Abstract
The Vibrio parahaemolyticus O3:K6 pandemic clonal strain was first observed in southern Chile in 2004 and has since caused approximately 8,000 seafood-related diarrhea cases in this region. The massive proliferation of the original clonal population offers a unique opportunity to study the evolution of a bacterial pathogen in its natural environment by detection and characterization of emerging bacterial variants. Here, we describe a group of pandemic variants characterized by the presence of a 42-kb extrachromosomal DNA that can be recovered by alkaline extraction. Upon treatment with mitomycin C, these variants lyse with production of a myovirus containing DNA of equal size to the plasmid but which cannot be recovered by alkaline extraction. Plasmid and phage DNAs show similar restriction patterns corresponding to enzyme sites in a circular permutation. Sequenced regions showed 81 to 99% nucleotide similarity to bacteriophage VHML of Vibrio harveyi. Altogether these observations indicate that the 42-kb plasmid corresponds to a prophage, consisting of a linear DNA with terminal hairpins of a telomeric temperate phage with a linear genome. Bacteria containing the prophage were 7 to 15 times more sensitive to UV radiation, likely due to phage induction by UV irradiation as plasmid curing restored the original sensitivity. The enhanced UV sensitivity could have a significant role in reducing the survival and propagation capability of the V. parahaemolyticus pandemic strain in the ocean.
Vibrio parahaemolyticus, a diverse bacterial species that inhabits coastal waters, includes strains associated with seafood-borne gastroenteritis (1, 7-9, 11, 14). Since 1996, many infections worldwide have been associated with isolates of a serovar O3:K6 group belonging to a clonal complex originally observed in Southeast Asia (28), now called the V. parahaemolyticus O3:K6 serovar pandemic clone (11, 22, 28, 37). The massive and rapid expansion of this clone has resulted in the clone diversity and the emergence of identifiable variants. At least 21 serovariants have been identified thus far (3, 5; also reviewed in reference 24). Other variants have been reported with minor differences in their restriction fragment length polymorphism-pulsed-field gel electrophoresis (PFGE) or arbitrarily primed PCR patterns (5, 29); lack of orf8 (5), which is characteristic of the pandemic clone (25); and presence of plasmids of different sizes (21). However, the method with the highest resolution to distinguish variants seems to be multiple locus analysis of variable-number tandem repeats, which allowed the differentiation of each of the 28 isolates belonging to the pandemic clone (20). In spite of these observations, the genome of the pandemic strain seems to be very conserved; of 4,832 open reading frames (ORFs) found in V. parahaemolyticus strain RIMD2210633, which was isolated in 1996, all except 10 or fewer were found to be absent in two pandemic strains isolated in 2001 and compared by microarray (17). The V. parahaemolyticus pandemic clone was first detected in the region of Puerto Montt in southern Chile in 2004, where it has proliferated and caused approximately 8,000 clinical cases since that year (6, 11, 12). This rapid and massive proliferation of the clonal pandemic population in Chile offers a unique opportunity to study the evolution of a bacterial pathogen in its natural environment by detection and characterization of emerging bacterial variants. Here, we describe V. parahaemolyticus pandemic clone variants that contain a 42-kb extrachromosomal element corresponding to a linear plasmid prophage that enhances sensitivity to UV light and likely impairs their survival in the ocean. The increased sensitivity is probably due to phage replication induction, which may have an important role in bacterial mortality in coastal waters (36). The prophage found in the pandemic strain variants is closely related to the bacteriophage VHML of Vibrio harveyi (26) and VP882 of V. parahaemolyticus (unpublished data) (sequence accession number EF057797), and it likely corresponds to a new group of temperate phages with a linear genome with cohesive ends and prophages consisting of linear plasmids with terminal hairpins (i.e., telomeres). This group includes phages ΦHAP-1 of the marine Halomonas aquamarina (23), N15 of Escherichia coli (33), PY54 of Yersinia enterocolitica (15), and pKO2 of Klebsiella oxytoca (4, 15, 33).
MATERIALS AND METHODS
Strains, growth, and characterization.
Bacterial strains, except those from the summer of 2008, have been previously described (10-12). Thirteen pandemic V. parahaemolyticus strains isolated in 2008 were included in this study: eight clinical isolates and five isolates from shellfish. Isolation growth and characterization were performed as previously described (10).
DNA analysis.
Presence or absence of the 42-kb plasmid was assessed by two methods: PFGE and alkaline DNA extraction. PFGE was performed as described previously (13), except that digestion with restriction enzymes was omitted. DNA alkaline extraction was performed using a Plasmid Mini Kit I (Omega Bio Tek, GA). Direct genome restriction analysis (DGREA) was performed as described previously (11). The presence or absence of the 42-kb plasmid DNA from different isolates was determined based on the restriction pattern from digestion with HaeIII (Promega, WI), performed as described by the manufacturer. Analysis of the presence or absence of the 42-kb plasmid in the prototype strain of other DGREA groups of V. parahaemolyticus was performed by PCR amplification using primers for ORF 1, ORF 24, and ORF 34 of VHML. Viral DNA was prepared from cell lysates obtained after mitomycin C induction at 30 ng/ml, as described previously (26). After centrifugation at 5,000 × g for 10 min, the supernatant was passed through a 0.2-μm-pore-size filter, and the filtrate was centrifuged at 140,000 × g for 50 min in a rotor 55.2 Ti ultracentrifuge (Beckman, CA). The pellet was suspended in synthetic seawater (23.4 g/liter NaCl, 24.7 g/liter MgSO4·7H2O, 1.5 g/liter KCl, and 1.43 g/liter CaCl2·2H2O, pH 6.5; 100 μl per 40 ml of lysate) and then incubated with DNase (2 μg/ml) and RNase (100 μg/ml) for 2 h at 37°C. For DNA extraction, the phage particle suspension was treated with 500 μg/ml proteinase K for 15 min at 37°C, sodium dodecyl sulfate was added at a final concentration of 0.5% (wt/vol), and the solution was incubated for 45 min at 65°C to break down the phage coat. The solution was then extracted twice with phenol-chloroform. Finally, the DNA was precipitated by adding 1/10 volume of 3 M sodium acetate, pH 5.0, and 2 volumes of absolute ethanol at −20°C. After the pellet was washed with 70% ethanol, it was dissolved in TE buffer (0.01 M Tris, 0.001 M EDTA, pH 7.5).
Cloning, PCR amplification, and sequencing.
Cloning was performed with a TOPO Shotgun Subcloning Kit (Invitrogen) using the 42-kb extrachromosomal DNA obtained after alkaline DNA extraction. Clones were sequenced by Macrogen (Seoul, Korea) using M13 forward and reverse primers. PCR amplification of VHML genes was performed with the primers listed in Table 1, which were designed according to the published sequence for this phage (26). For the amplification reactions, approximately 10 ng of bacterial DNA was incubated with a 0.2 mM concentration of each deoxynucleotide triphosphate, 2.5 mM MgCl2, and 0.05 U/μl of Taq polymerase. The thermal cycling consisted of one cycle of 94°C for 5 min, followed by 30 cycles of amplification at 94°C, 60°C, and 72°C for 1 min each and a final cycle of 72°C for 5 min. Amplicons were directly sequenced by Macrogen using forward and reverse primers.
TABLE 1.
Primers for PCR amplification of ORFs found in VHML and in clones of the 42-kb plasmid
| Primer | Sequence |
|---|---|
| VHML ORF 1f | GGCGAACACATCAAGGAACT |
| VHML ORF 1r | TTTCCACCTTGGAATCAAGC |
| VHML ORF 23f | CAAAGGGGGTATTCGTTTCA |
| VHML ORF 23r | CCGACAGAACTGGTGAATGA |
| VHML ORF 24f | CGGTGATTACAAAGCGGATT |
| VHML ORF 24r | CGGGTCAGTGGTATGGTTTT |
| VHML ORF 30f | TATGCAGGGGCTCCAAATAG |
| VHML ORF 30r | ATTTTTGCGGCAGTCACATC |
| VHML ORF 34f | CTTTTGGCGACATGGCTTAC |
| VHML ORF 34r | GCTTTCCATAACTCCGCATC |
| VHML ORF 39f | AATACCTACACGGCGCAGAA |
| VHML ORF 39r | GAAGGCGGCTGGATAATATG |
| VHML ORF 40f | AATCCGAGCAATGAACCTGT |
| VHML ORF 40r | CCCGGTCGGATTTTTAGTCT |
| VHML ORF 52f | AGCATTGTTCCACTCGCTTC |
| VHML ORF 52r | GGCCGCAAACTGGTTATTTA |
| 42-kb Seq 37f | CATTTTATCGGCGGCATAGT |
| 42-kb Seq 37r | CTTAAAGTGCGGGGTCTGAT |
| 42-kb Seq 22f | GCCTATTCCTTCCAGCCTTA |
| 42-kb Seq 22r | AGCATTGGCGAGATTTCATC |
UV irradiation.
For UV induction, 50 ml of exponentially growing bacteria at an A600 of 0.2 was irradiated in a petri dish under mild reciprocal shaking with a 310-nm wavelength 6-W lamp for 12.5 min at a distance of 25 cm inside a biological safety cabinet. Specific death rates were calculated from survival curves obtained after UV irradiation under the same conditions, except that cells were diluted to 104 CFU/ml in synthetic seawater. Intensity of the UV radiation in the petri dish was 0.18 W/m2, as measured with a iodide/iodate actinometer as previously described (32). The same conditions were used for solar irradiation, except that the culture was exposed at midday in an open environment. To avoid contamination, an ascending filtered airflow was maintained during irradiation. Water that evaporated during irradiation, assessed based on the decrease in volume observed on the dish walls, was replaced with distilled water. The number of CFU was determined by plating in soft agar by a modified pour-plate method (2): 1 ml of the appropriate dilution was mixed with 9 ml of LB medium with 3% NaCl and 1.5% agar at 45°C and immediately poured in a petri dish. Once hardened, the agar was covered with 10 ml of LB medium with 3% NaCl and 1.5% agar at 45°C so that nonsubmerged bacteria could not spread on the surface of the agar. Once the top agar had hardened, plates were incubated overnight at 37°C.
Other procedures.
Induction with mitomycin C was performed as described previously (26). Curing of the 42-kb plasmid was performed by growing the cells in LB medium with 3% NaCl containing 150 μg/ml of acrydine orange (Merck) as described previously (19); 10 colonies were subjected to PCR analysis for five plasmid genes (protelomerase, capsid protein, portal protein, tail contractile sheath protein, and baseplate spike protein), and two genes were found to lack the plasmid. Cured cells were also obtained by analysis of surviving bacteria after UV irradiation at a 0.05% survival. Electron microscopy was performed using the lysates of mitomycin C-induced bacteria after elimination of cell debris by centrifugation at 10,000 × g for 10 min and filtration through 0.2-μm-pore-size filters. After sample deposition on pioloform-carbon-coated 400-mesh copper grids, staining was performed with 2% aqueous uranyl acetate in HEPES buffer. Samples were then analyzed using a JEM-1010 transmission electron microscope (JOEL, Tokyo, Japan) at 80 kV.
Restriction site mapping was performed by digestion with restriction enzymes NheI, HindIII, and SphI from New England Biolabs (Massachusetts), according to the manufacturer's instructions. Samples were electrophoresed on a 0.4% agarose gel (Winkler) for 15 h at 20 V using a high-molecular-weight DNA marker (Invitrogen, CA) to distinguish fragments larger than 10 kb and on a 1% agarose gel for 2 h at 70 V using the GeneRuler 1-kb DNA ladder (Fermentas) as a marker for fragments of less than 10 kb.
Nucleotide sequence accession numbers.
Sequences of the phage genes have been deposited at GenBank under accession numbers EU647248 to EU647255.
RESULTS
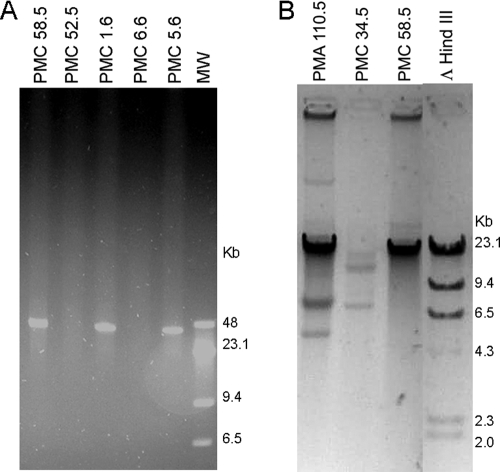
Analysis of 112 strains of V. parahaemolyticus O3:K6 pandemic clonal complex obtained from clinical and shellfish samples during the Puerto Montt outbreaks each summer between 2004 and 2008 revealed the emergence of variants containing a 42-kb extrachromosomal DNA in 2005. This DNA was detected after both PFGE of bacterial DNA (Fig. 1A) and conventional agarose gel electrophoresis of DNA extracted by alkaline lysis (Fig. 1B). Strains from Southeast Asia and the initial outbreaks in Antofagasta in 1998 lacked this plasmid. The percentage of bacterial isolates containing the prophage increased from 23% in 2005 to 33% in 2006 decreased to an undetectable level in 2007 but reappeared in 2008 (Table 2).
FIG. 1.
Detection of extrachromosomal DNA in strains of the O3:K6 pandemic clonal complex isolated in Chile. (A) PFGE of bacterial DNA from strains with (PMCs 58.5, 1.6, and 5.6) and without (PMCs 52.5 and 6.6) the extrachromosomal DNA. Lane MW, λ DNA and λ HindIII DNA molecular weight markers. (B) Gel electrophoresis of DNA extracted after alkaline hydrolysis from strains with (PMCs 110.5 and 58.5) and without (PMC 34.5) the extrachromosomal DNA. Lane ΛHindIII, λ HindIII DNA molecular weight marker.
TABLE 2.
Presence of the 42-kb extrachromosomal DNA in V. parahaemolyticus strains of the pandemic clonal complex from clinical and shellfish samples
| Origin | No. of isolates by sample type
|
Total for sample group (no. of isolates)
|
% Positive in sample group | ||||
|---|---|---|---|---|---|---|---|
| Clinical
|
Shellfish
|
||||||
| Analyzed | Positive | Analyzed | Positive | Analyzed | Positive | ||
| Southeast Asia | 12 | 0 | 0 | 0 | 12 | 0 | 0 |
| Antofagasta 1998 | 9 | 0 | 4 | 0 | 13 | 0 | 0 |
| Puerto Montt 2004 | 9 | 0 | 4 | 0 | 13 | 0 | 0 |
| Puerto Montt 2005 | 9 | 1 | 4 | 2 | 13 | 3 | 23 |
| Puerto Montt 2006 | 11 | 6 | 10 | 1 | 21 | 7 | 33 |
| Puerto Montt 2007 | 27 | 0 | 0 | 0 | 27 | 0 | 0 |
| Puerto Montt 2008 | 8 | 2 | 5 | 0 | 13 | 2 | 15 |
Restriction fragment length polymorphism analysis (with HaeIII) of the plasmid from different isolates showed identical patterns in every isolate analyzed (data not shown). Five clones obtained by random cloning of the 42-kb DNA extracted from the chosen prototype isolate (PMC 58.5) were sequenced, and the insert sequences revealed 81 to 99% sequence similarity with VHML, a phage found in V. harveyi (26). PCR amplification with primers designed according to the VHML nucleotide sequence revealed the presence of five key putative genes of VHML encoding protelomerase, capsid protein, portal protein, tail contractile sheath protein, and baseplate spike protein. Sequencing of three of these amplicons showed 93 to 98% similarity with the corresponding VHML genes (Table 3).
TABLE 3.
Similarity of the 42-kb extrachromosomal DNA with bacteriophage VHML
| Clone or PCR amplicon | ORF(s) in VHML | Putative function | Identity (no. of identical residues/total no. of residues [%]) |
|---|---|---|---|
| 58p10-8b | 37 | Transposase | 291/355 (81) |
| 58p10-8d | 53 | Unknown | 217/236 (92) |
| 58p10-9 | 10 | Recombination protein | 480/482 (99) |
| 58p10-11h | 4 | DNA primase | 581/629 (92) |
| 58p10-12f | 21, 22 | Terminase small and large subunits | 584/602 (97) |
| Amplicon 1 | 1 | Protelomerase | 508/542 (93) |
| Amplicon 2 | 24 | Capsid protein | 549/559 (98) |
| Amplicon 3 | 34 | Tail fiber protein | 283/297 (95) |
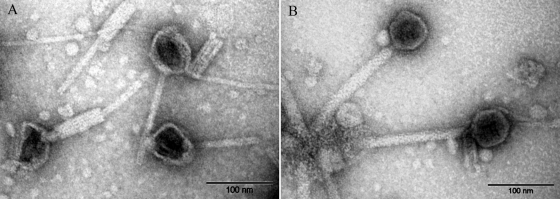
Mitomycin C treatment of strains that contained the 42-kb plasmid resulted in cell lysis within 1 h after treatment and the production of phage particles showing the morphology of myoviruses: 60-nm icosahedrical heads with a straight tail 160 nm long and 6 nm wide, enclosed by an apparent sheath 15 nm wide (Fig. 2). The amount of 42-kb DNA recovered from the induced lysate particles corresponded to approximately 200 phages per cell, or 20 to 100 times the amount of plasmid DNA observed after PFGE or alkaline DNA extraction in noninduced cells. Infective phages (up to 109 PFU/ml) were observed upon infection with the mitomycin C lysate in some but not all pandemic isolates lacking the 42-kb plasmid. Plaque-forming units were also observed in plasmid-cured PMC 58.5 strains (see below). Resistant clones that appeared after infection contained the 42-kb plasmid when examined by DNA alkaline extraction (results not shown). Altogether these observations show that the 42-kb plasmid corresponds to a temperate phage with a plasmid prophage.
FIG. 2.
Negative staining of phage particles observed after induction with mitomycin C of V. parahaemolyticus strain PMC 58.5.
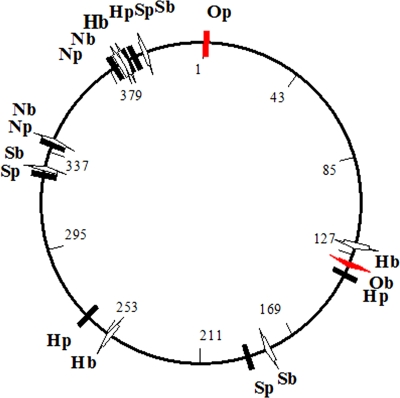
The presence of a putative protelomerase gene suggested that the prophage could consist of a linear DNA with covalently closed terminal hairpin ends instead of a closed circular DNA, as originally assumed because of the recovery of the plasmid after alkaline lysis. Terminal hairpins ends (telomeres) are observed in the prophages of the telomeric temperate phage group (33). These linear plasmids are recovered upon extraction with plasmid purification kits based on alkaline denaturation as the telomeres join the complementary strands and allow rapid renaturation after alkaline denaturation, which occurs with circular closed DNA. The failure to recover the phage DNA after alkaline lysis occurs because the encapsulated DNA of the telomeric phage group is a linear molecule that lacks telomeric ends. Genes in phage and plasmid DNA are circularly permuted. Restriction site mapping of both phage and prophage DNA with NheI, HindIII, and SphI showed the linear nature and circular permutation of the pandemic V. parahaemolyticus plasmid and phage DNA. The schematic shown in Fig. 3 is predicted if both linear DNAs are generated from a common circular molecule by a single cut at the indicated positions, a replication mechanism observed with telomeric phages (33).
FIG. 3.
Restriction maps of plasmid and bacteriophage DNA obtained from V. parahaemolyticus strain PMC 58.5. N, H, and S correspond to NheI, HindIII, and SphI restriction enzymes sites, respectively. A hypothetical circular map is shown in the middle, with the possible ends shown with a red arrow and the restriction sites with a black line. b, bacteriophage; p, plasmid.
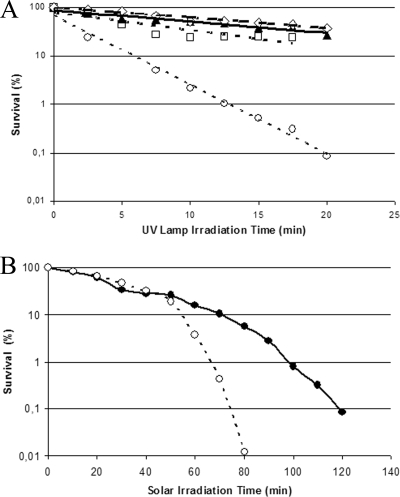
UV light irradiation may induce prophage replication, resulting in bacterial lysis. Like mitomycin C, UV irradiation of PMC 58.5 cultures as described in Materials and Methods induced lysis and phage particle production within 1 h (data not shown). In view of these results, we explored the possibility that the presence of the inducible prophage might increase the sensitivity of V. parahaemolyticus pandemic strains to UV light and solar radiation. Survival curves of V. parahaemolyticus pandemic strains obtained after UV irradiation either from a UV lamp or the sun showed a specific death rate 7 to 15 times higher in strains containing the 42-kb plasmid (Fig. 4). A single representative experiment is shown in Fig. 4A though several experiments were done in each case; the average specific death rates from four independent experiments were −0.175 ± 0.029 min−1 and 0.027 ± 0.011 min−1 for PMC 58.5 and PMA 37.5, respectively. Plasmid curing of PMC 58.5 by incubation with acrydine orange decreased the death rate to 0.019 ± 0.006 min−1. Also, survivors without the plasmid obtained after UV irradiation of PMC 58.5 showed a lower sensitivity with a death rate of 0.014 ± 0.005 min−1. Bacteria maintained under irradiation conditions, but protected from UV with a 6-mm-thick glass plate, showed 100% survival. Specific death rates measured under solar irradiation in different experiments (Fig. 4B) could not be directly compared due to the inability to reproduce solar irradiation conditions; however, in every experiment when the survival of PMA 37.5 was 10%, the survival of PMC 58.5 was between 0.2% and 1%.
FIG. 4.
Effect of irradiation from a 310-nm UV lamp (A) and from the sun (B) on the survival of strains of the O3:K6 pandemic clonal complex with and without the 42-kb DNA plasmid, PMC 58.5 (○) and PMA 37.5 (•), respectively. ⧫, PMC 58.5 cured with acrydine orange; ⋄, PMC 58.5 spontaneously cured after UV irradiation.
The presence of the 42-kb plasmid appeared to be specific to the O3:K6 pandemic clonal group. We analyzed prototype strains from the other DGREA groups isolated in Chile by PCR amplification for three genes in the 42-kb plasmid, and no strain contained the plasmid (10; also E. Harth, L. Matsuda, C. Hernández, M. L. Rioseco, J. Romero, N. González-Escalona, and R. T. Espejo, submitted for publication).
DISCUSSION
Our findings revealed the emergence of a variant from the pandemic V. parahaemolyticus O3:K6 clonal complex in its natural environment. This variant is characterized by the presence of a 42-kb plasmid. Mitomycin C and UV irradiation induction of infectious phage containing DNA with the same sequences of the plasmid indicates that the plasmid corresponds to a prophage. The different characteristics of prophage and phage DNA upon alkaline extraction and the circular permutation of restriction enzyme sites indicate that the prophage likely belongs to a group of temperate phages with a linear genome, with or without cohesive ends, whose prophages are linear plasmids with terminal hairpins (i.e., telomeres) (4, 15, 33). The presence of terminal hairpins (with covalent bonds between complementary strands) in the prophage would allow rapid renaturation after alkaline denaturation of the complementary strands, as it occurs with circular closed DNA. Linear phage DNA would not rapidly renature after denaturation and would not be recovered upon alkaline extraction.
The high sequence similarity of the 42-kb plasmid to bacteriophage VHML of V. harveyi also suggested that the 42-kb DNA was a prophage (26). Although VHML DNA contains putative genes characteristic of telomeric phages (23), the plasmidial nature of the prophage has not been shown; instead, integration of the prophage has been claimed (35). Unfortunately, VHML seems to have been lost (23), and we have been unable to obtain a starting culture for comparison in the detailed characterization of the 42-kb plasmid that is currently under way.
The absence of the 42-kb prophage in the examined Southeast Asian and early Chilean strains suggests that this DNA was introduced in part of the pandemic clone population along the Chilean coast. It is unlikely that a single cell infected with the phage propagated more rapidly than uninfected cells to reach 33% of the population, as observed in 2006. Instead, it seems more likely that the temperate phage propagated in the pandemic clonal population. The presence of sensitive and resistant strains within the clonal pandemic population suggests that a resistant population may be being selected. This phage lyses most infected bacteria but also lysogenizes some bacteria that become resistant when challenged with the same phage (data not shown). Although we can only speculate on the observed increase and decrease in the number of isolates with the 42-kb prophage, this fluctuation could be associated with the interaction between host and virus and their mutual dependence, as postulated by Jensen et al. (18) for V. cholerae. This case, however, adds further complexity because of death by UV solar radiation of lysogenized cells. The presence of the 42-kb prophage has a notable effect on the sensitivity of the pandemic strain to UV irradiation, including solar UV irradiation, due to prophage induction and cell lysis. This phenomenon is not unprecedented as prophage activation is the major lethal factor in the highly UV-sensitive Shewanella oneidensis MR1 (31). Death of lysogenic cells by temperate phage induction is one mode of bacterial death due to infection by bacteriophages (16, 36) and a substantial cause of marine bacterial mortality (34). The importance of prophages in marine bacteria proliferation has been discussed in a recent review (30). A similar observation on the presence of a temperate prophage in a pathogenic strain of V. parahaemolyticus (WP1) and its potential association with higher sensitivity to UV radiation was reported 18 years ago (27). Infection by the phage and the higher sensitivity of lysogenized pandemic strains to solar radiation shown here may have critical consequences on the persistence and propagation of the strain in open seawater and could be one cause for the apparent changes in the bacterial load of pandemic V. parahaemolyticus strains in seafood associated with diarrhea outbreaks.
Acknowledgments
We thank Stefan Hertwig for helpful comments on the manuscript and Jochen Reetz for electron microscopy of phage particles (both from the Federal Institute for Risk Assessment, Berlin, Germany). We also thank Natalia Peñaloza and Rafael Torres for performing the inactivation curves and measurement of UV fluence.
B. Zabala and K. García acknowledge scholarships from Deutscher Akademischer Austausch Diemst and Consejo Nacional de Ciencia y Tecnología, respectively. This work was supported in part by a grant from FONDECYT (no. 1070658).
Footnotes
Published ahead of print on 16 January 2009.
REFERENCES
- 1.Alam, M. J., S. Miyoshi, and S. Shinoda. 2003. Studies on pathogenic Vibrio parahaemolyticus during a warm weather season in the Seto Inland Sea, Japan. Environ. Microbiol. 5:706-710. [DOI] [PubMed] [Google Scholar]
- 2.Berney, M., H. U. Weilenmann, J. Ihssen, C. Bassin, and T. Egli. 2006. Specific growth rate determines the sensitivity of Escherichia coli to thermal, UVA, and solar disinfection. Appl. Environ. Microbiol. 72:2586-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhuiyan, N. A., M. Ansaruzzaman, M. Kamruzzaman, K. Alam, N. R. Chowdhury, M. Nishibuchi, S. M. Faruque, D. A. Sack, Y. Takeda, and G. B. Nair. 2002. Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40:284-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens, S. R., E. B. Gilcrease, W. M. Huang, K. L. Bunny, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186:1818-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhury, A., M. Ishibashi, V. D. Thiem, D. T. N. Tuyet, T. Van Tung, B. T. Chien, L. von Seidlein, D. G. Canh, J. Clemens, D. D. Trach, and M. Nishibuchi. 2004. Emergence and serovar transition of Vibrio parahaemolyticus pandemic strains isolated during a diarrhea outbreak in Vietnam between 1997 and 1999. Microbiol. Immunol. 48:319-327. [DOI] [PubMed] [Google Scholar]
- 6.Cordova, J. L., J. Astorga, W. Silva, and C. Riquelme. 2002. Characterization by PCR of Vibrio parahaemolyticus isolates collected during the 1997-1998 Chilean outbreak. Biol. Res. 35:433-440. [DOI] [PubMed] [Google Scholar]
- 7.DePaola, A., C. A. Kaysner, J. Bowers, and D. W. Cook. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66:4649-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePaola, A., J. L. Nordstrom, J. C. Bowers, J. G. Wells, and D. W. Cook. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69:1521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePaola, A., J. Ulaszek, C. A. Kaysner, B. J. Tenge, J. L. Nordstrom, J. Wells, N. Puhr, and S. M. Gendel. 2003. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69:3999-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuenzalida, L., L. Armijo, B. Zabala, C. Hernandez, M. L. Rioseco, C. Riquelme, and R. T. Espejo. 2007. Vibrio parahaemolyticus strains isolated during investigation of the summer 2006 seafood related diarrhea outbreaks in two regions of Chile. Int. J. Food Microbiol. 117:270-275. [DOI] [PubMed] [Google Scholar]
- 11.Fuenzalida, L., C. Hernandez, J. Toro, M. L. Rioseco, J. Romero, and R. T. Espejo. 2006. Vibrio parahaemolyticus in shellfish and clinical samples during two large epidemics of diarrhoea in southern Chile. Environ. Microbiol. 8:675-683. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Escalona, N., V. Cachicas, C. Acevedo, M. L. Rioseco, J. A. Vergara, F. Cabello, J. Romero, and R. T. Espejo. 2005. Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg. Infect. Dis. 11:129-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Escalona, N., J. Romero, and R. T. Espejo. 2005. Polymorphism and gene conversion of the 16S rRNA genes in the multiple rRNA operons of Vibrio parahaemolyticus. FEMS Microbiol. Lett. 246:213-219. [DOI] [PubMed] [Google Scholar]
- 14.Hara-Kudo, Y., K. Sugiyama, M. Nishibuchi, A. Chowdhury, J. Yatsuyanagi, Y. Ohtomo, A. Saito, H. Nagano, T. Nishina, H. Nakagawa, H. Konuma, M. Miyahara, and S. Kumagai. 2003. Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan. Appl. Environ. Microbiol. 69:3883-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertwig, S., I. Klein, V. Schmidt, S. Beck, J. A. Hammerl, and B. Appel. 2003. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 331:605-622. [DOI] [PubMed] [Google Scholar]
- 16.Hewson, I., and J. A. Fuhrman. 2007. Characterization of lysogens in bacterioplankton assemblages of the southern California borderland. Microb. Ecol. 53:631-638. [DOI] [PubMed] [Google Scholar]
- 17.Izutsu, K., K. Kurokawa, K. Tashiro, S. Kuhara, T. Hayashi, T. Honda, and T. Iida. 2008. Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilobase pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect. Immun. 76:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, M. A., S. M. Faruque, J. J. Mekalanos, and B. R. Levin. 2006. Modeling the role of bacteriophage in the control of cholera outbreaks. Proc. Natl. Acad. Sci. USA 103:4652-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keyhani, J., E. Keyhani, F. Attar, and A. Haddadi. 2006. Sensitivity to detergents and plasmid curing in Enterococcus faecalis. J. Ind. Microbiol. Biotechnol. 33:238-242. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, B., Y. Sekine, H. Takahashi, Y. Tanaka, H. Obata, A. Kai, S. Morozumi, and T. Fujii. 2008. Multiple-locus variable-number of tandem-repeats analysis distinguishes Vibrio parahaemolyticus pandemic O3:K6 strains. J. Microbiol. Methods 72:313-320. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Urtaza, J., A. Lozano-Leon, A. Depaola, M. Ishibashi, K. Shimada, M. Nishibuchi, and E. Liebana. 2004. Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J. Clin. Microbiol. 42:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto, C., J. Okuda, M. Ishibashi, M. Iwanaga, P. Garg, T. Rammamurthy, H. C. Wong, A. Depaola, Y. B. Kim, M. J. Albert, and M. Nishibuchi. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mobberley, J. M., R. N. Authement, A. M. Segall, and J. H. Paul. 2008. The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J. Virol. 82:6618-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair, G. B., T. Ramamurthy, S. K. Bhattacharya, B. Dutta, Y. Takeda, and D. A. Sack. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasu, H., T. Iida, T. Sugahara, Y. Yamaichi, K. S. Park, K. Yokoyama, K. Makino, H. Shinagawa, and T. Honda. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oakey, H. J., B. R. Cullen, and L. Owens. 2002. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J. Appl. Microbiol. 93:1089-1098. [DOI] [PubMed] [Google Scholar]
- 27.Ohnishi, T., and K. Nozu. 1986. Induction of phage-like particles from a pathogenic strain of Vibrio parahaemolyticus by mitomycin C. Biochem. Biophys. Res. Commun. 141:1249-1253. [DOI] [PubMed] [Google Scholar]
- 28.Okuda, J., M. Ishibashi, E. Hayakawa, T. Nishino, Y. Takeda, A. K. Mukhopadhyay, S. Garg, S. K. Bhattacharya, G. B. Nair, and M. Nishibuchi. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okura, M., R. Osawa, A. Iguchi, E. Arakawa, J. Terajima, and H. Watanabe. 2003. Genotypic analyses of Vibrio parahaemolyticus and development of a pandemic group-specific multiplex PCR assay. J. Clin. Microbiol. 41:4676-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul, J. H. 2008. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2:579-589. [DOI] [PubMed] [Google Scholar]
- 31.Qiu, X., J. M. Tiedje, and G. W. Sundin. 2005. Genome-wide examination of the natural solar radiation response in Shewanella oneidensis MR-1. Photochem. Photobiol. 81:1559-1568. [DOI] [PubMed] [Google Scholar]
- 32.Rahn, R. O., J. Bolton, and M. I. Stefan. 2006. The iodide/iodate actinometer in UV disinfection: determination of the fluence rate distribution in UV reactors. Photochem. Photobiol. 82:611-615. [DOI] [PubMed] [Google Scholar]
- 33.Rybchin, V. N., and A. N. Svarchevsky. 1999. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol. 33:895-903. [DOI] [PubMed] [Google Scholar]
- 34.Suttle, C. A. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801-812. [DOI] [PubMed] [Google Scholar]
- 35.Vidgen, M., J. Carson, M. Higgins, and L. Owens. 2006. Changes to the phenotypic profile of Vibrio harveyi when infected with the Vibrio harveyi myovirus-like (VHML) bacteriophage. J. Appl. Microbiol. 100:481-487. [DOI] [PubMed] [Google Scholar]
- 36.Weinbauer, M. G., and C. A. Suttle. 1996. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the Gulf of Mexico. Appl. Environ. Microbiol. 62:4374-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong, H. C., S. H. Liu, T. K. Wang, C. L. Lee, C. S. Chiou, D. P. Liu, M. Nishibuchi, and B. K. Lee. 2000. Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl. Environ. Microbiol. 66:3981-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]