Abstract
rRNA genes are attractive targets for developing PCR assays targeting human fungal pathogens. Most studies have focused on the 18S rRNA gene, internal transcribed spacers, and the 5′ end of the 28S rRNA gene. An approximately 2,900-bp region of the 28S rRNA gene remains largely unexplored because sequences of many medically relevant fungi are either unavailable or undefined in genomic databases. The internal transcribed spacers and 28S rRNA gene of nine medically and phylogenetically important fungi were sequenced. In addition, 42 sequences from this region were acquired from public databases, resulting in an alignment of 51 fungal sequences spanning 30 fungal genera. For the nearly 3,950-bp region from the 3′ end of 18S rRNA gene to the 3′ end of the 28S rRNA gene, 27 broad-range PCR primers were designed such that their sequence homology with the human rRNA gene was minimal. All 62 possible amplicons in the size range from 75 to 400 bp from 27 primers were screened using fungal genomic DNA from 26 species spanning 14 genera. Eleven of the 62 amplicons did not cross-react with 1 μg/PCR human DNA but simultaneously amplified 10 fg of fungal DNA. Phylogenetic distance matrices were calculated for regions covered by these 11 amplicons based on 51 fungi. Two of these 11 amplicons successfully amplified 30 fg of fungal DNA from 25 of 26 fungi and provided the most phylogenetic information for species identification based on the distance matrices. These PCR assays hold promise for detection and identification of fungal pathogens in human tissues.
Rapid detection and accurate identification of fungal pathogens can be critical for initiating treatment in the earliest stages of infection and for guiding antifungal therapy. Culture-based and histological analyses often have poor diagnostic sensitivity, and histopathological findings frequently do not distinguish among fungal genera or species (15, 18, 26). Moreover, some molecular diagnostic tests, such as the galactomannan antigen assay, reliably detect only pathogens belonging to the genus Aspergillus, and the beta-glucan assay does not detect fungi in the zygomycete and basidiomycete taxa (11, 15, 16, 26). Such shortcomings may lead to more empirical and unnecessary antifungal therapy because a fungal infection is not completely excluded by negative results in either of these assays.
PCR assays for the detection of fungal pathogens are an appealing approach due to their potential for rapid, sensitive, and accurate diagnosis of fungal infections. rRNA genes are particularly attractive targets because multiple copies are present in each genome and the genes have conserved regions for designing broad-range primers and have more variable regions for identifying fungi. Most studies have focused on regions within the 18S rRNA gene (4, 23), internal transcribed spacers (ITS1 and ITS2), the 5.8S rRNA gene (2, 3, 8, 9, 14, 22, 25), and the 5′ end of the 28S rRNA gene (D1-D2 hypervariable region) (1, 5, 8, 13, 17, 20, 24) for developing broad-range PCR primers targeting human fungal pathogens. The remainder of the 28S rRNA gene spanning nearly 2,900 bp remains largely unexplored. Sequences of the 28S rRNA gene beyond the D1-D2 hypervariable region for many important human fungal pathogens are not available or are undefined in public databases.
For most broad-range fungal PCR assays described in the literature, the focus is on identifying fungi in pure cultures, or their performance in the presence of large quantities of human DNA is unknown. From a clinical diagnostic standpoint, it is critical to be able to apply broad-range fungal PCR assays to DNA extracted from human tissue samples without interference from or interaction with human DNA.
We sought to develop new broad-range fungal PCR assays suitable for use with human tissues. Recently, several complete fungal genomes have been sequenced, providing an opportunity to explore additional rRNA operons for the design of PCR assays. To create a database of fungal sequences comprising ITS1, 5.8S rRNA genes, ITS2, and 28S rRNA genes, we sequenced de novo clinically and phylogenetically relevant fungi and derived sequences from fungal genomic databases or the GenBank database. Optimal PCR assays were selected based on their ability to detect phylogenetically diverse fungi and amplify small quantities of fungal DNA in the presence of a relatively large quantity of human DNA.
MATERIALS AND METHODS
Fungi.
Table 1 lists nine clinically or phylogenetically relevant fungal pathogens whose ITS, 5.8S rRNA genes, and 28S rRNA genes were subjected to sequencing. Table S1 in the supplemental material lists 42 fungi from which the same gene sequences were obtained from publicly available genomic databases or the GenBank database, as well as specific references for sequence data obtained from these databases for the fungi. Genomic DNA of the following organisms was used for analytical sensitivity testing to screen broad-range fungal primers: Aspergillus candidus ATCC 20022, Aspergillus flavus ATCC MYA-3631, Aspergillus fumigatus ATCC MYA-1163, Aspergillus oryzae ATCC 20719, Aspergillus terreus ATCC 10070, Aspergillus ustus ATCC 20063, Candida albicans ATCC 90028, Candida dubliniensis ATCC MYA-580, Candida glabrata ATCC 90876, Candida guilliermondii ATCC 90877, Candida kefyr ATCC 28838, a Candida krusei clinical isolate, Candida lusitaniae ATCC 42720, a Candida tropicalis clinical isolate, Rhizopus oryzae ATCC 10260, Saccharomyces cerevisiae (Novagen, Madison, WI), and Cryptococcus neoformans ATCC 28958D-5. In addition, the genomic DNA of all nine organisms listed in Table 1 was tested.
TABLE 1.
Fungi for which rRNA gene sequences were generated de novo
| Species | Strain | GenBank accession no. | Length of sequence (bp) |
|---|---|---|---|
| Absidiacorymbifera | ATCC 14058 | FJ345350 | 3,733 |
| Cunninghamellabertholletiae | ATCC 42115 | FJ345351 | 4,035 |
| Fusarium solani | ATCC 56480 | FJ345352 | 3,830 |
| Mucorracemosus | ATCC 42647 | FJ345353 | 3,999 |
| Paecilomycesvariotii | ATCC 10865 | FJ345354 | 3,972 |
| Penicilliumchrysogenum | ATCC 10108 | FJ345355 | 3,916 |
| Rhizomucormiehei | ATCC 46345 | FJ345356 | 3,983 |
| Rhodotorulaglutinis | ATCC 16726 | FJ345357 | 3,971 |
| Scedosporiumapiospermum | ATCC 28206 | FJ345358 | 4,907 |
Obtaining rRNA gene sequences from fungal genomic databases.
The fungal rRNA operon is a continuous sequence made up of the 18S, ITS1, 5.8S, ITS2, and 28S subunit regions (9). For most fungi whose genomes are publicly available (see Table 1 in the supplemental material), we obtained the rRNA gene sequence using the following protocol. The 18S subunit and/or ITS1, 5.8S rRNA gene, or ITS2 subunit sequence of a specific fungus was first obtained through GenBank. This section of the sequence was then used to perform a BLASTn search within the genome. Six kilobase pairs of sequence was obtained on either side of the match in the genome. Well-defined rRNA gene sequences of S. cerevisiae and C. albicans as well as other smaller sequence subunits (ITS1, 5.8S rRNA gene, ITS2 region, and D1-D2 region of the 28S rRNA gene) of each fungus, if available through GenBank, were used to map and trim this large contig to derive the rRNA gene sequences of interest. A combination of multiple-alignment and sequence confirmation tools in the Accelrys Gene software (Accelrys, San Diego, CA) were used for processing the larger contigs of sequences downloaded from public databases.
Primers for sequencing and broad-range fungal assays.
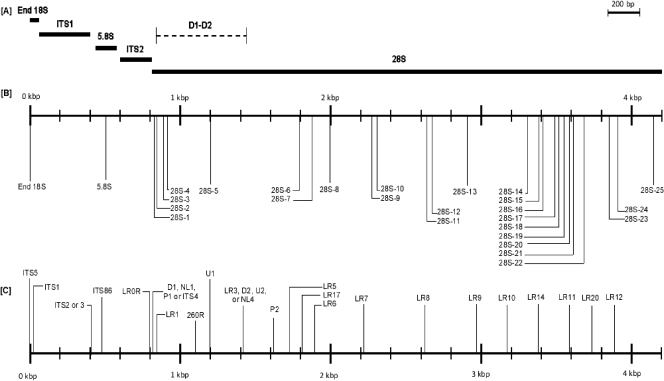
Primers which could be used for either sequencing or broad-range PCR were designed based on the multiple-sequence alignment of about 40 fungal rRNA operons. Maximizing nucleotide differences with the human rRNA gene sequence was an important criterion in designing primers. Primers that met this criterion are listed in Table 2, and the locations of these primers are shown on a map of the S. cerevisiae rRNA gene operon in Fig. 1B. During the initial stages of sequencing, several primers from the Vilgalys Lab website (R. Vilgalys, Duke University, Durham, NC [http://www.biology.duke.edu/fungi/mycolab/primers.htm]) were also used. These primers had either complete homology with or were only 1 bp different from the human rRNA gene sequence and therefore were not considered further for broad-range PCR development. They are mapped below the primers developed in this study in Fig. 1C. In addition, Fig. 1C shows several widely used broad-range fungal primers from the literature which target ITS1, ITS2, the 5.8S rRNA gene, and the D1-D2 region of the 28S rRNA gene.
TABLE 2.
Broad-range fungal rRNA gene primers
| Primer | Sequence (5′-3′)a | No. of mismatches with human rRNA gene |
|---|---|---|
| End18S | GTA AAA GTC GTA ACA AGG TTT C | 7 |
| 5.8S | GTG AAT CAT CGA RTC TTT GAA C | 9 |
| 28S-1 | TAC CCG CTG AAC TTA AGC ATA | 2 |
| 28S-2 | GCA TAT CAA TAA GCG GAG GAAA | 3 |
| 28S-3 | AGT ARC GGC GAG TGA AGC GG | 2 |
| 28S-4 | AGC TCA AAT TTG AAA SCT GG | 6 |
| 28S-5 | CTT CCC TTT CAA CAA TTT CAC RT | 6 |
| 28S-6 | GAG GTA AAG CGA ATG ATT AG | 2 |
| 28S-7 | CTT GTT RCT TAR TTG AAC GTG | 8 |
| 28S-8 | ACC ACA AAA GGT GTT AGT WCA TC | 5 |
| 28S-9 | GAA GTG GGG AAA GGT TCC | 2 |
| 28S-10 | GAC ATG GGT TAG TCG ATC CTA | 4 |
| 28S-11 | TCG TAC TCA TAA CCG CAG C | 3 |
| 28S-12 | GTT GAT AGA AYA ATG TAG ATA AGG | 5 |
| 28S-13 | CAA GGG GAA TCT GAC TGT C | 3 |
| 28S-14 | TTT ACT TAW TCA ATG AAG CGG | 6 |
| 28S-15 | CCG GGT TGA WGA CAT TGT CA | 7 |
| 28S-16 | GCT GGG GCG GCA CAT CTG TT | 4 |
| 28S-17 | GAA CAA AAG GGT AAA AGT CCC | 5 |
| 28S-18 | TTT GAT TTT CAG TGT GAA TAC AAA CCA | 5 |
| 28S-19 | ATG AAA GTG TGG CCT ATC G | 5 |
| 28S-20 | GAG GCT AGA GGT GCC AGA A | 5 |
| 28S-21 | AGG GAT AAC TGG CTT GTG GC | 0 |
| 28S-22 | ACC GAA GCA GAA TTC GGT AAG | 5 |
| 28S-23 | GAT AAT TGG TWT TTG CGG CTG | 7 |
| 28S-24 | GCT GAA CGC CTC TAA GTC AGA | 1 |
| 28S-25 | TCG TAR CAA CAA GGC TAC T | 7 |
Primers were used in either the forward or reverse orientation, but the forward orientation is shown.
FIG. 1.
Broad-range PCR primers for sequencing and PCR assay development. (A) Map of fungal rRNA from the 3′ end of the 18S rRNA gene to the 3′ end of the 28S rRNA gene based on the S. cerevisiae sequence. (B) Twenty-seven newly designed broad-range fungal primers based on differences with the human rRNA gene (this study). (C) Previously described broad-range fungal primers, including ITS5, ITS1, ITS2, ITS3, and ITS4 (8, 22, 25), ITS86 (22), NL1, NL4, and 260R (13, 24), D1 and D2 (8), U1, U2, P1, and P2 (20), and LR (http://www.biology.duke.edu/fungi/mycolab/primers.htm) primers. The LR series of primers have been recommended for sequencing of the 28S rRNA gene but have not been widely used for broad-range fungal PCR diagnostics.
Sequencing of rRNA operons. (i) PCR amplification.
Each 50-μl PCR mixture contained 1.5 U of PfuTurbo Hotstart polymerase, 1× PfuTurbo 10× PCR buffer (Stratagene, La Jolla, CA), 0.8 mM of the GeneAmp deoxynucleoside triphosphate blend (Applied Biosystems, Foster City, CA), 0.4 μM each of forward and reverse primers selected from Table 1, and 20 ng of extracted fungal genomic DNA based on a previously described protocol (12). The volume was brought up to 50 μl with DNA-grade water that was filtered through an Amicon Ultra-15 30-kDa centrifugal filter unit (Millipore Corporation, Billerica, MA) and UV irradiated at 240 mJ/cm2 (Spectrolinker, Westbury, NY). The PCR cycling conditions consisted of initial denaturation for 2 min at 95°C and then 30 cycles of 95°C for 30 s (denaturation), a temperature between 50 and 60°C for 30 s (annealing), and 72°C for 2 min (extension) and a final extension at 72°C for 10 min. The annealing temperature selected was the lower value of the melting temperatures of the two primers chosen for the PCR.
(ii) Sequencing of amplicon.
PCR products were cleaned with Montage PCR filters (Millipore Corporation, Billerica, MA), eluted with 30 μl of DNA-grade water, and frozen at −20°C until they were used. Sequencing was done with Big Dye terminators and an Applied Biosystems capillary sequencer (Applied Biosystems, Foster City, CA). Overlapping reads were generated from all amplicons using multiple sequencing primers. In addition to the primers used to amplify the original PCR product, one or two other primers that were expected to be contained in the amplicon were also used to sequence each product. Accelrys Gene software was used to assemble the sequence reads into the larger rRNA gene operon.
Broad-range PCR amplicon selection and screening criteria.
A matrix of all possible amplicon lengths for the 27 broad-range primers was generated in Microsoft Excel. Of the 351 possible amplicons using all combinations of these primers, 62 were chosen for screening based on an amplicon size ranging from 75 to 400 bp. Endpoint PCR was used to assess successful amplification of each fungal target, the impact of human genomic DNA on fungal amplification, and the cross-reactivity of human DNA in the fungal PCR assays. In addition, the ability of each amplicon to identify and differentiate fungal species was analyzed using distance matrices.
(i) Endpoint PCR.
Each 50-μl PCR mixture contained 1× buffer A, 3 mM of MgCl2, 1 mM of the GeneAmp deoxynucleoside triphosphate blend (12.5 mM with dUTP), 2.2 U of AmpliTaq Gold DNA polymerase, and 0.05 U AmpErase uracil N-glycosylase (all from Applied Biosystems, Foster City, CA), as well as 0.6 μM each of forward and reverse primers and 0.002% Triton X-100. The PCR cycling conditions consisted of uracil N-glycosylase activation for 2 min at 50°C, initial denaturation for 10 min at 95°C, and then 40 cycles of 15 s at 95°C (denaturation), 30 s at 55°C (annealing), and 40 s at 72°C (extension) and a final extension at 72°C for 7 min.
(ii) Analytical sensitivity and cross-reactivity testing.
The analytical sensitivity for amplicon screening was assessed by testing extracted fungal genomic DNA. Cross-reactivity was assessed in the presence of human genomic DNA (Roche Applied Sciences, Indianapolis, IN). A preliminary screen of all 62 amplicons involved amplification of 1,000 pg, 10 pg, and 30 fg of C. albicans genomic DNA and 30 fg of C. albicans genomic DNA in the presence of 100 ng of human genomic DNA. Based on an estimate of the number of rRNA operon copies present in a diploid C. albicans genome, 30 fg of genomic DNA was equivalent to approximately 94 copies (10). The final screen involved analytical sensitivity testing with 30 fg of genomic DNA from 26 different fungal species spanning 14 genera. Cross-reactivity testing was assessed using 10 fg of A. fumigatus genomic DNA in the presence of 1 μg of human genomic DNA.
(iii) Data analysis.
Accelrys Gene software was used to generate multiple-sequence alignments, distance matrices, and phylogenetic trees. Distance matrices and the resulting phylogenetic trees were generated based on the following parameters: tree-building method (neighbor joining [19]), rooting (unrooted), distance (absolute nucleotide differences or Tajima-Nei [21]), tie breaking (systematic), gamma correction (not available), TS/TV ratio (off), mode (best tree), and treatment of gaps (distributed proportionally). The distance matrices and resulting phylogenetic trees were used to assess the potential of amplicons to distinguish between species.
Nucleotide sequence accession numbers.
The nucleotide sequences generated in this study have been deposited in the GenBank database under the accession numbers shown in Table 1.
RESULTS
Generation of new fungal rRNA gene sequences.
Sequence information for several medically or phylogenetically important fungi is not available in public databases, limiting the ability of workers to design broad-range fungal PCR assays. To address this limitation, we sequenced rRNA genes from nine phylogenetically and clinically relevant fungal species. Seven of these fungal species were missing rRNA sequences from the 3′ end of 18S rRNA gene to the 3′ end of the 28S rRNA gene. Table 1 shows the GenBank accession numbers for sequences deposited from this study and their sequence lengths. Our sequences may be up to 90 bp short of the true end of the 28S rRNA gene since we used a conserved primer (28S-25) at the 3′ end of the gene for both PCR and sequencing. The sequencing of zygomycetes like Rhizomucor miehei, Cunninghamella bertholletiae, and Mucor racemosus was relatively complicated due to significant divergence of these species from other fungi. Several custom primers had to be used to successfully complete sequencing for these species. In addition, Scedosporium apiospermum posed a significant sequencing challenge due to the presence of inserts in the rRNA operon, resulting in multiple bands during agarose gel electrophoresis of PCR products.
Selection of broad-range fungal rRNA gene primers.
A multiple-sequence alignment was created using the 9 new fungal rRNA operon sequences generated in this study (Table 1) and 42 publicly available sequences (see Table S1 in the supplemental material) representing 30 genera. The phylogenetic position of these fungi based on the alignment was used to further verify the identity of the fungal sequences. Twenty-seven broad-range fungal primers (Table 2) were designed by manually reviewing the alignment to select areas of sequence conservation among fungi that had multiple nucleotide differences with the human rRNA operon. Six primers (End18S, 5.8S, 28S-1, 28S-2, 28S-5, and 28S-24) overlap either completely or partially with primers found in the literature, and most of them are in the region spanning the 3′ end of the 18S rRNA gene, the 5.8S rRNA gene, and the 5′ end of the 28S rRNA gene (8, 20, 22; http://www.biology.duke.edu/fungi/mycolab/primers.htm). Twenty-one primers from the 5′ end of the 28S rRNA gene up to the 3′ end of this gene are novel to the best of our knowledge. The positions of these primers (Fig. 1B) and of some widely used broad-range fungal primers described in the literature (Fig. 1C) are shown in the rRNA gene map in Fig. 1A. Our broad-range primers, along with several primers from the website of the Vilgalys Lab at Duke University (http://www.biology.duke.edu/fungi/mycolab/primers.htm), were used for de novo sequencing of fungal rRNA genes. In addition, all primers listed in Table 2 were chosen as candidates for development of broad-range fungal PCR assays applicable to human tissue samples.
Screening of PCR amplicons based on analytical sensitivity and cross-reactivity.
Based on the 27 broad-range primers designed in this study, a total of 351 unique amplicons of various sizes could be generated. To develop broad-range PCR assays with maximum sensitivity, all 62 amplicons in the size range from 75 to 400 bp were selected for screening. Fifty-one amplicons were eliminated from further consideration because they amplified human genomic DNA and/or failed to amplify 30 fg of C. albicans DNA in the presence of 100 ng of human genomic DNA. The remaining 11 amplicons were subjected to extensive screening using analytical sensitivity testing with 30 fg of fungal genomic DNA from 26 fungi spanning 14 genera (Table 3 and Fig. 2). None of these top 11 broad-range fungal amplicons generated a product with 1 μg human genomic DNA or were inhibited from amplifying 10 fg of A. fumigatus DNA in the presence of 1 μg of human genomic DNA (Table 3). Five amplicons, ITS2(5.8SF-1R), 28S(9F-12R), 28S(10F-12R), 28S(18F-22R), and 28S(18F-23R), detected the widest range of fungi (Table 3). The ITS2(5.8SF-1R) amplicon detected all fungi tested, but some of the results were weak, as shown by relatively faint gel bands. The 28S(10F-12R) amplicon strongly detected 25 of the 26 fungi but could not detect Rhodotorula glutinis at the 30-fg level. In most cases when amplification was either unsuccessful or weak, there was a mismatch between the sequence of the specific organism and at least one of the primers.
TABLE 3.
Screening data for the top 11 fungal broad-range amplicons
| Species or parameter | Screening dataa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS1 (End18SF-5.8SR) | ITS2 (5.8SF-1R) | ITS2 (5.8SF-2R) | ITS2 (5.8SF-3R) | 28S (9F-12R) | 28S (10F-12R) | 28S (12F-13R) | 28S (15F-22R) | 28S (18F-22R) | 28S (18F-23R) | 28S (23F-25R) | |
| Absidiacorymbifera | − | + | − | − | +++ | +++ | +++ | − | ++ | +++ | + |
| Aspergillus candidus | +++ | +++ | + | + | +++ | +++ | +++ | +++ | +++ | +++ | + |
| Aspergillus flavus | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | ++ |
| Aspergillus fumigatus | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | + |
| Aspergillus oryzae | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | ++ |
| Aspergillus terreus | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| Aspergillus ustus | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | ++ |
| Candida albicans | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Candida dubliniensis | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Candida glabrata | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Candida guilliermondii | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Candida kefyr | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Candida krusei | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | +++ |
| Candida lusitaniae | +++ | +++ | +++ | +++ | +++ | +++ | + | +++ | +++ | ++ | − |
| Candida tropicalis | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Cryptococcus neoformans | +++ | +++ | + | ++ | +++ | +++ | ++ | +++ | +++ | ++ | − |
| Cunninghamellaberthollotiae | − | ++ | ++ | + | +++ | +++ | +++ | − | ++ | +++ | − |
| Fusarium solani | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| Mucorracemosus | +++ | ++ | ++ | +++ | +++ | +++ | +++ | − | +++ | +++ | + |
| Paecilomycesvariotii | − | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | + |
| Penicilliumnotatum | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Rhizomucormiehei | − | ++ | + | − | + | +++ | + | − | ++ | +++ | + |
| Rhizopusoryzae | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | +++ | ++ |
| Rhodotorulaglutinis | +++ | +++ | +++ | +++ | − | − | +++ | +++ | +++ | − | +++ |
| Saccharomyces cerevisiae | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Scedosporiumapiospermum | ++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | + |
| No. of +++ bands | 21 | 22 | 20 | 21 | 24 | 25 | 16 | 21 | 22 | 23 | 10 |
| No. of ++ bands | 1 | 3 | 2 | 1 | 0 | 0 | 8 | 0 | 3 | 2 | 6 |
| No. of + bands | 0 | 1 | 3 | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 7 |
| No. of with absent band | 4 | 0 | 1 | 2 | 1 | 1 | 0 | 5 | 1 | 1 | 3 |
| Effect of 1 μg human DNAb | None | None | None | None | None | None | None | None | None | None | None |
| Sum for distance matrixc | 642.5d | 1,055.8 | NDe | ND | ND | 310.8 | 470.6 | 67.5 | 74.5 | 117.3 | 358.3d |
| Sum for distance matrixf | 97,914d | 113,388 | ND | ND | ND | 82,452 | 61,321 | 17,522 | 9,629 | 28,716 | 66,445d |
| Amplicon lengthg | 295 ± 70d | 254 ± 43 | ND | ND | ND | 339 ± 7 | 198 ± 25 | 289 ± 1 | 147 ± 1 | 308 ± 46 | 263 ± 10d |
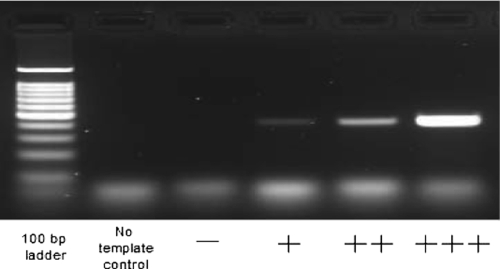
+++, ++, +, and − indicate the PCR yield reflected by the intensity level of product bands on a 1.5% agarose gel. +++, ++, and + indicate decreasing intensity levels, and − indicates no amplification. An example of a gel image is shown in Fig. 2.
Cross-reactivity was evaluated based on the ability to amplify 10 fg of A. fumigatus DNA in the presence of 1 μg of human genomic DNA.
Distances based on the Tajima-Nei algorithm and the neighbor-joining tree-building method. The sum for distance matrix was estimated from amplicons of 51 fungal species representing 30 genera. A higher value indicates greater sequence diversity among fungi for the selected amplicon.
Due to unavailability of some complete sequences spanning the ITS1(End18SF-5.8SR) and 28S(23F-25R) amplicons, 49 and 45 sequences, respectively, were used to estimate the sum of the distance matrix and mean amplicon length.
ND, distance not estimated because the amplicon overlaps significantly with another amplicon in the table which has a greater breadth of fungi detected.
Distances based on absolute nucleotide differences and the neighbor-joining tree-building method.
Means ± standard deviations for 51 fungal species representing 30 genera. The S. apiospermum sequence for the 28S(15F-22R), 28S(18F-22R), and 28S(18F-23R) amplicons was excluded from estimating the mean ± standard deviation due to the presence of a very large insert (nearly 400 bp) in the amplicon which disproportionately skewed the standard deviation estimate.
FIG. 2.
Intensity levels of PCR product bands in a 1.5% agarose gel. See Table 3.
Assessment of the potential for amplicons to distinguish among fungal species.
To evaluate the potential for accurate identification of species using the top 11 PCR amplicons, distance matrices were generated from the multiple-sequence alignment of 51 fungi for each amplicon. The sequences of the forward and reverse primers were excluded from the analysis. Distance matrices for three amplicons, ITS2(5.8SF-2R), ITS2(5.8SF-3R), and 28S(9F-12R), were not estimated due to significant sequence overlap with another amplicon that showed greater breadth of analytical sensitivity. The distance matrices based on sequences of common human fungal pathogens for the remaining top eight amplicons are shown in Tables S2 to S9 in the supplemental material. The sum for all the elements in the distance matrix was a quantity that reflected the magnitude of the ability to distinguish between species for each amplicon. For distances estimated using the Tajima-Nei algorithm, larger values reflect more nucleotide differences among fungi and therefore greater phylogenetic resolution for species identification. For example, the ITS2(5.8SF-1R) amplicon overlapping the ITS2 region, which was expected to have the highest level of sequence variation, had a distance matrix sum of 1,055.8, whereas the 28S(18F-22R) amplicon, which covers a highly conserved region of the 28S rRNA gene, had a distance matrix sum of only 74.5 (Table 3). A similar and more intuitive trend emerged when the sum of the distance matrix was estimated based on the absolute difference algorithm, which calculates the total number of base differences between pairs of fungal sequences in an alignment. In this case the ITS2(5.8SF-1R) amplicon also had the highest number of nucleotide differences, with a sum of 113,388, while the 28S(18F-22R) amplicon had the lowest sum of nucleotide differences, 9,629 (Table 3). Therefore, based on these genetic distances, the ITS2(5.8SF-1R) amplicon clearly had the greatest ability to distinguish between species. Combining information from the primer map (Fig. 1), sensitivity data (Table 3), and distance matrices (see Tables S2 to S9 in the supplemental material) provides useful data for selecting broad-range fungal PCR assays. Based on the ability to detect the widest range of fungi and simultaneously identify species, the ITS2(5.8SF-1R) and 28S(10F-12R) amplicon assays emerged as the top assays for broad-range fungal PCR.
An alternative perspective for analyzing the ability of amplicons to distinguish among species uses phylogenetic trees. Figures S1 and S2 in the supplemental material show phylogenetic trees for amplicons 28S(10F-12R) and ITS2(5.8SF-1R), respectively, based on 51 fungi. Note that the ITS2(5.8SF-1R) amplicon is highly polymorphic and lacks the property of a molecular clock, making it unreliable for inferring evolutionary relationships. The trees demonstrate that closely related fungi are resolved using the proposed amplicon sequences.
DISCUSSION
Fungal infections remain a major cause of morbidity and mortality in immunocompromised patients, such as those undergoing cancer chemotherapy, solid organ transplants, or hematopoietic cell transplants. Culture-based methods have poor diagnostic sensitivity for many fungal infections, which has led to the adoption of other diagnostic approaches, such as detection of fungal antigens. However, the galactomannan antigen-based assay and the glucan assay do not detect all fungal species (15, 16, 26). Accordingly, a negative antigen assay does not rule out fungal infection in a high-risk host, such as a stem cell transplant recipient with zygomycosis. In addition, the spectrum of fungal infections is likely to change with increasing use of antifungal medications for prophylaxis. The next generation of diagnostic tests will need to be capable of detecting emerging pathogens. Finally, pathogenic fungi in the same genus, such as C. albicans and C. krusei, may have different antifungal susceptibility profiles. Accurate identification of fungal species using molecular methods may allow more directed and therefore more successful antifungal therapy.
In this study we analyzed nearly 3,950 bp of sequence from the 3′ end of the 18S rRNA gene to the 3′ end of the 28S rRNA gene for the development of broad-range fungal PCR assays applicable to human tissue samples. Our focus was on selecting primers that had sequences broadly conserved among fungi and also significant sequence dissimilarity with human rRNA genes. A sequence alignment of 51 fungal rRNA genes was created by generating sequences de novo and deriving sequences available in public databases. Based on the ability to detect a broad range of fungi and the potential to distinguish between species, 62 amplicons in the size range from 75 to 400 bp generated from 27 unique primers were screened to obtain optimal amplicons for broad-range fungal PCR. Our analysis revealed that the 28S rRNA gene beyond the D1-D2 region is useful for designing broad-range fungal PCR assays with a good ability to distinguish between species. Our top assays have the potential to detect one-third of an A. fumigatus genome (10 fg) estimated to be equivalent to nine copies of the target rRNA gene in a background containing 1 μg of human DNA, representing a 100,000,000-fold excess of nonrelevant DNA.
Four of the nine fungi sequenced de novo were zygomycetes (R. miehei, C. bertholletiae, M. racemosus, and Absidia corymbifera). As shown by phylogenetic analysis of the large subunit of the rRNA gene, there is a large degree of sequence heterogeneity within the zygomycete taxon. Nevertheless, we were able to design broad-range PCR primers that amplify rRNA gene sequences from most zygomycetes without amplifying human DNA. The design of primers shown in Table 2 was constrained by the degree of similarity with the human rRNA gene sequence. For sequencing fungi in pure cultures, additional primers could be developed for highly conserved regions with more complete homology with fungal rRNA gene sequences. Although several sequencing primers from the website of the Vilgalys Lab at Duke University (http://www.biology.duke.edu/fungi/mycolab/primers.htm) used in this study have a high degree of homology with the zygomycete rRNA genes, most of them do not have mismatches with the human rRNA gene.
Distance matrices and resulting phylogenetic trees generated from sequence alignments of amplicons for a specific set of PCR primers reflect the nucleotide differences between fungi. Such analyses have been used to compare ITS1, ITS2, and the D1-D2 region of the 28S rRNA gene to determine their potential to distinguish between medically important fungal species (3, 6-8, 13, 17). The parameter in Table 3 which represents the sum of all elements of the distance matrix provides a global measure of nucleotide differences between fungal sequences for a specific amplicon. This distance matrix sum may not be a good measure of the ability of an amplicon to differentiate between species within a genus. In addition, an outlier sequence, i.e., a sequence with a large insert in the amplicon region [for example, 28S(18F-22R) of S. apiospermum], has the capacity to disproportionately increase the sum of all elements of the distance matrix, and as a consequence its impact may eclipse that of smaller yet more relevant nucleotide differences between species within a genus. Therefore, we provide distance matrices in Tables S2 to S9 in the supplemental material which display nucleotide differences for the most promising PCR amplicons and every unique combination of human fungal pathogens in this study. If species identification within a genus is desired, comparison of distance matrix data can help determine which amplicon would be able to better distinguish between the species of interest. For example, the ITS2(5.8SF-1R) and 28S(18F-22R) amplicons are comparable based on sensitivity testing, but when the distance matrix data are considered, it is evident that there are more nucleotide differences between any given combination of fungi for the ITS2(5.8SF-1R) amplicon.
Other studies have described the development of broad-range fungal primers targeting the rRNA operon, but there are limitations to many of these studies. First, it is unclear to what degree these previously described PCR primers interact with human DNA, and there are no quantitative data examining how human DNA impacts assay sensitivity and specificity for most studies. Second, in silico analysis of several previously described broad-range fungal primers shown in Fig. 1C (U2, D2, and NL4) reveals complete homology with their human counterparts, and for primer 260R and the human rRNA gene there was only one nucleotide difference. For other primers there were 3- to 9-bp mismatches with human rRNA genes, which is comparable to the findings for our primers. Third, variability in ITS lengths could result in inconsistent analytical sensitivity of the fungal PCR assay. For instance, an ITS assay may produce a 200-bp amplicon from one fungus and a 600-bp amplicon from a second fungus. The detection assay thresholds for these two fungi are unlikely to be the same. The 28S(10F-12R) amplicon, which had both excellent analytical sensitivity and the ability to distinguish between species, may help overcome the potential shortcomings of the ITS regions. Last, for PCR assays that use the amplicon length or melting temperature of the amplicons to distinguish between species, a single-amplicon approach may be insufficient. Therefore, use of more than one PCR target may be optimal (2, 6, 8, 14, 24). Our analysis of analytical sensitivity and the ability to distinguish between species revealed that a combination of the ITS2(5.8SF-1R) and 28S(10F-12R) amplicons was the most useful.
There are some limitations of our study. First, conventional endpoint PCR with gel electrophoresis was used to assess amplification. Quantitative PCR can be a better indicator of amplification efficiency, but analysis of PCR products by gel electrophoresis provides data on amplicon size, the generation of nonspecific amplification products, and product throughput (band intensity). Second, the 18S rRNA gene subunit was excluded from this analysis since it has been previously explored for broad-range fungal PCRs. There could be primers in the 18S rRNA gene that could be useful when they are used in conjunction with primers described in this study. Third, because the 28S rRNA gene is underrepresented in the GenBank database, the targets beyond the D1-D2 region may currently be less useful for species identification of novel fungi. In the future this issue could be mitigated by submission of more sequence information, as was the case in this study. Last, since only one sequence was used for each species, there could be polymorphic positions at primer sites or within the amplicons of interest that could impact the results. However, since primers were designed for highly conserved regions, the specificity of the primers is unlikely to be affected.
In conclusion, nine fungal rRNA operons from the 3′ end of the 18S rRNA gene to the 3′ end of 28S rRNA gene were sequenced de novo. A database of 51 rRNA gene sequences was created using additional sequences derived from publicly available genomes or other sequence databases. Our analysis of this region showed that there is nearly 2,900 bp of sequence beyond the D1-D2 region which is useful for the development of broad-range fungal PCR assays. The ITS2(5.8SF-1R) and 28S(10F-12R) rRNA gene PCR assays hold promise for detection and identification of fungal pathogens in human tissues using PCR.
Supplementary Material
Acknowledgments
This study was supported by NIH grant R01 AI054703 from the National Institute of Allergy and Infectious Diseases.
Sequence data for many fungi used in this study were obtained from the Fungal Genome Initiative, Broad Institute of Harvard and MIT (http://www.broad.mit.edu/node/304), or were produced by the Pathogen Genomics Group at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/Projects/Fungi/).
Footnotes
Published ahead of print on 9 January 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baskova, L., C. Landlinger, S. Preuner, and T. Lion. 2007. The Pan-AC assay: a single-reaction real-time PCR test for quantitative detection of a broad range of Aspergillus and Candida species. J. Med. Microbiol. 56:1167-1173. [DOI] [PubMed] [Google Scholar]
- 2.Bergman, A., V. Fernandez, K. O. Holmstrom, B. E. Claesson, and H. Enroth. 2007. Rapid identification of pathogenic yeast isolates by real-time PCR and two-dimensional melting-point analysis. Eur. J. Clin. Microbiol. Infect. Dis. 26:813-818. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. LaFe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evertsson, U., H. J. Monstein, and A. G. Johansson. 2000. Detection and identification of fungi in blood using broad-range 28S rDNA PCR amplification and species-specific hybridisation. APMIS 108:385-392. [DOI] [PubMed] [Google Scholar]
- 6.Henry, T., P. C. Iwen, and S. H. Hinrichs. 2000. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J. Clin. Microbiol. 38:1510-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinrikson, H. P., S. F. Hurst, L. De Aguirre, and C. J. Morrison. 2005. Molecular methods for the identification of Aspergillus species. Med. Mycol. 43(Suppl. 1):S129-S137. [DOI] [PubMed] [Google Scholar]
- 8.Hinrikson, H. P., S. F. Hurst, T. J. Lott, D. W. Warnock, and C. J. Morrison. 2005. Assessment of ribosomal large-subunit D1-D2, internal transcribed spacer 1, and internal transcribed spacer 2 regions as targets for molecular identification of medically important Aspergillus species. J. Clin. Microbiol. 43:2092-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwen, P. C., S. H. Hinrichs, and M. E. Rupp. 2002. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 40:87-109. [DOI] [PubMed] [Google Scholar]
- 10.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kedzierska, A., P. Kochan, A. Pietrzyk, and J. Kedzierska. 2007. Current status of fungal cell wall components in the immunodiagnostics of invasive fungal infections in humans: galactomannan, mannan and (1→3)-beta-d-glucan antigens. Eur. J. Clin. Microbiol. Infect. Dis. 26:755-766. [DOI] [PubMed] [Google Scholar]
- 12.Khot, P. D., D. L. Ko, R. C. Hackman, and D. N. Fredricks. 2008. Development and optimization of quantitative PCR for the diagnosis of invasive aspergillosis with bronchoalveolar lavage fluid. BMC Infect. Dis. 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau, A., S. Chen, T. Sorrell, D. Carter, R. Malik, P. Martin, and C. Halliday. 2007. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J. Clin. Microbiol. 45:380-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLintock, L. A., and B. L. Jones. 2004. Advances in the molecular and serological diagnosis of invasive fungal infection in haemato-oncology patients. Br. J. Haematol. 126:289-297. [DOI] [PubMed] [Google Scholar]
- 16.Ostrosky-Zeichner, L., B. D. Alexander, D. H. Kett, J. Vazquez, P. G. Pappas, F. Saeki, P. A. Ketchum, J. Wingard, R. Schiff, H. Tamura, M. A. Finkelman, and J. H. Rex. 2005. Multicenter clinical evaluation of the (1→3) beta-d-glucan assay as an aid to diagnosis of fungal infections in humans. Clin. Infect. Dis. 41:654-659. [DOI] [PubMed] [Google Scholar]
- 17.Rakeman, J. L., U. Bui, K. Lafe, Y. C. Chen, R. J. Honeycutt, and B. T. Cookson. 2005. Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J. Clin. Microbiol. 43:3324-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichenberger, F., J. Habicht, P. Matt, R. Frei, M. Soler, C. T. Bolliger, P. Dalquen, A. Gratwohl, and M. Tamm. 1999. Diagnostic yield of bronchoscopy in histologically proven invasive pulmonary aspergillosis. Bone Marrow Transplant. 24:1195-1199. [DOI] [PubMed] [Google Scholar]
- 19.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 20.Sandhu, G. S., B. C. Kline, L. Stockman, and G. D. Roberts. 1995. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 33:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tajima, F., and M. Nei. 1984. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1:269-285. [DOI] [PubMed] [Google Scholar]
- 22.Turenne, C. Y., S. E. Sanche, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Burik, J. A., D. Myerson, R. W. Schreckhise, and R. A. Bowden. 1998. Panfungal PCR assay for detection of fungal infection in human blood specimens. J. Clin. Microbiol. 36:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vollmer, T., M. Stormer, K. Kleesiek, and J. Dreier. 2008. Evaluation of novel broad-range real-time PCR assay for rapid detection of human pathogenic fungi in various clinical specimens. J. Clin. Microbiol. 46:1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 26.Yeo, S. F., and B. Wong. 2002. Current status of nonculture methods for diagnosis of invasive fungal infections. Clin. Microbiol. Rev. 15:465-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.