Abstract
The occurrence of genes conferring resistance to tetracyclines in the organic pig gut was assessed through the metagenomic approach. Of 9,000 bacterial artificial chromosome clones analyzed, 10 were identified as carrying the known tet(C), tet(W), and tet(40) genes, as well as novel genes encoding resistance to the tetracyclines minocycline and doxycycline. The latter are different from the known tet genes and are homologous to genes encoding UDP-glucose 4-epimerases, with the domain structure characteristic for these enzymes. The majority of the resistance genes were associated with putative mobile genetic elements. The sequence of a novel 9.7-kb plasmid carrying tet(W) and tet(40) was also identified. Conserved flanking regions identified around the tet(W) and tet(40) genes in our metagenomic library may play a role in genetic exchange of these genes. This is the first report describing the occurrence of tet(40) outside the human intestine. The maintenance of antibiotic resistance genes in apparently antibiotic-free animals is probably due to their presence on mobile genetic elements, the fitness cost of which for the cell is ameliorated during the previous antibiotic selection.
The broad-spectrum tetracycline (Tc) family of antibiotics, chlorotetracycline and oxytetracycline, were discovered in the late 1940s and by the mid 1950s they were introduced into clinical practice. At the same time, the growth promoting properties of Tcs in food animals were recognized, and they were also extensively used in agriculture. The nontherapeutic use of Tcs was extended to their use in aquaculture and horticulture (8). Expanded-spectrum Tcs, semisynthetic compounds such as doxycycline (Dx) and minocycline (Mn), were introduced in 1967 and 1972, respectively, in response to the growing problem of bacterial resistance to original Tcs. The usefulness of these Tcs in infection control has become increasingly limited in recent years due to the emergence of resistant pathogens (8, 25). Presently, about 40 distinct Tc resistance (Tcr) genes are known and they are usually associated with mobile genetic elements (http://faculty.washington.edu/marilynr/) (25). The most common forms of resistance are the active efflux of Tcs from the cell and the synthesis of ribosomal protection proteins that prevent the binding of Tcs to the ribosomes (8, 25). In a relatively few cases the resistance phenotype relies on mutations in the 16S rRNA gene or chemical inactivation of the antibiotic (26, 31). Tcr genes are very common in the environmental samples (22), and they are frequently isolated from pigs and soil and water samples collected from the areas surrounding pig farms (7, 16, 32, 33).
The question of how antibiotic resistance genes (ABRG) persist in the environment is contradictory (2). Selection and dissemination of ABRG is usually linked to the use of antibiotics, which imposes selective pressure specific for a given antibiotic (7, 17, 34). At the same time, ABRG may persist in apparently antibiotic-free environments (1, 13, 21, 35). As a precautionary measure, some countries enacted legislations limiting the nontherapeutic use of antibiotics, in particularly in food animals (http://ec.europa.eu/food/food/animalnutrition/feedadditives/index_en.htm). The question remains, however, how the discontinuation of antibiotic use may affect the pool of ABRG in food animals. To answer this, we constructed a fecal metagenomic library from pigs reared in an antibiotic-free environment and screened for resistance to the narrow- and expanded-spectrum Tcs. Besides the known Tcr genes, such as tet(C), tet(W), and tet(40), we also identified novel genes (galE1 and galE2) that confer resistance to Mn and Dx. Most of the genes resided on putative mobile genetic elements, which presumably contribute to the maintenance and dissemination of antibiotic resistance in antibiotic-free environments.
MATERIALS AND METHODS
Chemicals, bacterial strains, and growth.
All chemicals, unless otherwise stated, were purchased from Sigma-Aldrich. Escherichia coli ATCC 23724 (C600) and E. coli ATTC 33694 (HB101) were purchased from the American Type Culture Collection, E. coli HB101R (a spontaneous rifampin-resistant mutant) was obtained during this work. E. coli strains were routinely cultured in LB medium at 37°C. Enterococcus faecalis JH2-2 was grown on brain heart infusion medium at 37°C.
Total DNA isolation.
Fecal samples were collected from five organically reared adult pigs (farmed in Aberdeenshire, Scotland, United Kingdom) that did not receive antibiotics for growth promotion, and none had been used for disease treatment. Samples were transported from the farm in the Cary Blair transport medium for anaerobic organisms (Oxoid, United Kingdom). Fecal material, 5 g from each animal, was pooled, resuspended in 50 ml of phosphate-buffered saline buffer with 0.1% Tween 80, incubated for 1 h on ice with vigorous shaking, and subjected to low-speed centrifugation to remove the debris (700 × g, 5 min). Supernatant was saved on ice, and the same procedure was repeated with the pellet. Both supernatants were combined and centrifuged for 10 min at 10,000 × g. The pellet was washed twice with 100 ml of phosphate buffer and then washed twice with 100 ml of TES (Tris-EDTA-sucrose) buffer. The pellet was resuspended in 20 ml of lysis buffer (50 mM Tris [pH 8], 1.25 mM EDTA [pH 8], 0.5 M glucose, 20 mg of lysozyme/ml, 25 μg of lysostaphine/ml) and incubated for 1 h at 37°C. Proteinase K was added to a final concentration of 100 μg/ml, and the mix incubated for further 2 h at 55°C prior to centrifugation for 5 min at 14,000 × g. The pellet was resuspended in 5 ml of TE buffer, and n-lauryl sarcosine was added to a final concentration of 2.5%, followed by incubation for 10 min at 60°C. The sample then was subjected to two phenol-chloroform-isoamyl alcohol (25:24:1) extractions. The liquid phase was transferred to a sterile 15-ml Falcon tube and, after the addition of RNase A at 50 μg/ml, incubated for 30 min at 37°C. This was followed by a further phenol-chloroform-isoamyl alcohol extraction, and DNA was finally precipitated in ethanol, washed with 70% ethanol, air dried, and resuspended in 5 ml of TE buffer. High-molecular-weight DNA was additionally purified by using BD Chromaspin-1000 plus TE (BD Biosciences) columns according to the manufacturer's instructions.
Library construction.
A CopyControl BAC cloning kit (Epicentre) was used to generate metagenomic libraries. Total gut metagenomic DNA was partially digested with HindIII (Promega, United Kingdom), and the enzyme was heat inactivated at 65°C for 15 min. DNA was ethanol precipitated, air dried, and resuspended in 86 μl of deionized water. The insert DNA was ligated into the pCC1BAC vector, precut with HindIII. After overnight incubation at 16°C, the ligation mix was heat inactivated and desalted as recommended by the manufacturer. Then, 2 μl of ligation was electroporated into 50 μl of electrocompetent E. coli Transformax EPI300 cells (Epicentre). Electroporation was done in a 1.0-mm cuvette using a geneZAPPER 450/2500 apparatus (IBI) with the following settings: capacity, 21 μF; resistance, 200 Ω; and voltage, 2,500 V. Clones were selected on LB agar plates containing 12.5 μg of chloramphenicol/ml. A total of 9,000 colonies were picked up by using a BioRobotics BioPick colony picker (Genomic Solutions) and arrayed into a 384-well microtiter plate containing 70 μl of the cryoprotective solution (2× LB medium supplemented with 10% glycerol). The cells were grown overnight at 37°C and stored at −70°C. Screening for antibiotic resistance was performed by printing on LB agar plates containing 5 μg of Tc/ml, 3 μg of Mn/ml and 3 μg of Dx/ml. Antibiotics at these concentrations completely inhibited the growth of the negative control, host E. coli cells harboring an empty vector.
Transposon mutagenesis and sequencing.
In vitro random insertion transposon mutagenesis was carried out by using an EZ-Tn5 <Kan-2> insertion kit (Epicentre) according to the manufacturer's protocol. Bacterial artificial chromosome (BAC) DNA with transposon inserts was isolated by using a BACMAX DNA purification kit (Epicentre) as recommended by the supplier. Regions around the insertion sites were sequenced with KAN-2 FP1 forward and KAN-2 RP1 reverse primers supplied in the kit as described below.
Standard molecular biology procedures.
Standard PCR was performed with the BioTaq DNA (Bioline, United Kingdom) or GoTaq Flexi DNA (Promega) polymerase kits. Annealing temperatures and extension times were optimized depending on the melting temperatures of the primers and the expected sizes of the amplicons.
Restriction digests were carried out using restriction endonucleases (Promega), dephosphorylation reactions utilized shrimp alkaline phosphatase (Roche, United Kingdom), and ligations were performed with T4 DNA ligase (Roche).
Sequencing reactions were read on an automated eight-capillary Beckman sequencer (Beckman, United Kingdom). Chromatograms were assembled by using a Lasergene 6 package. Nucleotide and translated amino acid sequences were analyzed by using on-line BLAST (http://www.ncbi.nlm.nih.gov/blast), PFAM (http://www.sanger.ac.uk/Software/Pfam), and PSORT (http://psort.nibb.ac.jp) programs.
IC determination.
The inhibitory concentrations (ICs) of Dx and Mn were determined by inoculating 0.1 ml of overnight culture grown without antibiotics into 5 ml of LB medium containing serial dilutions of antibiotic (0 to 20 μg/ml) in triplicate. Tubes were incubated at 37°C for 16 h, and the growth was monitored spectrophotometrically at 650 nm (Novaspec II; LKB, Pharmacia, Sweden). The lowest concentration of antibiotic inhibiting growth by 50% compared to control gave the corresponding IC50.
Efflux pump inhibitor studies.
The effect of efflux pump inhibitor, phenyl-arginine-β-naphthylamide (PAβN), on the IC values was assessed as described above with the addition of PAβN at a final concentration of 25 μg/ml prior to overnight incubation.
Plasmid rescue.
Recombinant BAC DNA was isolated from clones 2 and 3 (Fig. 1), and the corresponding inserts were excised, self-ligated, and transformed into chemically competent E. coli JM109 cells according to the manufacturer's protocol (Promega). Inserts from clone 3 were also transformed into the electrocompetent E. coli ATCC 23724 and E. faecalis JH2-2 strains. Electrocompetent E. coli ATCC 23724 was prepared using the protocol described in reference 27 with the following modifications: bacteria were cultured in 2×YT medium, and 10% glycerol was used as a an electroporation solution. Electrocompetent E. faecalis JH2-2 cells were prepared as described previously (9).
FIG. 1.
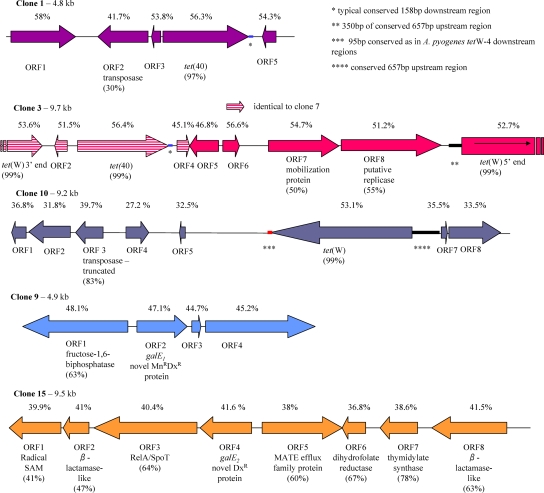
Structure of inserts conferring antibiotic resistance in the organic swine fecal metagenomic library. ORFs are represented by arrows. The shaded arrows within clone 3 indicate the area identical to the sequence of clone 7. The DNA %G+C content is shown above the ORFs, and the percentage of similarity to database entries (where appropriate) is in brackets below the ORFs. Conserved regions are marked with: *, 158-bp region downstream of tet(40); **, 350 bp within the core 657-bp region upstream of tet(W); ***, 95-bp region downstream of tet(W) as in A. pyogenes; ****, 657-bp core region upstream of tet(W).
Nucleotide sequence accession numbers.
Sequence data generated in the present study were deposited in GenBank under the following accession numbers: clone 7, FJ157999; clone 1, FJ158000; clone 2, FJ158001; clone 3, FJ158002; clone 9, FJ158003; clone 10, FJ158004; and clone 15, FJ158005.
RESULTS
Organic swine fecal metagenomic library.
Based on restriction digest of randomly chosen clones, the size of inserts ranged from 1.5 to 40 kb, with the average insert size in the library being ∼15 kb (data not shown). Thus, the library represented approximately 44 bacterial genomes (assuming an average bacterial genome size of 3 Mb). Ten resistant clones were isolated and purified on higher antibiotic concentrations, up to 10 μg/ml, to confirm the resistant phenotype. Most of the Tcr clones (designated 2, 3, 4, 5, 6, 7, and 10) were able to grown on 10 μg of all three antibiotics tested/ml. Clone 1 was able to grow on 10 μg of Tc/ml, but on only 5 μg of Mn and Dx/ml. Clones 9 and 15 were Tc sensitive but exhibited Mnr Dxr and Dxr phenotypes, respectively.
Recombinant DNA from resistant clones was initially end sequenced with vector-specific primers. Four of the ten clones (clones 2, 4, 5, and 6), with insert sizes of 9.3 kb, were identical and carried the tet(C) gene. Thus, only one of them (clone 2) was sequenced to completion. Two out of the six remaining resistant clones (3 and 7), with the insert sizes of 9.7 and 3.7 kb, respectively, encoded identical tet(W) gene sequences on one end of the inserts. Thus, the larger clone 3 was chosen for further sequence analyses by genome walking. End sequencing of the remaining clones (clones 1, 9, 10, and 15) did not reveal the presence of antibiotic resistance determinants, and transposon mutagenesis was used to identify the genes responsible for the resistant phenotype.
Antibiotic-sensitive mutants were sequenced from the ends of EZ-Tn5 <Kan-2>. In the 4.8-kb insert of clone 1 the tet(40) gene, encoding the efflux of Tc from cells, was found, and the 9.2-kb insert of clone 10 harbored the tet(W) gene. The sequences surrounding Tn5 in clones 9 (4.9 kb) and 15 (9.5 kb) had no resemblance to any known Tcr genes and presumably encoded novel mechanisms of resistance.
Characterization of novel ABRG.
In clones 9 and 15 the transposon interrupted open reading frame 2 (ORF2; galE1) and ORF4 (galE2), respectively (Fig. 1). The translated galE1 encoded a 345-amino-acid protein that shared 71 and 68% sequence identity with putative UDP-glucose 4-epimerases (UGEs) from Bacteroides vulgatus ATCC 8482 (YP_001301108) and Bacteroides thetaiotaomicron VPI-5482 (NP_809536), respectively. The protein product of galE1 was therefore called UGE-1. The translated 342-amino-acid sequence of galE2 from clone 15 shared 72 and 65% sequence identity with proposed UGEs from Clostridium sp. strain SS2/1 (ZP_02440543) and Bacillus subtilis subsp. subtilis strain 168 (NP_391765). The protein product of galE2 was therefore named UGE-2. galE1 and galE2 shared 56.2% sequence identity at the nucleotide level.
The full ORFs of galE1 and galE2, together with the upstream regions, were amplified by PCR and cloned into the pGEM-T Easy vector. Since no Mn and Dx resistant phenotypes were expressed in this high-copy-number vector, the inserts were recloned from pGEM-T Easy into the low-copy-number BAC vector. This time the antibiotic resistance phenotype was expressed, suggesting that the presence of a single ORF (the ORF of galE1 or galE2) was sufficient to confer antibiotic resistance. The IC50 for Mn and Dx in the case of galE1 was 11 μg/ml, and the IC50 for Dx in the case of galE2 was 11 μg/ml. At present, the mechanisms of resistance conferred by galE1 and galE2 are not clear since the experiments with efflux pump inhibitors and liquid chromatography-tandem mass spectrometry studies (done as described previously [15]) were not conclusive (data not shown).
The complete ORFs encoding the UGE genes from B. thetaiotamicron VPI-5482 and B. subtilis subsp. subtilis strain 168, together with the upstream regions, were amplified and cloned into pGEM-T Easy vector and subsequently recloned into the BAC vector, but no antibiotic resistant phenotype was observed.
Sequence analysis of resistant clones.
In clones 1, 3, 7, and 10, apart from the known Tcr genes, the majority of other ORFs demonstrated weak or no similarity with database sequences (Fig. 1). The easily identifiable ORFs exhibited similarity with various mobile elements. ORF1 in clone 1 was similar to a transposase (YP_001707264), with a conserved transposase DDE domain at the C terminus (Fig. 2). This D115(X77)D192(X47)E239 triad is also a conserved motif in transposases of the IS982 and IS4 families of insertion sequences (12, 24) (Fig. 2). In clone 3, ORF7 was similar to the mobilization protein from Clostridium leptum DSM753 (ZP_02081735), and ORF8 was similar to the putative replication protein from Staphylococcus sciuri (NP_899174). Clone 7 (3.7 kb) was contained within clone 3 (Fig. 1).
FIG. 2.
Translated transposase sequences in clones 1 (A) and 10 (B). The residues in the DDE motif are indicated in boldface, characteristic residues upstream and/or downstream of the triad are in boldface italics, and regions N2, N3, and C1 are underlined.
In clone 10, ORF3 was similar to the transposase from Lactobacillus helveticus DPC4571 (YP_001576992) and also contained the conserved DDE domain (12, 24) (Fig. 2). The transposase homologues encoded by ORF3 in clone 10 and by ORF2 in clone 1 shared only 20% identity.
The recently described tet(40) gene (15) was found in clones 1, 3, and 7 (Fig. 1), and the ORFs were 98% (clone 1) and 99% (clones 3 and 7) identical to Tet(40) (AJ295238 and AM419751). The previously described gene was located immediately downstream of the mosaic tet(O/32/O) gene (15), while in the present study tet(40) was present as a single determinant in clone 1 and 1.2 kb downstream from tet(W) in clones 3 and 7 (Fig. 1). In the latter clones, tet(W) had been interrupted during the cloning step and the Tcr phenotype was apparently achieved through the expression of tet(40) (Fig. 1).
The upstream regions of tet(40) were not conserved, while the 160-bp downstream region was 100% identical between the clones and database entries (AJ295238 and AM419751). Apart from this limited conservation, the genetic background of clones 1 and 3 is completely different: transposase-like ORF2 in the former and mobilization and replication protein-resembling ORFs in the latter suggest that tet(40) may reside on different mobile elements.
The tet(W) gene was found in its entirety in clone 10, as an interrupted ORF in clone 3, and as a partial sequence in clone 7 (Fig. 1). Structurally, these genes were 96 to 99% identical to the previously described tet(W) genes (3, 6, 14, 18, 29, 32). The 657-bp upstream region in clone 10 was 99% identical to the corresponding regions in Arcanobacterium pyogenes strains OX9 and OX4 (DQ519394 and DQ517519) (6) (Fig. 1), while the similarity with the corresponding regions in six different genera of gut commensals was lower at 93 to 96% (14). The 95-bp downstream region in clone 10 was 100% similar to A. pyogenes strains, OX4, OX9, and 5278-99 (DQ519395 [6]; Fig. 1) and 97% similar to tet(32) in Streptococcus salivarius strain FStet12 (DQ647324 [23]). No similarity to downstream regions conserved in human gut commensals (14) was detected. The G+C content of tet(W) was ∼53%, which is not significantly different from the other ORFs in clones 3 and 7 but substantially higher than in clone 10 ORFs, suggesting a possible horizontal gene transfer event in the latter case.
The downstream region of tet(W) in clone 3 was not conserved, but the 351-bp upstream region was 94 to 100% similar to the intestinal (14) and other bacteria carrying tet(W). The putative regulatory region containing a 14-amino-acid leader peptide involved in transcription attenuation was present in this region (18) (Fig. 1).
Clone 9 encodes four ORFs (Fig. 1), and one of them, ORF2, was identified as a resistance gene by transposon mutagenesis. ORF1 showed 63% conservation with fructose-1,6-biphosphatase (11) from Bacteroides fragilis YCH46 (YP_099100), while ORF3 and ORF4 displayed no discernible similarity to database entries.
Eight ORFs were identified in clone 15 with ORF4 encoding resistance to Dx. The closest similarity for ORFs 1, 2, 3, and 5 was observed for proteins from Clostridium phytofermentans ISDg: ORF1 was similar to the coproporphyrinogen III oxidase (YP_001557676 [30]), ORF2 was similar to the beta-lactamase domain protein (YP_001557675), ORF3 was similar to the (p)ppGpp synthetase I of SpoT/RelA family of proteins (YP_001557674), and ORF5 was similar to the MATE efflux family protein (YP_001559167). The order of the first three ORFs was also conserved similarly to C. phytofermentans ISDg. The closest matches for ORF6 to ORF8 were the hypothetical proteins from C. thermocellum ATCC 27405: ORF6 was similar to dihydrofolate reductase (YP_001037651), ORF7 was similar to thymidylate synthase (YP_001037652), and ORF8 was similar to metallo-beta-lactamase (YP_001037039).
Clones 2, 4, 5, and 6 were identical and 99.7% similar to the IncQ plasmid pSC101 from Salmonella enterica serovar Typhimurium (NC_002056 [5]). The differences included 28 single nucleotide polymorphisms, as well as the presence of a 54-bp insertion sequence between the coordinates 7952 and 8005 in clone 2 compared to pSC101.
Clone 3 also resembles a linearized plasmid: both insert ends carry parts of tet(W), interrupted by HindIII digest during the cloning step (Fig. 1). Thus, we attempted to rescue to restore the original structures in clones 2 and 3.
Plasmid rescue.
Clones 2 and 3 were partially digested by HindIII, and the appropriate bands were purified from agarose gels, self-ligated, and transformed into competent cells. A completely functional plasmid was recovered from clone 2 by transforming E. coli JM109, and the structure was verified by restriction and PCR analyses (data not shown).
In the case of clone 3 neither E. coli JM109 nor E. coli ATCC 23724 produced any Tcr transformants. Since sequence analysis had suggested that the plasmid may originate from a gram-positive host, we also tested a gram-positive recipient, E. faecalis JH2-2, but without success. Thus, this plasmid may have a different host range, precluding its replication and/or expression in the hosts we tested.
DISCUSSION
In this work, we constructed a fecal metagenomic library from organic pigs, with the total size of inserts representing about 44 bacterial genomes. Screening for Tcr revealed that even in this small-scale sampling test, we encountered tet(C) four times, tet(40) three times, and tet(W) two times and also observed single incidences of two novel resistant determinants, galE1 and galE2. Additionally, it is likely that not all Tcr genes were detected because of limitations imposed by heterologous gene expression in the E. coli host. In a parallel study, we constructed a BAC library from the fecal samples of conventional adult pigs and, among the 10,400 clones analyzed, we encountered 132 Tcr clones, suggesting that the transition to organic practice may decrease the pool of these genes by >100-fold (unpublished data). However, even in an apparently antibiotic-free environment, Tcr genes still persist, and we were interested to find out what genes and genetic mechanisms may contribute to their maintenance in antibiotic-free animals.
The most frequently encountered tet(C) gene was found within a sequence that displayed more than 99% nucleotide sequence identity to the well-characterized mobilizable IncQ plasmid pSC101 (5). This may explain the high frequency of the occurrence of the tet(C) gene in the metagenomic library. Moreover, we were able to reconstruct the insert as a fully functional, autonomously replicating plasmid. In our preliminary competition experiments in isogenic background the fitness cost of this plasmid was substantially ameliorated during antibiotic selection (unpublished data).
One of the most recently discovered Tcr genes, tet(40), was encountered on three occasions. In previous work, it was identified in a human bacterial isolate and in the human gut metagenomic library, and in both cases it was linked to another Tcr gene, the mosaic tet(O/32/O) (15), and present on the conjugative transposon TnK10 (19). The three separate incidences of its cloning in the pig gut metagenome suggest that the gene is common among pig intestinal bacteria as well. Moreover, we also encountered two different genetic contexts for this gene: one on a putative transposon and another on a putative plasmid. In the former case, it was present as a single Tcr gene, and in the latter it was linked to tet(W). Although we were unable to reconstruct clone 3 as an autonomously replicating plasmid, most probably due to the absence of a suitable host, its structure strongly suggests that originally it was a 9.7-kb plasmid, which was linearized during the cloning step. The location on mobile genetic elements may contribute to the dissemination and maintenance of tet(40) in different ecosystems. Also, the downstream region of tet(40) possessed a short 160-bp sequence that is perfectly conserved among all currently known tet(40) genes. The G+C content of the inserts with tet(40) was fairly high in the present study (45 to 58%) compared to C. saccharolyticum, a low G+C bacterium. Thus, it appears that tet(40) and its immediate flanking regions originated from a common source and disseminated to other bacteria in different environments. The short conserved sequence may serve as a hot spot for excision and integration of the gene thus contributing to its mobility.
tet(W) is one of the most widely disseminated Tcr genes (4, 22, 28, 29, 32, 37), and the conservation of its flanking regions was recently described in multiple species of gut bacteria (14). In the present study we found two incidences of tet(W): clone 3 tet(W), which had higher identity with the genes described in reference 14), and clone 10 tet(W), which was more closely related to the genes described earlier (6). The flanking sequences of the tet(W) genes described here were different. The DNA %G+C content of tet(W) in clone 10 is substantially higher (53.1%) than the rest of the clone (<37%), indicating a possible horizontal gene transfer event. Here, similarly to tet(40), the tet(W) flanking regions showed sequence conservation among the genes analyzed in the present study and of others in databases. This especially concerns tet(W)-4 from A. pyogenes strains OX9, OX4, and 5278-99 (GenBank accession numbers DQ519394, DQ517519, and DQ519395, respectively [6]). The presence of such conserved core sequences may be indicative of their role in horizontal gene transfer. Previous works have also suggested that tet(40) and tet(W) reside on mobile genetic elements (15, 19) and are capable of transfer (6, 14, 18, 19).
Two novel genes, galE1 and galE2, that confer resistance to the expanded-spectrum Tcs were encountered during our analysis of the organic pig fecal metagenomic library. Interestingly, the resistance phenotype was expressed only when a low copy number of the genes was present in the cell. The genes do not display any similarity to known Tcr genes, but they share 65 to 72% amino acid identity with UGEs and correspondingly possess the conserved GalE domain characteristic for these proteins (COG1087) (10, 36). The metabolic function of UGE is the interconversion of UDP-glucose and UDP-galactose, and the latter is used as a monomer in the biosynthesis of extracellular lipopolysaccharides and capsular polysaccharides. Interestingly, screening a sludge metagenomic library for resistance to menadione resulted in the cloning of the similar gene (20). Structurally, menadione is very different from Mn and Dx, and it is unlikely that the resistance mechanism to such structurally different compounds may involve a similar enzymatic modification mechanism. Besides, using liquid chromatography-tandem mass spectrometry, we were unable to demonstrate any modification of Dx upon incubation with recombinant E. coli cells expressing galE1 or galE2 (data not shown). Further work is required for more detailed characterization of the resistance mechanisms encoded by these genes.
In conclusion, we demonstrated the occurrence of known and novel Tcr genes in an apparently antibiotic-free environment. The source of these genes cannot be currently identified. The regulations governing organic farm operations prohibit the use of any antibiotic in these animals and, since the sows were also born and raised on farms without any antibiotic usage, it is unlikely that they are the source of antibiotic-resistant microbiota. However, there is a possibility that the corresponding resistant microbiota was brought to the farm in the beginning of operation and thus became endemic. Another possibility could be the acquisition from the environmental sources (2). Factors contributing to the maintenance of these genes under nonselective conditions may include the location on mobile genetic elements, with the reduced fitness cost achieved during the initial selection process with antibiotics. Small conserved flanking sequences found in tet(W) and tet(40) may act as mini-elements contributing to the dissemination and maintenance of antibiotic resistance genes as hypothesized previously (14). The metagenomic approach, therefore, could be a valuable tool to reveal potential resistance and transfer mechanisms that may interfere with the efficiency of antibiotics for the clinical and veterinary use.
Acknowledgments
We thank P. Young and G. Campbell for excellent technical support.
This study and K.A.K. were supported by the DEFRA project OD2014. The Rowett Institute of Nutrition and Health of the University of Aberdeen is supported by the Scottish Government Rural and Environment Research and Analysis Directorate.
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Alonso, A., P. Sanchez, and J. L. Martinez. 2001. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 3:1-9. [DOI] [PubMed] [Google Scholar]
- 2.Aminov, R. I., and R. I. Mackie. 2007. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271:147-161. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa, T. M. 1998. Tetracycline resistance transfer among obligate anaerobes form the ruminant gut. Ph.D. thesis. University of Aberdeen, Aberdeen, United Kingdom.
- 4.Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 5.Bernardi, A., and F. Bernardi. 1984. Complete sequence of pSC101. Nucleic Acids Res. 12:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billington, S. J., and B. H. Jost. 2006. Multiple genetic elements carry the tetracycline resistance gene tet(W) in the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 50:3580-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Rodz, A. L., and M. S. Gilmore. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152-154. [DOI] [PubMed] [Google Scholar]
- 10.Fry, B. N., S. Feng, Y. Y. Chen, D. G. Newell, P. J. Coloe, and V. Korolik. 2000. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita, Y., K. I. Yoshida, Y. Miwa, N. Yanai, E. Nagakawa, and Y. Kasahara. 1998. Identification and expression of the Bacillus subtilis fructose-1,6-bisphosphatase gene (fbp). J. Bacteriol. 180:4309-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 13.Karami, N., F. Nowrouzian, I. Adlerberth, and A. E. Wold. 2006. Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrob. Agents Chemother. 50:156-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazimierczak, K. A., H. J. Flint, and K. P. Scott. 2006. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob. Agents Chemother. 50:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazimierczak, K. A., M. T. Rincon, A. J. Patterson, J. C. Martin, P. Young, H. J. Flint, and K. P. Scott. 2008. A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O), in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob. Agents Chemother. 52:4001-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koike, S., I. G. Krapac, H. D. Oliver, A. C. Yannarell, J. C. Chee-Sanford, R. I. Aminov, and R. I. Mackie. 2007. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a three-year period. Appl. Environ. Microbiol. 73:4813-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy, S. B., and M. Nelson. 1998. Reversing tetracycline resistance. A renaissance for the tetracycline family of antibiotics. Adv. Exp. Med. Biol. 456:17-25.:17-25. [PubMed] [Google Scholar]
- 18.Melville, C. M., R. Brunel, H. J. Flint, and K. P. Scott. 2004. The Butyrivibrio fibrisolvens tet(W) gene is carried on the novel conjugative transposon TnB1230, which contains duplicated nitroreductase coding sequences. J. Bacteriol. 186:3656-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melville, C. M., K. P. Scott, D. K. Mercer, and H. J. Flint. 2001. Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob. Agents Chemother. 45:3246-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori, T., H. Suenaga, and K. Miyazaki. 2008. A metagenomic approach to the identification of UDP-glucose 4-epimerase as a menadione resistance protein. Biosci. Biotechnol. Biochem. 72:1611-1614. [DOI] [PubMed] [Google Scholar]
- 21.Pallecchi, L., C. Lucchetti, A. Bartoloni, F. Bartalesi, A. Mantella, H. Gamboa, A. Carattoli, F. Paradisi, and G. M. Rossolini. 2007. Population structure and resistance genes in antibiotic-resistant bacteria from a remote community with minimal antibiotic exposure. Antimicrob. Agents Chemother. 51:1179-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson, A. J., R. Colangeli, P. Spigaglia, and K. P. Scott. 2007. Distribution of specific tetracycline and erythromycin resistance genes in environmental samples assessed by macroarray detection. Environ. Microbiol. 9:703-715. [DOI] [PubMed] [Google Scholar]
- 23.Patterson, A. J., M. T. Rincon, H. J. Flint, and K. P. Scott. 2007. Mosaic tetracycline resistance genes are widespread in human and animal fecal samples. Antimicrob. Agents Chemother. 51:1115-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rezsohazy, R., B. Hallet, J. Delcour, and J. Mahillon. 1993. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 9:1283-1295. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 26.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Scott, K. P., T. M. Barbosa, K. J. Forbes, and H. J. Flint. 1997. High-frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 63:3405-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speer, B. S., L. Bedzyk, and A. A. Salyers. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol. 173:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanton, T. B., J. S. McDowall, and M. A. Rasmussen. 2004. Diverse tetracycline resistance genotypes of Megasphaera elsdenii strains selectively cultured from swine feces. Appl. Environ. Microbiol. 70:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stine, O. C., J. A. Johnson, A. Keefer-Norris, K. L. Perry, J. Tigno, S. Qaiyumi, M. S. Stine, and J. Morris. 2007. Widespread distribution of tetracycline resistance genes in a confined animal feeding facility. Int. J. Antimicrob. Agents 29:348-352. [DOI] [PubMed] [Google Scholar]
- 34.Swartz, M. 2002. Human diseases caused by foodborne pathogens of animal origin. Clin. Infect. Dis. 34:S111-S122. [DOI] [PubMed] [Google Scholar]
- 35.Thakur, S., D. A. Tadesse, M. Morrow, and W. A. Gebreyes. 2007. Occurrence of multidrug resistant Salmonella in antimicrobial-free (ABF) swine production systems. Vet. Microbiol. 125:362-367. [DOI] [PubMed] [Google Scholar]
- 36.Thoden, J. B., A. D. Hegeman, G. Wesenberg, M. C. Chapeau, P. A. Frey, and H. M. Holden. 1997. Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 36:6294-6304. [DOI] [PubMed] [Google Scholar]
- 37.Villedieu, A., M. L. Diaz-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]