Abstract
Gluconacetobacter diazotrophicus utilizes plant sucrose with a constitutively expressed levansucrase (LsdA), producing extracellular levan, which may be degraded under energetically unfavored conditions. Reverse transcriptase-PCR analysis revealed that lsdA and the downstream exolevanase gene (lsdB) form an operon. lsdB transcription was induced during growth with low fructose concentrations (0.44 to 33 mM) and repressed by glucose. Transport of LsdB to the periplasm involved N-terminal signal peptide cleavage. Type II secretion mutants failed to transfer LsdB across the outer membrane, impeding levan hydrolysis.
Bacterial levan [β(2,6)-linked polyfructan] and inulin [β(2,1)-linked polyfructan] are synthesized from extracellular sucrose to play important habitat-related functions and occasionally serve as energy reserves (5). The regulation of fructan metabolism is best studied with gram-positive bacteria. In the soil resident Bacillus subtilis, the genes for simultaneous levan synthesis (sacB, encoding levansucrase) and degradation (levB, encoding endolevanase) are cotranscribed via a sucrose-inducible antitermination mechanism (9, 21), whereas exolevanase (sacC) transcription is induced by fructose and controlled by CcpA-dependent carbon catabolite repression (CCR) (16, 17). The oral species Streptococcus mutans and Actinomyces naeslundii produce inulin or levan by use of constitutively expressed fructosyltransferases (6, 14). Fructose activates S. mutans exofructanase transcription, but glucose suppresses the activation via a CcpA-independent CCR mechanism (26, 27). A. naeslundii secretes a sucrose-inducible exolevanase and a fructose-inducible fructanase (7). In gram-negative bacteria, the levansucrase genes so far characterized are expressed constitutively, but the conditions controlling levan degradation remain unknown. No experimental data are available on the secretion process of bacterial fructanases, except for B. subtilis SacC, which follows a signal-peptide-dependent route (15).
The gram-negative, nitrogen-fixing endophyte Gluconacetobacter diazotrophicus secretes a constitutively expressed levansucrase (LsdA), allowing sucrose utilization and levan production (10, 12). LsdA is transferred to the periplasm via a signal-peptide-dependent pathway (11) and requires the type II machinery for extracellular release (4). The gene downstream of lsdA encodes a fructose-releasing exolevanase (LsdB), as determined in recombinant experiments (19, 20). In this study, we investigated the catabolic and genetic requirements for LsdB synthesis and secretion as a step forward in elucidating the role of levan metabolism in G. diazotrophicus.
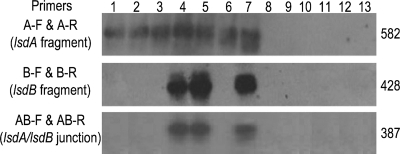
The transcriptional linkage of the chromosomal lsdAB gene cluster (Fig. 1) was examined by reverse transcriptase (RT) PCR in G. diazotrophicus SRT4 grown to early exponential phase in LGIE medium (3) containing sucrose (1%), glycerol (1%), or fructose (0.5 and 1%) as the sole carbon source and two sugar combinations (fructose [0.1%] plus glycerol [0.5%] or fructose [0.1%] plus glucose [0.5%]). Total RNA samples (1 μg) treated with RQ1 DNase (Promega) were reacted with three set of primers designed to identify messengers from lsdA, lsdB, and/or the intercistronic region (Table 1) using the Access RT-PCR system (Promega). As shown in Fig. 2, the reaction with the lsdA primer pair amplified the internal 582-bp fragment for all assayed substrate conditions, confirming the constitutive character of lsdA expression (12). In contrast, the expected 428-bp lsdB fragment was observed exclusively in cells cultivated with lower fructose concentrations, either alone (0.5%) or combined (0.1%) with glycerol to support growth. The replacement of glycerol by glucose suppressed lsdB transcription, as inferred from the absence of a reaction product. lsdA and lsdB were cotranscribed in the two induced treatments as the 387-bp fragment spanning the intergenic region was generated. The three RT-PCR products matched in size the corresponding fragments amplified from chromosomal DNA. No amplification was observed in the RT-untreated RNA samples. Southern hybridizations with 32P-labeled DNA probes containing lsdA, lsdB, or the intergenic region from the plasmid pALS5 (3) confirmed the specificity of the RT-PCRs. Further RT-PCR experiments were performed for detection of bicistronic lsdAB messengers in cells grown on increasing amounts of fructose (0.002 to 0.8%). The lowest concentration needed to induce lsdB transcription was 0.008% (0.44 mM), whereas the threshold value for repression was 0.6% (33.3 mM). Similarly, fructose acts as an inducer at low concentrations and as a corepressor at high concentrations to regulate the expression of exofructanases in the fructan-producing species B. subtilis (16), S. mutans (27), and A. naeslundii (7).
FIG. 1.
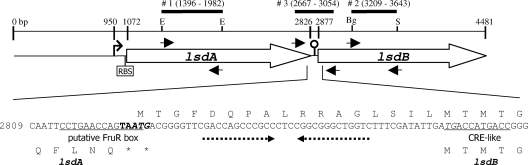
Organization of Gluconacetobacter diazotrophicus lsdAB operon and relevant features of the intergenic sequence. Numbers match the nucleotide position for strain SRT4 sequence in DDBJ/EMBL/GenBank databases under accession number L41732. The lsdA (1072 to 2826) and lsdB (2877 to 4481) coding regions are represented by thick arrows. The bent arrow indicates the predicted transcription start site (T residue at position 950). RBS denotes the ribosome binding site AGGAGGA (1061 to 1067). The LsdA C-terminal and LsdB N-terminal sequences are in single-letter code. The two in-frame lsdB ATG codons are in italics. The lsdA stop codons are labeled with asterisks. The overlapping stop-start codon TAATG is in boldface. The putative FruR box and CRE-like sequences are underlined. The 12-bp inverted-repeat sequence is indicated by opposing dashed arrows and corresponds to the hairpin structure in the map. Small arrowheads below and above lsdA and lsdB refer the location of the primers used for RT-PCRs. The Southern hybridization probes are represented by thick lines and correspond to the 583-bp EcoRI fragment (#1, lsdA), the 434-bp BglII-SacII fragment (#2, lsdB), and the 387-bp PCR band (#3, intergenic region). Restriction sites are as follows: E, EcoRI; Bg, BglII; and S, SacII.
TABLE 1.
Primers used in RT-PCRs
| Primer | Sequence | Applicationa |
|---|---|---|
| A-F | 5′GAATTCCCTGCCGGCGCAACTGACC | Sense lsdA (1395) |
| A-R | 5′GTTCTGCGCACCGTTCTGATACAGC | Antisense lsdA (1977) |
| B-F | 5′GAGGAGATCTTTTCCGGAAGCCTGG | Sense lsdB (3204) |
| B-R | 5′GTCCAGCTTCAGCGGAACCAGTGTG | Antisense lsdB (3632) |
| AB-F | 5′CCTGGGCGGCTATGGCGATATTCCG | Sense lsdA/lsdB junction (2667) |
| AB-R | 5′CGTTCGGATCGTTCATCCAGTACGC | Antisense lsdA/lsdB junction (3054) |
Each number in parentheses refers to the position of the primer's 5′ end in the SRT4 lsdAB operon sequence as available in the DDBJ/EMBL/GenBank databases under accession number L41732.
FIG. 2.
RT-PCR-Southern blot results for transcription of lsdA, lsdB, and the lsdA/lsdB junction. Lanes 1 to 6 correspond to RT-PCRs with RNA samples from cells grown on 1% sucrose, 1% glycerol, 1% fructose, 0.5% fructose, 0.1% fructose plus 0.5% glycerol, and 0.1% fructose plus 0.5% glucose, respectively. Lane 7 corresponds to PCRs with chromosomal DNA (positive control). Lanes 8 to 13 correspond to PCRs with RT-untreated RNAs for lines 1 to 6 (negative controls). The fragment sizes (bp) on the right are assigned according to the operon sequence and matched with the migration of an unlabeled 1-kb DNA ladder (Promega) used as molecular weight markers. Hybridization probes #1 for lsdA, #2 for lsdB, and #3 for the lsdA/lsdB junction are represented in Fig. 1.
The operon transcription start site was predicted at the T residue 122 bp upstream of lsdA with a score of 0.92 using a neural-network method (22). Since no potential ribosome-binding site was detected in the 50-bp intergenic region, the sequence AGGAGGA centered 8 nucleotides upstream of lsdA should mediate translation of the bicistronic mRNA. Translational coupling has been documented for intercistronic distances of even 60 nucleotides (18). Alternatively, lsdAB translation may be coupled at the UAAUG overlapping stop-start fusion codon (Fig. 1).
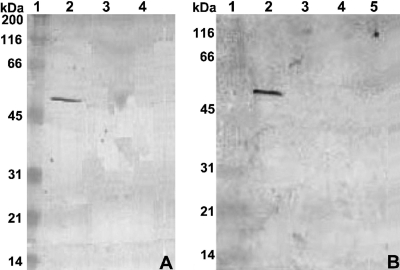
In Western blotting (Fig. 3A), a rabbit polyclonal antibody against LsdB produced in Escherichia coli (20) reacted with the expected 54-kDa protein in the culture supernatant of wild-type SRT4 grown to stationary phase with 0.1% fructose plus 1% glycerol but not when glycerol was replaced by glucose to suppress lsdB transcription. The protein was not detected in the lsdA::nptII mutant AD1 previously constructed by allelic exchange mutagenesis (3), indicating that insertional disruption of lsdA resulted in the inactivation of the gene downstream. This characteristic is consistent with that of polar mutation within an operon.
FIG. 3.
Western blot analysis of LsdB secretion by G. diazotrophicus. Cells were grown with 0.1% fructose plus 1% glycerol, except in blot A, lane 4. Supernatant proteins separated by SDS-PAGE were probed with anti-LsdB polyclonal antibody. Blot A lanes: 2, wild-type SRT4; 3, lsdA::nptII-ble mutant AD1; 4, SRT4 with 0.5% glucose replacing glycerol (negative control). Blot B lanes: 2, SRT4; 3, gspG::nptII-ble mutant AD6; 4, gspO::nptII-ble mutant AD8; 5, gspF::nptII mutant AD9. Lane 1 in both blots corresponds to the prestained broad-range protein marker from New England Biolabs.
G. diazotrophicus mutants AD6, AD8, and AD9, unable to secrete LsdA due to nptII insertions in the type II secretion (gsp) operon (4), were grown with 0.1% fructose plus 1% glycerol and assayed for LsdB secretion. None of the mutants released extracellular LsdB, as judged by the lack of levan-hydrolyzing activity (Table 2) or antibody recognition (Fig. 3B) in the culture supernatants. In contrast, LsdB activity in the soluble extracts of sonicated cells increased about threefold for the mutants in comparison with that of SRT4 (Table 2). Neither strain showed levanase activity in washed cell debris. Mature LsdB is unlikely to attach cell membranes, considering that no transmembrane regions were predicted using the method described in reference 13). The three mutants failed to hydrolyze 1-kestose [β-d-fructofuranosyl-(2→1)-β-d-fructofuranosyl-(2→1)-α-d-glucopyranoside], inulin, or levan added to liquid LGIE medium with glycerol as a growth-supporting carbon source.
TABLE 2.
Enzyme assay of LsdB secretion in G. diazotrophicusa
| Strain/mutant/transconjugant | Intracellular LsdB activity (U mg−1) | Extracellular LsdB activity (U mg−1) |
|---|---|---|
| SRT4 | 0.25 ± 0.2 | 14.24 ± 0.9 |
| AD6 | 0.75 ± 0.2 | 0.33 ± 0.3 |
| AD8 | 0.73 ± 0.1 | 0.52 ± 0.2 |
| AD9 | 0.69 ± 0.3 | 0.71 ± 0.3 |
| SRT4(pALS198) | 1.97 ± 0.8 | 40.53 ± 2.2 |
| AD8(pALS198) | 2.75 ± 0.9 | 1.61 ± 0.6 |
One unit is defined as the amount of LsdB releasing 1 μmol of fructose per min based on initial velocity measurements under the following reaction conditions: 1% levan in 0.1 M sodium acetate buffer, pH 5, at 30°C. Activity is given per mg of total soluble proteins, as measured by the Bradford procedure (8). Levan from Erwinia herbicola (Sigma) was used as the substrate. The values are means ± standard deviations (n = 5).
To characterize the intracellular and extracellular LsdB forms, the broad-range plasmid pALS198, carrying lsdB fused to a C-terminal polyhistidine tag between the constitutive nptII promoter and the rrnBt1-t2 terminator, was constructed and mobilized into SRT4 and the nonsecreting mutant AD8 via conjugative mating using a described procedure (3). His6-tagged LsdB (LsdB-His6) in the culture supernatant of SRT4(pALS198) and LsdB-His6 in the soluble cell extract of AD8(pALS198) were purified to homogeneity by Ni affinity chromatography using 0.1 M sodium phosphate buffer (pH 7) for polyhistidine tag binding and 0.15 M imidazole in 0.1 M sodium acetate (pH 5) for protein elution. The N terminus of purified intracellular and extracellular LsdB-His6 was sequenced by automated Edman degradation, yielding the sequence ADTPQWRPVL in both cases. This sequence perfectly matched residues 37 to 46 of encoded LsdB, ascertaining the removal of the predicted N-terminal Sec-dependent signal peptide during transfer across the cytoplasmic membrane (19). The specific activity was determined to be 53 ± 5 U mg−1 for extracellular LsdB-His6 and 51 ± 3 U mg−1 for the periplasmic form. Since the two values are similar, LsdB likely adopts its final conformation during transit through the periplasm. LsdB has a β-propeller fold, as established for members of glycoside hydrolase family 32 (1). This structural feature serves as a recognition signal for targeting of substrate periplasmic proteins to the type II secretion apparatus (24). LsdB transfer through the outer membrane appears to limit the secretion rate, considering that the plasmid pALS198 increased LsdB activity inside and outside the SRT4 cells about eight- and twofold, respectively (Table 2).
G. diazotrophicus metabolizes fructose through cytoplasmic reactions (2), but levan hydrolysis occurs extracellularly. A search in the type strain PAl5 genome sequence (GenBank accession number AM889285) identified two gene clusters presumably involved in fructose import and conversion to fructose-1P (loci GDI1186 to -1190) and fructose-6P (loci GDI0702 and -0703), as well as genes for transforming fructose-6P to either glucose-6P (locus GDI0286) or fructose-1,6-biphosphate (locus GDI1032). The four phosphorylated forms were produced by G. diazotrophicus cell extracts (2) and might represent intracellular signals operating to activate and suppress lsdB transcription. Another identified chromosomal gene (locus GDI2661) appears to encode a fructose repressor protein (FruR) with relatively strong identities (29 to 38% identical to 100 most-similar orthologs) to transcriptional regulators of the GalR-LacI family. The sequence CCTGAACCAGTAAT (mismatches underlined) overlapping the lsdA stop codon (Fig. 1) resembles the E. coli FruR-binding consensus sequence RSTGAAWCSNTHHW (23) and might serve as the cis-acting element mediating the fructose-dependent regulation of lsdB transcription. Alternatively, lsdB expression may be controlled by an antitermination mechanism, considering that an intercistronic stem-loop structure with free energy (ΔG0) of −18.3 kcal mol−1 and characteristics of Rho-independent terminators (Fig. 1) was predicted using the MFOLD program (28). However, BLAST searches failed to identify G. diazotrophicus homologs of the transcriptional antiterminators BglG from E. coli and SacY from B. subtilis, and no putative ribonucleic antiterminator site was detected surrounding or overlapping the stem-loop structure. Fructose activation of lsdB transcription was inhibited by glucose, indicating a dual transcriptional control mechanism governed by CCR. The inhibitory mechanism may involve the sequence TGATGACCATGACC, overlapping the lsdB translation start (Fig. 1), which resembles the consensus cis-acting factor TGWNANCGNTNWCA, controlling CCR in B. subtilis (25).
The discovery of the carbon catabolic conditions determining the transcription profile of the lsdAB operon and the identification of a common secretion route for the two enzymes should contribute to uncovering the role of levan metabolism in G. diazotrophicus and other plant-interactive bacteria. On this basis, it will be noteworthy to elucidate the molecular mechanisms controlling the fructose-mediated induction of lsdB expression and its fast repression by preferred energy sources.
Footnotes
Published ahead of print on 9 January 2009.
REFERENCES
- 1.Alberto, F., C. Bignon, G. Sulzenbacher, B. Henrissat, and M. Czjzek. 2004. The three-dimensional structure of invertase β-fructosidase from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J. Biol. Chem. 279:18903-18910. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, B., and G. Martínez-Drets. 1995. Metabolic characterization of Acetobacter diazotrophicus. Can. J. Microbiol. 41:918-924. [Google Scholar]
- 3.Arrieta, J., L. Hernández, A. Coego, V. Suárez, E. Balmori, C. Menéndez, M. F. Petit-Glatron, R. Chambert, and G. Selman-Houssein. 1996. Molecular characterization of the levansucrase gene from endophytic sugar cane bacterium Acetobacter diazotrophicus SRT4. Microbiology 142:1077-1085. [DOI] [PubMed] [Google Scholar]
- 4.Arrieta, J. G., M. Sotolongo, C. Menéndez, D. Alfonso, L. E. Trujillo, M. Soto, R. Ramírez, and L. Hernández. 2004. A type II protein secretory pathway required for levansucrase secretion by Gluconacetobacter diazotrophicus. J. Bacteriol. 186:5031-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banguela, A., and L. Hernández. 2006. Fructans: from natural sources to transgenic plants. Biotecnol. Apl. 23:202-210. [Google Scholar]
- 6.Bergeron, L. J., E. Morou-Bermudez, and R. A. Burne. 2000. Characterization of the fructosyltransferase gene of Actinomyces naeslundii WVU45. J. Bacteriol. 182:3649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergeron, L. J., and R. A. Burne. 2001. Roles of fructosyltransferase and levanase-sucrase of Actinomyces naeslundii in fructan and sucrose metabolism. Infect. Immun. 69:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Crutz, A. M., M. Steinmetz, S. Aymerich, R. Richter, and D. Le Coq. 1990. Induction of levansucrase in Bacillus subtilis: an antitermination mechanism negatively controlled by the phosphotransferase system. J. Bacteriol. 172:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernández, L., J. Arrieta, C. Menéndez, R. Vazquez, A. Coego, V. Suárez, G. Selman-Houssein, M. F. Petit-Glatron, and R. Chambert. 1995. Isolation and enzymatic properties of levansucrase secreted by Acetobacter diazotrophicus SRT4, a bacterium associated with sugar cane. Biochem. J. 309:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández, L., J. Arrieta, L. Betancourt, V. Falcón, J. Madrazo, A. Coego, and C. Menéndez. 1999. Levansucrase from Acetobacter diazotrophicus SRT4 is secreted via periplasm by a signal-peptide-dependent pathway. Curr. Microbiol. 39:146-152. [DOI] [PubMed] [Google Scholar]
- 12.Hernández, L., M. Sotolongo, Y. Rosabal, C. Menéndez, R. Ramírez, J. Caballero-Mellado, and J. Arrieta. 2000. Structural levansucrase gene (lsdA) constitutes a functional locus conserved in the species Gluconacetobacter diazotrophicus. Arch. Microbiol. 174:120-124. [DOI] [PubMed] [Google Scholar]
- 13.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 14.Kiska, D. L., and F. L. Macrina. 1994. Genetic regulation of the fructosyltransferase in Streptococcus mutans. Infect. Immun. 62:124-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leloup, L., J. Le Saux, M. F. Petit-Glatron, and R. Chambert. 1999. Kinetics of the secretion of Bacillus subtilis levanase overproduced during the exponential phase of growth. Microbiology 145:613-619. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Verstraete, I., J. Stulke, A. Klier, and G. Rapoport. 1995. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177:6919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Verstraete, I., J. Deutscher, and A. Galinier. 1999. Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signaling pathway for the Bacillus subtilis levanase operon. J. Bacteriol. 181:2966-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy, J. E., and C. Gualerzi. 1990. Translational control of prokaryotic gene expression. Trends Genet. 6:78-85. [DOI] [PubMed] [Google Scholar]
- 19.Menéndez, C., L. Hernández, G. Selman, M. F. Mendoza, P. Hevia, M. Sotolongo, and J. G. Arrieta. 2002. Molecular cloning and expression in Escherichia coli of an exo-levanase gene from the endophytic bacterium Gluconacetobacter diazotrophicus SRT4. Curr. Microbiol. 45:5-12. [DOI] [PubMed] [Google Scholar]
- 20.Menéndez, C., L. Hernández, A. Banguela, and J. País. 2004. Functional production and secretion of the Gluconacetobacter diazotrophicus fructose-releasing exo-levanase (LsdB) in Pichia pastoris. Enzyme Microb. Technol. 34:446-452. [Google Scholar]
- 21.Pereira, Y., M. F. Petit-Glatron, and R. Chambert. 2001. yveB, encoding endolevanase LevB, is part of the sacB-yveB-yveA levansucrase tricistronic operon in Bacillus subtilis. Microbiology 147:3413-3419. [DOI] [PubMed] [Google Scholar]
- 22.Reese, M. G., N. L. Harris, and F. H. Eeckman. 1996. Large scale sequencing specific neural networks for promoter and splice site recognition, p. 737-738. In L. Hunter and T. E. Klein (ed.), Biocomputing proceedings of the 1996 Pacific Symposium. World Scientific Publishing Co., Singapore.
- 23.Saier, M. H., Jr., and T. M. Ramseier. 1996. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 178:3411-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 25.Weickert, M. J., and G. H. Chambliss. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:6238-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen, Z. T., and R. A. Burne. 2002. Analysis of cis- and trans-acting factors involved in regulation of Streptococcus mutans fructanase gene (fruA). J. Bacteriol. 184:126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng, L., Z. T. Wen, and R. A. Burne. 2006. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol. Microbiol. 62:187-200. [DOI] [PubMed] [Google Scholar]
- 28.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]