farm were excluded from any analysis with a time or harvest stage component.
The environmental samples collected included preload nursery transport trailer swabs (five swabs per truck), prefill finisher pen drag swabs (10 pens per barn), and finisher lagoon water samples (inlet, outlet, and pooled samples for each farm site). Swab samples were also collected from trucks that transported pigs from the nursery to the finishing farms and also from the finishing farms to the slaughterhouse. One swab sample per truck, pooled from each corner and the center of the truck floor, was collected. For the swine waste lagoons at farms and truck wash stations, lagoon outlet, lagoon inlet, and pooled lagoon samples were collected.
Furthermore, a cross section of samples from four truck wash stations that served the whole production system was collected based on the types of disinfectants used and the sources of water (fresh or recycled lagoon) and examined for Salmonella contamination. There was no known specific relationship between the stations and the farm flows described above. Trucks that were sampled were not identified individually, and no records of their visits were accessible. Therefore, it is possible that the same truck may have serviced more than one of the farm flows included in the study. There were no trucks that were dedicated to specific production stages. Therefore, it was also possible that the same truck(s) might have transported pigs from the nursery site to the finishing site or from the finishing site to slaughter. However, each of the trucks was cleaned and disinfected before its assigned activities. Swab samples were collected before washing and after washing to determine the effectiveness of the truck wash sanitization systems. Truck wash station A used recycled lagoon water with Virkon-S disinfectant (Antek International, Sudbury, United Kingdom). Truck wash stations B and D used recycled lagoon water with a phenol disinfectant. Truck wash station C used freshwater followed by soap and a phenol disinfectant.
Salmonella isolation and identification.
All samples were transported to the laboratory on the same day and processed for Salmonella isolation and identification immediately upon arrival. Isolation and identification of Salmonella were performed using conventional methods as described previously (9, 10). From each of the positive specimens, up to five colonies were preserved for further characterization.
Serogrouping was performed for 173 isolates selected systematically as described below for the amplified fragment length polymorphism (AFLP) analysis. Some of the isolates were also serotyped to further delineate phenotypes. Serogrouping was performed using commercially available polyvalent O and group-specific antisera (Mira Vista, Copenhagen, Denmark). Serotyping was performed for 96 Salmonella isolates from the environment (n = 48) and animals (n = 48) at the National Veterinary Services Laboratory in Ames, IA. These isolates were selected based on the fact that they exhibited the two most predominant antimicrobial resistance phenotypes (ACSSuT and SSuT, where A indicates resistance to ampicillin, C indicates resistance to chloramphenicol, S indicates resistance to streptomycin, Su indicates resistance to sulfisoxazole, and T indicates resistance to tetracycline).
Antimicrobial susceptibility testing.
The antimicrobial susceptibility of Salmonella isolates from the various samples was tested using the Kirby-Bauer disk diffusion method as recommended by the Clinical Laboratory Standards Institute (formerly NCCLS), (23, 24) and as described previously (9, 10). Of the 1,640 Salmonella isolates whose antimicrobial susceptibility was tested, 1,405 were from farms (animal and environmental isolates) and 235 were from the cross-sectional sampling of truck wash stations (see Tables 2 and 3). The following antimicrobials and amounts on disks were used: ampicillin (A), 10 μg; amoxicillin-clavulanic acid (Ax), 30 μg; amikacin, 30 μg; ceftriaxone (Cro), 30 μg; cephalothin (Cf), 30 μg; chloramphenicol (C), 30 μg; ciprofloxacin, 5 μg; gentamicin (Gm), 10 μg; kanamycin (K), 30 μg; streptomycin (S), 10 μg; sulfisoxazole (Su), 250 μg; and tetracycline (T), 30 μg. Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 25923, and Pseudomonas aeruginosa ATCC 27853 were used as quality control organisms.
TABLE 2.
Comparison of predominant antimicrobial resistance phenotypes of Salmonella isolates from pigs (fecal, cecal, or mesenteric lymph node samples) and the environment (floor swab, truck swab, lagoon, or lairage swab samples)a
| Resistance pattern | No. of isolates | No. (%) positive on:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Farm 1
|
Farm 2
|
Farm 3
|
Farm 4
|
All farms
|
|||||||
| Animal | Environment | Animal | Environment | Animal | Environment | Animal | Environment | Animal | Environment | ||
| ACSSuT | 812 | 151 (52) | 44 (26) | 266 (83) | 69 (39) | 56 (67) | 7 (50) | 202 (86) | 17 (20) | 675 (71) | 137 (30) |
| SSuT | 226 | 86 (30) | 45 (27) | 1 (0.3) | 57 (32) | 1 (1) | 1 (7) | 35 (40) | 88 (9) | 138 (30) | |
| Pansusceptible | 71 | 25 (9) | 25 (15) | 6 (2) | 7 (3) | 8 (9) | 38 (4) | 33 (7) | |||
| T | 59 | 2 (1) | 17 (10) | 2 (1) | 32 (18) | 4 (5) | 1 (0.4) | 1 (1) | 9 (1) | 50 (11) | |
| ACSSuTAxCfCroKGm | 44 | 7 (2) | 17 (5) | 5 (3) | 10 (4) | 5 (6) | 34 (4) | 10 (2) | |||
| ST | 34 | 4 (1.5) | 8 (5) | 3 (1) | 3 (2) | 12 (14) | 1 (7) | 3 (3) | 19 (2) | 15 (3) | |
| A(ST)AxCfCro | 18 | 9 (3) | 4 (2) | 5 (36) | 9 (1) | 9 (2) | |||||
| STK | 17 | 12 (7) | 5 (2) | 5 (0.5) | 12 (3) | ||||||
| SuT | 19 | 8 (3) | 7 (4) | 4 (5) | 8 (1) | 11 (2) | |||||
| CSSuT | 10 | 10 (12) | 10 (2) | ||||||||
| SSuTKGm | 9 | 6 (2) | 3 (1) | 9 (1) | |||||||
| ASSuTKGm | 11 | 8 (5) | 3 (1) | 3 (0.3) | 8 (2) | ||||||
| S | 7 | 5 (2) | 1 (0.6) | 1 (1) | 6 (0.6) | 1 (0.2) | |||||
| AT | 4 | 3 (4) | 1 (0.4) | 4 (0.4) | |||||||
| Others | 64 | 12 (4) | 7 (4) | 18 (6) | 8 (4.5) | 7 (8) | 8 (3) | 2 (2) | 45 (5) | 19 (4) | |
| Total | 1405 | 292 (21) | 167 (12) | 341 (24) | 185 (13) | 84 (6) | 14 (1) | 235 (17) | 87 (6) | 952 (68) | 453 (32) |
Statistically significant associations (P < 0.05) between sample origin (animal or environment) and predominant resistance patterns, including ACSSuT (odds ratio, 5.6), SSuT (odds ratio, 0.23), T (odds ratio, 0.08), and pansusceptible (odds ratio, 0.53), were found.
TABLE 3.
Antimicrobial resistance phenotypes of 235 Salmonella isolates that originated from truck wash stations A, B, C and D in a cross-sectional sampling analysis
| Resistance pattern | No. of isolates | No. (%) positive for:
|
||||
|---|---|---|---|---|---|---|
| Truck wash station A | Truck wash station B | Truck wash station C | Truck wash station D | All truck wash stations | ||
| ACSSuT | 38 | 9 (10) | 21 (28) | 7 (14) | 1 (8) | 38 (16) |
| SSuT | 49 | 29 (31) | 10 (13) | 9 (17) | 1 (8) | 49 (21) |
| Pansusceptible | 56 | 27 (28) | 5 (7) | 21 (40) | 3 (23) | 56 (17) |
| T | 39 | 19 (20) | 13 (17) | 6 (12) | 1 (8) | 39 (24) |
| ACSSuTAxCfCroKGm | 16 | 16 (21) | 16 (7) | |||
| ST | 4 | 1 (1) | 3 (4) | 4 (2) | ||
| A(ST)AxCfCro | 3 | 3 (3) | 3 (1) | |||
| STK | 2 | 1 (1) | 1 (2) | 2 (1) | ||
| SuT | 13 | 4 (4) | 2 (3) | 5 (10) | 2 (15) | 13 (6) |
| S | 3 | 1 (1) | 2 (4) | 3 (1) | ||
| Others | 12 | 1 (1) | 5 (7) | 1 (2) | 5 (38) | 12 (5) |
| Total | 235 | 95 (40) | 75 (32) | 52 (22) | 13 (6) | 235 |
AFLP genotyping.
Genotypic analysis using AFLP fingerprinting was carried out for a total of 176 isolates. Isolates were selected as follows. The isolates were first stratified by farm and within each farm, and they were then further stratified by sample type (animal and environment). Random samples of animal and environmental isolates were then selected for each farm using a random number generator function of SPSS software (SPSS Inc., Chicago, IL). The isolates included 114 isolates of animal origin (49 isolates of fecal origin, 36 isolates from mesenteric lymph nodes, and 29 isolates from cecal contents at slaughter) and 62 isolates from various environmental sources associated with the farm and slaughter (40 isolates from truck swabs), 12 isolates from lairage swabs, 5 isolates from floor swabs, 3 isolates from barn flush samples, and 2 isolates from lagoon water).
The AFLP genotyping protocol used was the protocol described previously by Vos et al., with slight modifications (6, 34). Briefly, DNA was purified using a Qiagen DNeasy tissue kit (Qiagen, Valencia, CA), and a total of 100 ng of genomic DNA was digested with two restriction enzymes, EcoRI and MseI (New England BioLabs, Beverly, MA), at 37°C for 2 h. Adapter oligonucleotides unique to each end were ligated using T4 DNA ligase (New England Biolabs, Beverly, MA) by overnight incubation at 25°C. Fragments diluted 1:9 were then amplified using EcoRI (5′-GACTGCGTACCAAATC) and MseI (5′-GATGAGTCCTGAGTAA) (Integrated DNA Technologies, Coralville, IA) primers. The amplification protocol was 20 cycles at 94°C for 30 s, 56°C for 1 min, and 72°C for 1 min. The amplified products were diluted 1:9 using molecular-grade water and subjected to a final selective amplification using the MseI primer (5′-GATGAGTCCTGAGTAA) and the well-red dye-labeled EcoRI primer with an additional adenine at the 3′ end, EcoRI-A (5′-GACTGCGTACCAAATCA) (Integrated DNA Technologies, Coralville, IA). The conditions used for the final selective amplification were one cycle of denaturation at 94°C for 30 s, primer annealing at 65°C for 30 s and extension at 72°C for 1 min, followed by 12 cycles in which the initial annealing temperature (65°C) was decreased by 0.7°C per cycle while the denaturation and extension temperatures and times were kept the same. This was followed by 23 cycles of final denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min. The fragments were separated with a capillary electrophoresis-based system using a CEQ 8000 genetic analyzer (Beckman Coulter, CA). Amplified fragments between 50 bp and 600 bp long were scored using the AFLP dominant scoring algorithm (Beckman Coulter, CA), and the data were imported into BioNumerics software v4.6 (Applied Maths, Kortrijk, Belgium) for further analysis. A cluster analysis was done using Pearson pairwise correlation, and the unweighted-pair group method with arithmetic mean was used to construct a dendrogram. A threshold genetic similarity level of 79% was used as a cutoff coefficient to cluster genotypes. This cutoff was selected based on previous recommendations and its plausibility using various criteria, particularly epidemiological and phenotypic characteristics (antimicrobial susceptibility and serogrouping) (7). A larger group of clades was also formed at a genetic similarity of 67% conservatively to facilitate classification of the clusters.
Statistical analysis.
Statistical analysis was done using the SAS statistical software package (SAS Institute Inc., Cary, NC). The nonparametric Page test (12) was used for the prevalence-over-time data. Tukey grouping was used to compare levels of contamination between truck washes. To assess the degree of deviation among different phenotypes, odds ratios and 95% confidence intervals (CI) were calculated using the formula lnψ ± (1.96)√[(1/A1) + (1/A0) + (1/B1) + (1/B0)] and the chi-square test statistic. Results with a type I error P value of ≤0.05 were considered statistically significant.
RESULTS
Salmonella prevalence in pigs and associated environments.
Salmonella prevalence significantly increased with age from late nursery to slaughter, and this was consistent for all three cohorts that were followed from farm to slaughter. The Salmonella prevalence increased from 6.4% to 56.7%, from 5% to 65%, and from 0% to 50% on farms 1, 2, and 4, respectively (P = 0.007) (Table 1).
TABLE 1.
Occurrence of Salmonella in pigs at different stages of production, in truck swabs, and in lagoon water samples on four swine farms
| Samples | No. from each farm | No. (%) positive for:
|
||||
|---|---|---|---|---|---|---|
| Farm 1 | Farm 2 | Farm 3 | Farm 4 | Total | ||
| Fecal | ||||||
| Nursery | 60 | 4 (6.4) | 3 (5.0) | 13 (21.7) | 5 (8.3) | 25 (10.4) |
| Finisher 1 | 60 | 9 (15.0) | 3 (4.8) | 16 (26.7) | 28 (11.6) | |
| Finisher 2 | 60 | 7 (11.7) | 21 (33.9) | NDa | 5 (8.3) | 33 (18.3) |
| Slaughter | ||||||
| Cecal contents | 60 | 38 (63.3) | 29 (48.3) | ND | 15 (25.0) | 82 (45.5) |
| Mesenteric lymph nodes | 60 | 34 (56.7) | 39 (65.0) | ND | 30 (50.0) | 103 (57.2) |
| Environment | ||||||
| Nursery truck swabs | 1 | 1 | 1 | 1 | 1 | 4 |
| Finisher pen swabs | 10 | 1 (10.0) | ND | 1 (10.0) | 2 (20.0) | 4 (13) |
| Lagoon inlet | 1 | ND | 1 | 1 | ||
| Lagoon outlet | 1 | 1 | ND | 1 | 2 | |
| Pooled lagoon | 1 | 1 | ND | 1 | 1 | 3 |
| Barn flush | 1 | 1 | ND | 1 | 1 | 3 |
| Preload truck swabs | 3 | 3 | 3 | ND | 2 | 8 |
| Postload truck swabs | 3 | 3 | 3 | ND | 2 | 8 |
| Holding pen swabs | 3 | 3 | 3 | ND | 0 | 6 |
ND, not determined.
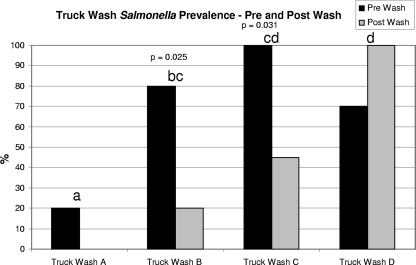
Cross-sectional sampling of the four truck wash stations showed that there was a reduction in the level of contamination after sanitization, except for one station, truck wash station D. The extent of reduction greatly varied among the stations, and the postwash Salmonella prevalence value were 0%, 20%, 45%, and 100% for truck wash stations A, B, C, and D, respectively (Fig. 1). At station D, the level of recovery of Salmonella after washing increased to 100% (10/10) from 70% (7/10) before washing. At only two of the truck wash stations, stations B and C, was there a statistically significant decrease in the contamination levels (P < 0.05).
FIG. 1.
Prevalence of Salmonella as determined by prewash and postwash swabbing of trailers at truck wash stations. Ten prewash samples and 10 postwash samples were collected at each of the stations. Truck wash station A used recycled lagoon water and Virkon-S; truck wash station B used recycled lagoon water and phenol; truck wash station C used freshwater, soap, and phenol; and truck wash station D used recycled water and phenol (this station handled trucks from farms with outbreaks). Different letters indicate that values are statistically significantly different (P ≤ 0.05) as determined by Tukey grouping. The P values above the bars indicate that the decrease or increase in the prevalence of Salmonella for pre- and postwash samples is significant. The letters represent a Tukey grouping for truck wash effect on Salmonella.
On the farms, all trucks (one truck per farm, four farms) that were sampled at nursery barns before the pigs were loaded to be relocated to the finishing sites were positive for Salmonella. When the empty pens in the receiving finishing barns were sampled before the pigs were unloaded from the trucks at the finishing sites (10 pens/site, four sites), 10% to 20% of the floor swab samples were positive for Salmonella. Salmonella was detected in lagoon samples collected from the inlet, the outlet, and flushing and from pooled samples (a total of four samples per site) (Table 1).
At the time of slaughter, trucks were sampled before the pigs were loaded (preload). The preload truck swab Salmonella prevalence ranged from 67.7% to 100%. All the trucks were sampled again after the pigs were unloaded at the slaughter facility, and the prevalence remained unchanged. The holding pens (lairage) at the slaughter facility were also sampled before the pigs entered for the required rest period. In two instances all three floor swabs of holding pen samples were positive, and in one instance none of the pens was Salmonella positive.
Serogroups and serotypes.
A total of 173 isolates were tested to determine the serogroups. The most common serogroup was serogroup B, with a frequency of 79.7%, followed by serogroup E (10.4%). Serogroups C (n = 8) and D1 (n = 1) were also detected. The remaining eight isolates were not typeable using the available polyvalent and group-specific antisera. Serotyping of 96 isolates exhibiting the two most predominant resistance patterns also corroborated the findings that the majority (78 of 96) of the serotypes were also members of serogroup B. The most common serotypes were serotypes Typhimurium (n = 46) and Derby (n = 32). In addition, serotypes Muenchen (n = 7), London (n = 4), and Mbandaka (n = 3) that belong to serogroups C and E1 were found.
Antimicrobial resistance patterns.
For the Salmonella isolates (n = 1,640) of farm (animal and environmental) and truck wash origin, the resistance ranged from pansusceptible (127/1,640, 7.7%) to panresistant (1/1,640). Four predominant phenotypes (R-types) were detected based on antimicrobial susceptibility (Tables 2 and 3). For all of the isolates from the farms (animal and environment) (Table 2) and the truck washes (Table 3), the predominant antibiotypes included ACSSuT (with or without AxCf) (51.8%), SSuT (16.8%), T (6%), and pansusceptible (7.7%). At the farm level (Table 2), the ACSSuT phenotype was 5.6 times more likely to be found in the animals than in the environment (95% CI, 4.4 to 7.2) (P < 0.05). The SSuT, T, and pansusceptible phenotypes were 0.23 times (95% CI, 0.17 to 0.31 times), 0.08 times (95% CI, 0.04 to 0.16 times), and 0.53 times (95% CI, 0.33 to 0.86 times) more likely to be isolated from the animals than from the environment, respectively (P < 0.05). The ACSSuT(AxCf) phenotype accounted for 70.9% of the total animal isolates that were obtained from fecal samples on farms and from gastrointestinal tract samples (cecal contents and mesenteric lymph nodes) at slaughter.
Genotyping of isolates.
Salmonella isolates (n = 176) obtained from each of the production flows, including 49 isolates of fecal origin, 36 isolates from mesenteric lymph nodes, 29 isolates from cecal contents at slaughter, and 62 isolates from various environmental sources associated with the farm (5 isolates from floor swabs, 3 isolates from barn flush samples, and 2 isolates from lagoon water) and slaughter (40 isolates from truck swabs and 12 isolates from lairage swabs), were genotyped using AFLP DNA fingerprinting.
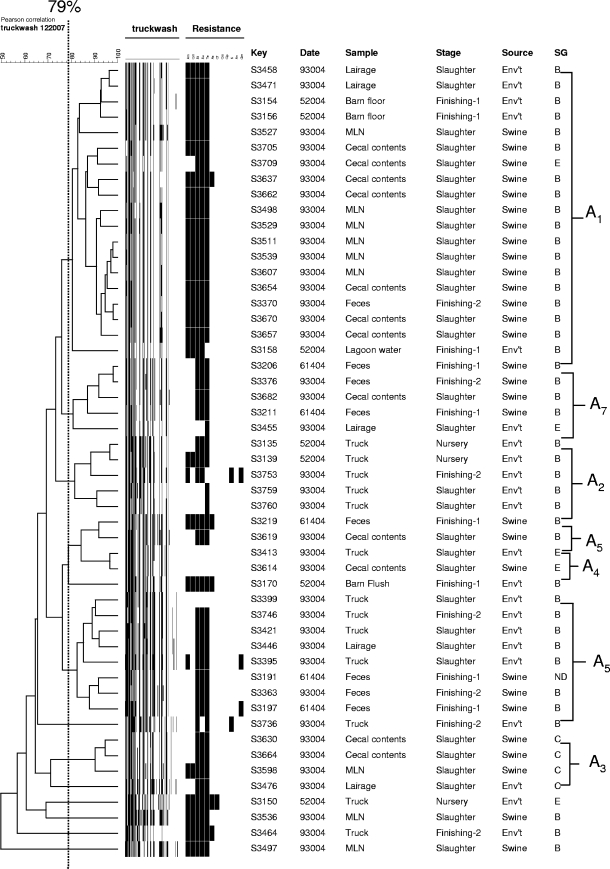
Overall, five major clades with a total of 11 genotypic clusters were identified (Fig. 2). Five of the 11 clusters (45%) were unique to either the environment or animals. Genotypic clusters unique to the environmental sources (clusters A2, B1, and C1) and to the animals (clusters D and E) were identified. The remaining six clusters (55%) contained both animal- and environment-derived isolates. Cluster A1 was the largest cluster, with 77 isolates. This genotypic cluster appeared to be ubiquitous and occurred regardless of the origin (farm flow, production stage, or sample type). The remaining five genotypic clusters (clusters A3, A4, A5, A6, and A7) are perhaps the most significant from the food safety standpoint. All five of these clusters contained isolates that originated from trucks and lairage swabs initially and were subsequently identified in cecal contents and/or mesenteric lymph nodes. More interestingly, genotypic clusters A3, A4, and A6 (but not clusters A5 and A7) were not detected on the farms and were isolated from the trucks and lairage swabs before subsequent isolation from the cecal contents and/or mesenteric lymph nodes.
FIG. 2.
Dendrogram based on AFLP fragments of Salmonella isolates recovered from swine (cecal, mesenteric lymph node, and fecal samples) and the environment (truck, lairage, and lagoon samples). The vertical dotted line indicates a genetic similarity threshold of 79%, which was used to separate the genotypic clusters.
Despite the fact that the four production flows were independent (there was no comingling of pigs), only 3 of the 11 genotypic clusters (27%) (clusters A2, A3, and E) were unique to a cohort. The remaining eight clusters (73%) were detected in more than one farm flow. Three genotypic clusters were unique to environmental sources (clusters A2, B1, and C1), and genotypic clusters D and E were specific to animal sources but were not found in the environment (Fig. 2).
For comparative purposes, a dendrogram depicting the genotypic diversity of production flow 1 is shown on Fig. 3. In this cohort, a total of seven genotypic clusters were identified. All of the clusters were in major clade “A.” Four of the seven clusters were obtained only from the environment or pigs (fecal, cecal, or mesenteric lymph node samples). Clusters A1 and A5 were the predominant clusters in this production flow, and both of them were composed predominantly of isolates that originated from slaughtered pigs and environmental sources (mainly trucks and lairage).
FIG. 3.
Genotyping of isolates obtained from various stages for one of the cohorts (farm flow 1) and the associated slaughter using AFLP. The vertical dotted line indicates a genetic similarity threshold of 79%, which was used to separate the genotypic clusters.
DISCUSSION
The findings of this study arguably show that the current truck-washing and disinfection process used in commercial swine production systems is not adequate to completely prevent Salmonella contamination. There was great variation in the truck disinfection practices, some of which resulted in significant reductions in contamination. Salmonella contamination that originates from environmental sources, including trucks and lairage, could be a significant concern for food safety, as the phenotypic and genotypic findings showed that distinct clonally related strains that were obtained from the environment were commonly detected in pigs or gastrointestinal tract-associated samples from the pigs (cecal contents and mesenteric lymph nodes).
In the current work, phenotypic (prevalence, serogroups, and antimicrobial resistance patterns) and genotypic (AFLP DNA fingerprinting) analyses were used. AFLP genotyping is one of the most discriminatory methods with high reproducibility. Previously, our investigation team conducted a study of comparative genotyping of Salmonella isolates from the same geographic location using three approaches: pulsed-field gel electrophoresis (PFGE), AFLP, and repetitive extragenic palindromic (7). As described in our previous work, while PFGE and AFLP were found to be highly discriminatory, AFLP was found to be preferable for large-scale epidemiologic investigations. In addition to its high discriminatory power, this method was also found to be cost-effective and efficient in terms of turnaround time. Other studies also compared AFLP and PFGE and reported that AFLP had a greater ability to discriminate among isolates than PFGE (29) and was valid for epidemiologic studies (33).
The increase in the prevalence of Salmonella as the age of the cohorts increases is a very important finding since the impact on food safety is greatest when Salmonella is detected at market age. A high prevalence of Salmonella among finishing herds has been reported previously (27). The current study was done using one large integrated swine production system. It is interesting that the increase in prevalence in older groups was consistently found in all three cohorts that were followed to slaughter (the fourth cohort was lost to follow up). While the findings have internal validity, it may not be possible to extrapolate them to swine farming systems in general. Findings that are similar to or were in contrast to the findings of the current study were reported previously. The results of a recent study done by Rodríguez-Buenfil et al. were similar to the results of this study in that the prevalence of Salmonella was the highest when the pigs were 117 days old compared to the data for several sampling times when the pigs were younger (28). In another study, the prevalence of Salmonella was found to be higher on sow farms than in finishing units (22). Although the latter study did not include nursery sampling, the pattern of higher prevalence in older age groups is consistent with the findings of the current study. In contrast, there have also been studies which reported a decline in the prevalence of Salmonella as pigs reached market age (17, 30). The increase in prevalence at slaughter may be a result of cross contamination at the periharvest stage (35), including contamination from trucks, and to some extent due to stress shedding induced by the trucking process (14) and cross contamination during transport and holding.
Of particular interest in this study was the role of trucks and truck wash protocols that were identified as potential sources of Salmonella. Three of the four disinfection protocols resulted in a reduction in the Salmonella level after sanitization of trailers. While the majority of the practices reduced the contamination level, trucks from only one of the wash stations were totally negative after cleaning and disinfection. In one instance, the cleaning and disinfection procedure increased the Salmonella contamination level, in contrast to the expected outcome of disinfection. A recent study conducted in pig barns showed that the cleaning and disinfection protocol used effectively reduced the level of Salmonella; however, there was not complete elimination of Salmonella using cleaning and disinfection procedures in the barns. This finding suggests that there is a need for multiple intervention procedures at different stages or perhaps more stringent disinfection protocols in swine production environments (18). An in vitro trial in our laboratory showed that some strains of Salmonella may develop tolerance to biocides, including commonly used disinfectants (unpublished data). At the field level, crucial factors in the efficacy of disinfectants have previously been shown to be even distribution on the surface, formation of biofilms, contact and drying times, and other factors associated with the application protocol (19). In addition to the truck trailers that were positive soon after disinfection, the current study also showed that all of the trucks that transported pigs from nurseries to finishing units were also Salmonella positive. These trucks could be potential sources of contamination of the pigs and could contribute to the increased prevalence of Salmonella in finishing units and to pork safety.
The other significant environmental samples were the positive lairage (holding pen) swabs at the slaughter plant. On average, pigs stay in holding pens for about 3 h, which could be sufficient time for Salmonella contamination and establishment in the gastrointestinal tract and associated lymphatic tissues. A previous study by Hurd et al. identified the lairage as a critical point of entry for herd-level Salmonella contamination (13). The findings of the current study corroborate the findings of the previous report by Hurd et al. as some of the genotypic clusters (such as clusters A4 and A5) that originated from the lairage were subsequently obtained from the cecal contents or mesenteric lymph node samples.
Multidrug-resistant Salmonella isolates were found in both animal and environmental samples. However, distinctly different phenotypes were associated with the different types of samples. We found that isolates with the ACSSuT resistance pattern were more likely to be isolated from animals (fecal or cecal samples or mesenteric lymph nodes) than from the environment (odds ratio, 5.6; P < 0.05). This finding was consistent across all cohorts, as shown in Table 2. While this finding shows that there is a strong correlation between sample origin and antimicrobial resistance pattern, it is not clear why the highly multidrug-resistant strains are more common in the gastrointestinal tract, while several other strains with less multidrug resistance are commonly present in the environment. This finding is interesting since it may have important implications for the variation in the selective pressure within the gastrointestinal tract compared to the environment outside the host. It could also be due to a unique selective pressure in the outside environment that results in better survival of strains that have the SSuT phenotype and other phenotypes commonly found in the environment (Table 2). Further investigation of both of these hypotheses is currently under way. Previously, we found phage type DT104 with the ACSSuT resistance type commonly in swine herds in the same geographic area (8). The common pentaresistance pattern in DT104 is known to be encoded chromosomally on two integrons (1, 6, 25, 26). We also reported that a lower proportion of DT104, which is commonly found in the environment, had only the SSu phenotype and carried only one integron (6).
The AFLP genotyping results in the current study appear to be consistent with the phenotypic findings. One predominant genotypic cluster (cluster A1) was found to contain more than 40% of the isolates (77 of 176), whereas some of the genotypic clusters were found to be specific to either the environment or the pig (fecal samples or samples collected at slaughter). Recently, in an ecologic study of Salmonella in swine production systems, Weigel et al. reported that there was no apparent separation of genotypes by host and environmental compartment, and they found only one farm where the same biotic compartment (swine or floor samples) was associated with a tight genotypic cluster (36). The predominantly nonclonal nature of genotypes in a production system that was found in the current study was also consistent with the findings of Weigel et al. (36). The most interesting genotyping finding was that 5 of the 11 genotypic clusters were found in the environment (truck and lairage swabs) initially and were subsequently detected in cecal content and mesenteric lymph node samples. This suggests that the food safety significance of contamination during transportation of pigs to slaughter or in holding pens is very high. A previous study conducted by Swanenburg and colleagues (32) indicated that while farms and holding pens could be important sources, their level of significance varied depending on the type of tissue examined (liver, tongue, or lymph nodes) and also the status of the herd on the farm (32).
Overall, the findings of this study have important implications. First, they show that various environmental factors could play a significant role in disseminating Salmonella to pork. Second, distinct strains (genotypic clusters) could be found in specific niches, and thus, not all strains present in a specific environmental niche may be harbored in the porcine gastrointestinal tract. In addition, the findings strongly showed that the current cleaning and disinfection practices for trucks in commercial swine production systems are not adequate to completely eliminate Salmonella contamination.
Acknowledgments
This work was funded by North Carolina Pork Council grant 04M30.MF2 to W.A.G.
We thank Heather Lowman for technical assistance.
Footnotes
Published ahead of print on 9 January 2009.
REFERENCES
- 1.Briggs, C., and P. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies, P., F. Bovee, J. Funk, W. Morrow, F. Jones, and J. Deen. 1998. Isolation of Salmonella serotypes from feces of pigs raised in a multiple-site production system. J. Am. Vet. Med. Assoc. 212:1925-1929. [PubMed] [Google Scholar]
- 3.Funk, J., P. Davies, and W. Gebreyes. 2001. Risk factors associated with Salmonella enterica prevalence in three-site swine production systems in North Carolina, USA. Berl. Muench. Tieraerztl. Wochenschr. 114:335-338. [PubMed] [Google Scholar]
- 4.Futagawa-Saito, K., S. Hiratsuka, M. Kamibeppu, T. Hirosawa, K. Oyabu, and T. Fukuyasu. 2008. Salmonella in healthy pigs: prevalence, serotype diversity and antimicrobial resistance observed during 1998-1999 and 2004-2005 in Japan. Epidemiol. Infect. 136:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebreyes, W., and S. Thakur. 2005. Multidrug-resistant Salmonella enterica serovar Muenchen from pigs and humans and potential interserovar transfer of antimicrobial resistance. Antimicrob. Agents Chemother. 49:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gebreyes, W., and C. Altier. 2002. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 40:2813-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebreyes, W., C. Altier, and S. Thakur. 2006. Molecular epidemiology and diversity of Salmonella serovar Typhimurium in pigs using phenotypic and genotypic approaches. Epidemiol. Infect. 134:187-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebreyes, W., S. Thakur, P. Davies, J. Funk, and C. Altier. 2004. Trends in antimicrobial resistance, phage types and integrons among Salmonella serotypes from pigs, 1997-2000. J. Antimicrob. Chemother. 53:997-1003. [DOI] [PubMed] [Google Scholar]
- 9.Gebreyes, W., P. Davies, W. Morrow, J. Funk, and C. Altier. 2000. Antimicrobial resistance of Salmonella isolates from swine. J. Clin. Microbiol. 38:4633-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebreyes, W., P. Davies, P. Turkson, W. Morrow, J. Funk, and C. Altier. 2004. Salmonella enterica serovars from pigs on farms and after slaughter and validity of using bacteriologic data to define herd Salmonella status. J. Food Prot. 67:691-697. [DOI] [PubMed] [Google Scholar]
- 11.Gebreyes, W., P. Davies, P. Turkson, W. Morrow, J. Funk, C. Altier, and S. Thakur. 2004. Characterization of antimicrobial-resistant phenotypes and genotypes among Salmonella enterica recovered from pigs on farms, from transport trucks, and from pigs after slaughter. J. Food Prot. 67:698-705. [DOI] [PubMed] [Google Scholar]
- 12.Hollander, M., and D. A. Wolfe. 1999. Nonparametric statistical methods. John Wiley & Sons, Inc., New York, NY.
- 13.Hurd, H., J. McKean, I. Wesley, and L. Karriker. 2001. The effect of lairage on Salmonella isolation from market swine. J. Food Prot. 64:939-944. [DOI] [PubMed] [Google Scholar]
- 14.Isaacson, R., L. Firkins, R. Weigel, F. Zuckermann, and J. DiPietro. 1999. Effect of transportation and feed withdrawal on shedding of Salmonella Typhimurium among experimentally infected pigs. Am. J. Vet. Res. 60:1155-1158. [PubMed] [Google Scholar]
- 15.Jindal, A., S. Kocherginskaya, A. Mehboob, M. Robert, R. Mackie, L. Raskin, and J. Zilles. 2006. Antimicrobial use and resistance in swine waste treatment systems. Appl. Environ. Microbiol. 72:7813-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, J., A. Rajic, and L. McMullen. 2005. Antimicrobial resistance of selected Salmonella isolates from food animals and food in Alberta. Can. Vet. J. 46:141-146. [PMC free article] [PubMed] [Google Scholar]
- 17.Kranker, S., L. Alban, J. Boes, and J. Dahl. 2003. Longitudinal study of Salmonella enterica serotype Typhimurium infection in three Danish farrow-to-finish swine herds. J. Clin. Microbiol. 41:2282-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannion, C., F. Leonard, P. Lynch, and J. Egan. 2007. Efficacy of cleaning and disinfection on pig farms in Ireland. Vet. Rec. 161:371-375. [DOI] [PubMed] [Google Scholar]
- 19.McDonnell, G., and A. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDowell, S., R. Porter, R. Madden, B. Cooper, and S. Neill. 2007. Salmonella in slaughter pigs in Northern Ireland: prevalence and use of statistical modelling to investigate sample and abattoir effects. Int. J. Food Microbiol. 118:116-125. [DOI] [PubMed] [Google Scholar]
- 21.Mead, P., L. Slutsker, V. Dietz, L. McCaig, J. Bresee, C. Shapiro, P. Griffin, and R. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mejía, W., J. Casal, D. Zapata, G. Sánchez, M. Martín, and E. Mateu. 2006. Epidemiology of Salmonella infections in pig units and antimicrobial susceptibility profiles of the strains of Salmonella species isolated. Vet. Rec. 159:271-276. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disc and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A2, 2nd ed. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 24.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing; 12th informational supplement. M100-S12. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 25.Recchia, G., and R. Hall. 1997. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 5:389-394. [DOI] [PubMed] [Google Scholar]
- 26.Recchia, G., and R. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez, A., P. Pangloli, H. Richards, J. Mount, and F. Draughon. 2006. Prevalence of Salmonella in diverse environmental farm samples. J. Food Prot. 69:2576-2580. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez-Buenfil, J., M. Alvarez-Fleites, and J. Segura-Correa. 2006. Incidence of salmonellosis and identification of serogroups and serotypes in a pig commercial farm in Yucatan. Rev. Latinoam. Microbiol. 48:10-13. [PubMed] [Google Scholar]
- 29.Ross, I., and M. Heuzenroeder. 2005. Use of AFLP and PFGE to discriminate between Salmonella enterica serovar Typhimurium DT126 isolates from separate food-related outbreaks in Australia. Epidemiol. Infect. 133:635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowe, T., F. Leonard, G. Kelly, P. Lynch, J. Egan, A. Quirke, and P. Quinn. 2003. Salmonella serotypes present on a sample of Irish pig farms. Vet. Rec. 153:453-456. [DOI] [PubMed] [Google Scholar]
- 31.Small, A., C. James, S. James, R. Davies, E. Liebana, M. Howell, M. Hutchison, and S. Buncic. 2006. Presence of Salmonella in the red meat abattoir lairage after routine cleansing and disinfection and on carcasses. J. Food Prot. 69:2342-2351. [DOI] [PubMed] [Google Scholar]
- 32.Swanenburg, M., B. Berends, H. Urlings, J. Snijders, and F. van Knapen. 2001. Epidemiological investigations into the sources of Salmonella contamination of pork. Berl. Muench. Tieraerztl. Wochenschr. 114:356-359. [PubMed] [Google Scholar]
- 33.Torpdahl, M., M. N. Skov, D. Sandvang, and D. L. Baggesen. 2005. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J. Microbiol. Methods 63:173-184. [DOI] [PubMed] [Google Scholar]
- 34.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, and M. Kuiper. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wegener, H., and D. Baggesen. 1996. Investigation of an outbreak of human salmonellosis caused by Salmonella enterica ssp. enterica serovar Infantis by use of pulsed field gel electrophoresis. Int. J. Food Microbiol. 32:125-131. [DOI] [PubMed] [Google Scholar]
- 36.Weigel, R., D. Nucera, B. Qiao, B. Teferedegne, D. Suh, D. Barber, P. Bahnson, R. Isaacson, and B. White. 2007. Testing an ecological model for transmission of Salmonella enterica in swine production ecosystems using genotyping data. Prev. Vet. Med. 81:274-289. [DOI] [PubMed] [Google Scholar]
- 37.Zhao, S., P. McDermott, D. White, S. Qaiyumi, S. Friedman, J. Abbott, A. Glenn, S. Ayers, K. Post, W. Fales, R. Wilson, C. Reggiardo, and R. Walker. 2007. Characterization of multidrug resistant Salmonella recovered from diseased animals. Vet. Microbiol. 123:122-132. [DOI] [PubMed] [Google Scholar]