Abstract
Populations of Escherichia coli from juvenile and adult ring-billed gulls, juvenile common terns, and adult Canada geese were sampled over 6 years at five locations on Lake Superior (Duluth, MN, and Wisconsin) and Lake Michigan (Wisconsin, Illinois, and Indiana) to determine the extent of spatial and temporal variability in E. coli strains. Strain identity was determined using horizontal fluorophore-enhanced repetitive element palindromic DNA fingerprinting. Multivariate statistics were used to determine if spatial or temporal changes in E. coli populations occurred in waterfowl species. Pairwise multivariate analyses of variance revealed that E. coli populations of adult gulls from three regions of Lake Michigan and the Wisconsin shore of Lake Superior were similar to one another but different from an E. coli population of gulls from the Duluth region of Lake Superior. Juvenile and adult gulls from the Duluth area harbored different E. coli populations. The E. coli strains from juvenile gulls, however, were similar to those found in juvenile terns obtained from the same island rookery. Temporal changes in E. coli populations from several waterfowl species were also demonstrated for this site. Although portions of source tracking databases might be successfully used in other geographic regions, it is clear that juvenile birds should not be the sole source of E. coli strains used for source tracking databases, and multiple-year libraries should be constructed in order to identify the potential sources of E. coli in the environment.
The presence of Escherichia coli in recreational waters is commonly used as an indicator of recent fecal contamination. Recreational waters and beaches contaminated with fecal bacteria may contain pathogens that pose a health risk to humans (9). Monitoring fecal indicator bacteria in recreational waters and developing reliable methods to identify their possible sources require great effort and expense (42). While many studies have attempted to determine sources of E. coli in the environment (4, 19, 28, 35, 43), questions concerning host specificity, spatial and temporal differences, and environmental sources of E. coli have recently been raised by scientists, regulators, and managers of waste and water treatment facilities (1, 11, 13, 16, 21, 27, 33, 37, 44).
The use of molecular and phenotypic methods to determine the potential sources of fecal bacteria has been referred to as microbial source tracking (MST) (34, 36). Several library-independent and library-dependent methods have been developed for MST studies (17, 34, 36, 38, 39, 42, 46). Library-independent MST methods employ host-specific markers, such as host-specific PCR primers (8, 23, 24, 26) or gene probes (14). Both phenotypic characteristics (e.g., antibiotic resistance profiles, carbon utilization patterns) and genotypic characteristics (e.g., DNA fingerprint patterns) of microorganisms have been used for library-dependent MST methods. One of these methods, the horizontal fluorophore-enhanced repetitive element palindromic PCR (HFERP) DNA fingerprinting method, has been used widely and is currently one of the best available library-dependent MST methods because it is relatively quick, easy, and inexpensive to perform and has high discriminatory power (34, 36, 46).
When bacteria are used for library-dependent MST studies, a large database is necessary to represent the diverse phenotypic or genetic characteristics of target microorganisms, such as E. coli, that are found in multiple hosts. A fundamental assumption of MST studies, regardless of the methods used, is that strains or ecotypes of E. coli and other fecal bacteria are differentially distributed among animal and human hosts (17, 34, 42). If E. coli is not unique to a host source group, then the efficacy of MST studies is compromised (42). Several studies have shown that E. coli populations from swine (23), ducks and geese (14), and cattle (40) contain host-specific DNA sequences.
While these studies suggest that some populations of E. coli display some level of host specificity, some hosts can also harbor ephemeral (cosmopolitan) E. coli strains (1, 21, 42). Gordon and Lee (13) used multilocus enzyme electrophoresis to characterize enteric bacteria and reported that only 6% of the E. coli genetic similarity in 10 mammalian families could be explained by taxonomic classification. Anderson et al. (1) reported that many E. coli strains do not persist for more than 1 month in a given host. Moreover, some E. coli strains in humans (5), steers (21), and feral mice (11) have been shown to be shared and are continually being introduced and extirpated (5).
The population structure of E. coli is also influenced by the host's diet (16, 27, 33). Since different geographic regions provide different resources and diets for animals, populations of E. coli may display spatial divergence (42). Previous studies examining the spatial stability of E. coli populations have reported conflicting results. For example, Hartel et al. (15) reported that the percentage of shared E. coli ribotypes in cattle and horses decreased as the distance between the hosts increased, while ribotypes of E. coli isolated from chickens and swine did not show this geographic effect. Conversely, these authors reported that identical strains of E. coli could be found as far as 2,900 km apart. Other studies examining E. coli in animals and humans found identical strains across large geographic ranges (6, 28, 32, 45), perhaps due to the itinerant nature of humans (12) and the migratory behavior of animals (30).
Host-specific MST libraries are laborious to develop, so it is important for the workers engaged in MST studies to know if their databases reflect spatial and temporal differences in microbial population structure (38). We previously reported that wild waterfowl are a major source of E. coli in recreational waters and on beaches in Lake Superior (19, 25). Thus, it is important to understand the host specificity and the spatial and temporal dynamics of E. coli populations in wild waterfowl in the Great Lakes. In the study reported here, we examined the spatial and temporal structure of E. coli populations in samples from juvenile ring-billed gulls and juvenile common terns that inhabited the same island rookery in four different years, from Canada goose populations from the same area over 2 years, and from adult gull populations from Lake Michigan and Lake Superior that were sampled over a 6-year period.
MATERIALS AND METHODS
Isolation of E. coli from waterfowl and site descriptions.
E. coli was isolated from birds in Duluth, MN, from cloacal swabs for captured birds or from swab samples of freshly voided fecal material (BBL Culture Swabs; Becton Dickinson Microbiology Systems, Cockeysville, MD) and culturing on mFC agar as previously described (20). In 2006, E. coli strains were isolated from freshly voided fecal material on plastic sheets from adult ring-billed gulls (Larus delawarensis) at Southworth Marsh on Park Point and from the Blatnik Bridge site near Interstate Island in Duluth, MN (Table 1 and Fig. 1B). Southworth Marsh (46°44′26.4‴N, 92°03′42.8‴W) is a public beach adjacent to a popular city park in the Duluth-Superior harbor. The Blatnik Bridge site (46°45′04.7‴N, 92°06′11.0‴W) is a public boat landing in the Duluth-Superior harbor and is located about 3 km west of Southworth Marsh. E. coli isolates from juvenile (i.e., flightless) ring-billed gulls and juvenile common terns (Sterna hirundo) that inhabited Interstate Island were collected over multiple years from cloacal swabs. Interstate Island is a protected common tern rookery in the Duluth-Superior harbor (Fig. 1B) and is also home to 10,000 nesting pairs of ring-billed gulls (F. Strand, personal communication). E. coli strains were obtained from juvenile ring-billed gulls in 2002, 2003, 2005, and 2006 and from juvenile common terns in 2002, 2003, and 2006 (Table 1). E. coli strains from Canada geese (Branta canadensis) were isolated in 2005 from fecal material freshly excreted onto an asphalt parking lot at Park Point in 2005 and directly from the cloacae of captured geese in 2006.
TABLE 1.
Numbers of E. coli strains collected from adult and juvenile birds at different locations and on different dates in the Great Lakes
| Locationa | Site | Sampling dates | Avian species | Bird ageb | No. of E. coli isolates |
|---|---|---|---|---|---|
| DLH | Southworth Marsh | 2006 | Gull | Adult | 32 |
| Blatnik Bridge | 2006 | Gull | Adult | 59 | |
| Interstate Island | 2002, 2003, 2005, 2006 | Gull | Juvenile | 164 | |
| Interstate Island | 2002, 2003, 2006 | Tern | Juvenile | 92 | |
| Park Point | 2005 | Geese | Adult | 48 | |
| Park Point | 2006 | Geese | Adult | 74 | |
| SS | Cornucopia, WI | July and August 2004 | Gull | Adult | 18 |
| Ashland, WI | July and August 2004 | Gull | Adult | 7 | |
| Saxon Harbor, WI | July and August 2004 | Gull | Adult | 23 | |
| Madeline Island, WI | August 2004 | Gull | Adult | 8 | |
| Washburn, WI | August 2004 | Gull | Adult | 1 | |
| DC | Egg Harbor, WI | June 2005 | Gull | Adult | 4 |
| Jacksonport, WI | June to August 2005 | Gull | Adult | 16 | |
| Baileys Harbor, WI | June to August 2005 | Gull | Adult | 12 | |
| Sturgeon Bay, WI | June 2005 | Gull | Adult | 3 | |
| LC | Lake County, IL | February and March 2003 | Gull | Adult | 31 |
| IN | West Beach, IN | June 1998 to August 2001 | Gull | Adult | 19 |
DLH, Duluth-Superior harbor, Lake Superior; SS, south shore of Lake Superior, Wisconsin; DC, Door County, WI, Lake Michigan; LC, Lake County, IL, Lake Michigan; IN, Indiana, Lake Michigan.
Flightless birds were considered juveniles.
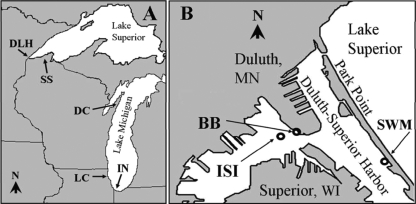
FIG. 1.
Maps of study areas, showing sampling site locations. (A) Map of Lakes Superior and Michigan, showing sites in the Duluth-Superior harbor, Lake Superior (DLH); the south shore of Lake Superior (SS); Door County, WI, Lake Michigan (DC); Lake County, IL, Lake Michigan (LC); and Indiana, Lake Michigan (IN). (B) Map of Duluth-Superior harbor, showing the locations of Interstate Island (ISI), the Blatnik Bridge site (BB), and the Southworth Marsh site (SWM).
E. coli strains were also isolated by other researchers from adult ring-billed gulls in four other regions of Lake Superior and Lake Michigan (Table 1 and Fig. 1A). Gull E. coli strains were isolated from fresh fecal material on the ground at five different sites along Lake Superior's Wisconsin shoreline in 2004 (G. Kleinheinz, personal communication), from five Lake Michigan sites in Door County, WI, in 2005, from Illinois public beaches on Lake Michigan during 2003 (W. Ting, personal communication), and from Lake Michigan in Indiana during 1998 and 2001 (Fig. 1A).
Confirmation of E. coli identity.
The identities of E. coli strains were verified by using the microbiological and biochemical criteria described by Ishii et al. (20), with the following changes. Bacterial colonies that were dark blue on mFC agar (Difco, Detroit, MI) were transferred to MacConkey agar (Difco), and pink to red colonies were tested on CHROMagar ECC (CHROMagar Microbiology, Paris, France). The identities of bacterial colonies that were blue or white on CHROMagar ECC agar were verified with the IMVIC series of tests as described by Ishii et al. (20), and the organisms were stored in 50% glycerol at −80°C until they were used. E. coli strains from Wisconsin were obtained from Greg Kleinheinz and were isolated on modified m-TEC agar (41). Between 1 and 5% of these isolates were also verified to be E. coli by using API 20E biochemical test strips (G. Kleinheinz, personal communication). Gull E. coli isolates from Illinois and Indiana were obtained from W. T. Evert Ting (Purdue University, Calumet). The gull E. coli strains collected in Indiana were identified using the BBL Crystal identification system (Becton Dickinson Microbiology Systems, Cockeysville, MD) (W. Ting, personal communication), while the identities of E. coli strains from Illinois were verified by the Illinois Department of Health.
DNA fingerprinting and statistical analyses.
DNA fingerprinting of E. coli isolates was performed by using HFERP DNA fingerprinting and the BOXA1R primer as previously described (20, 22). Electrophoresis, visualization, and analyses of bands were done as previously described (20). HFERP DNA fingerprints were analyzed using BioNumerics v.4.5 software (Applied Maths, Sint-Martens-Latem, Belgium). Dendrograms were constructed using the curve-based Pearson's product-moment correlation coefficient and the unweighted-pair group method with arithmetic means clustering method (20, 22), and they were analyzed by comparing the proportions of subclusters that were assembled exclusively from one E. coli population (29). A subcluster was defined as a group of two or more E coli strains that were not clones and were ≥80% similar. E. coli strains were considered clones if the similarity of their HFERP DNA fingerprints was ≥92%, a value based on the work of Johnson et al. (22). Identical E. coli clones obtained from the same animal were removed from the analyses to reduce bias (22).
Spatial, temporal, and host-specific relationships of E. coli populations were analyzed by using the multivariate analysis of variance (MANOVA) and cluster analysis subroutines of the BioNumerics software. Bonferroni corrections were employed for MANOVA that compared three or more populations.
RESULTS AND DISCUSSION
Adult ring-billed gull E. coli populations from different regions of Lake Superior and Lake Michigan.
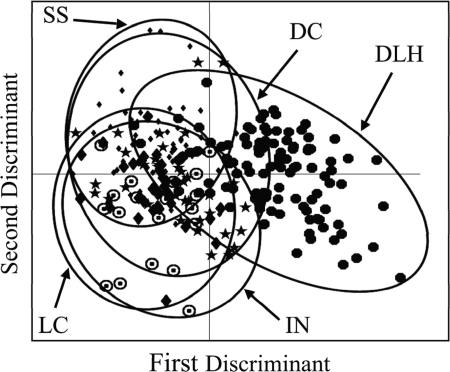
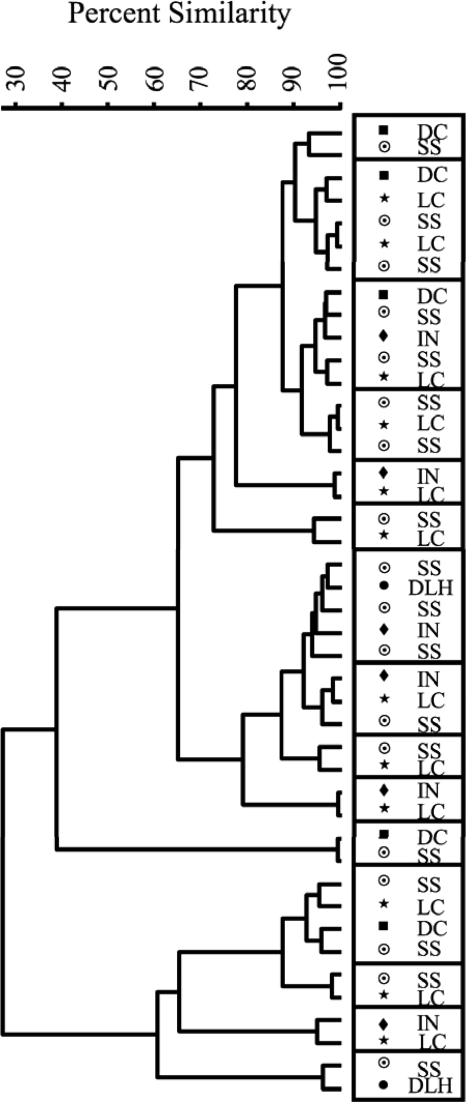
Pairwise MANOVA revealed that the HFERP patterns of E. coli populations in adult ring-billed gulls from three regions of Lake Michigan and the Wisconsin shoreline of Lake Superior were similar to one another (P < 0.01) but different from the HFERP pattern of the E. coli population in gulls from the Duluth region of Lake Superior (Fig. 2 and Table 2). Seventy-six percent of the subclusters formed in the cluster analysis were comprised of E. coli from gulls from multiple regions. Ten E. coli strains were found in two different regions, four strains were found in three different regions, and one strain was found in four different regions (Fig. 3).
FIG. 2.
Plot of the first two discriminants from a MANOVA comparing E. coli populations in adult ring-billed gulls from Minnesota, Wisconsin, Illinois, and Indiana. DLH, Duluth-Superior harbor (solid circles); SS, south shore of Lake Superior (small filled diamonds); DC, Door County, WI (filled stars); LC, Lake County, IL (large filled diamonds); IN, Indiana (circles with dots).
TABLE 2.
Similarity of E. coli strains isolated from adult ring-billed gulls from different regions of the Great Lakes based on the similarity of their HFERP DNA fingerprint patterns
| Locationa | No. of E. coli isolates |
P valueb
|
|||
|---|---|---|---|---|---|
| DLH | SS | DC | LC | ||
| DLH | 91 | ||||
| SS | 57 | <0.001 | |||
| DC | 35 | <0.001 | 0.366 | ||
| LC | 31 | <0.001 | 0.331 | 0.910 | |
| IN | 19 | <0.001 | 0.622 | 0.924 | 0.997 |
DLH, Duluth-Superior harbor, Lake Superior; SS, south shore of Lake Superior, Wisconsin; DC, Door County, WI, Lake Michigan; LC, Lake County, IL, Lake Michigan; IN, Indiana, Lake Michigan.
P values for comparisons tested by MANOVA.
FIG. 3.
Dendrogram of E. coli strains in adult ring-billed gulls from sites in Lake Superior and Lake Michigan. E. coli isolates in boxes have an HFERP fingerprint similarity of ≥92% and were considered clones. The dendrogram was generated from HFERP DNA fingerprints using Pearson's product-moment correlation coefficient and the unweighted-pair group method with arithmetic means clustering method. DLH, Duluth-Superior harbor (•); SS, south shore of Lake Superior (⊙); DC, Door County, WI (▪); LC, Lake County, IL (★); IN, Indiana (⧫).
These results indicate that E. coli populations from adult gulls can be highly related over large geographic distances. One reason for this pattern may be the migratory habits and dispersal patterns of gulls. Ring-billed gulls from the Great Lakes migrate to the Gulf Coast of the United States and winter mainly in Florida (10), where they intermingle. After studying the migration and dispersal patterns of Great Lakes ring-billed gulls, Gabrey (10) concluded that movement of gulls was extensive, particular Great Lakes and colonies do not define distinct populations, and the dispersal of young birds plays a significant role in establishing and maintaining colonies. Similarly, it appears that E. coli strains in ring-billed gulls from most areas of Lake Michigan and Lake Superior do not define distinct regional ecotypes or populations. It is interesting, however, that the significance (i.e., P values) in the pairwise MANOVA increased as the distance between gull E. coli populations decreased in Lake Michigan (Table 2). Taken together, these results suggest that portions of source tracking databases constructed for one area might be successfully used for studies done in other Great Lakes regions. However, the variety and geographic similarity of E. coli strains in wild animal species have not been well studied. Like the study reported by Gabrey (10) for gull populations in the Great Lakes, new research on E. coli population dynamics could help clarify the scales over which distinct bacterial populations and metapopulations form in wild animal species and the probable success of transferring and using MST databases in other geographic regions.
There are several possible explanations for the difference between the E. coli populations in ring-billed gulls from the Duluth site and the E. coli populations from other regions of Lake Superior and Lake Michigan. The food resources available to gulls in an urban area like Duluth, MN, may support different E. coli populations than the food resources available in less urban areas of the Great Lakes support. At more remote sites, the diets of ring-billed and herring gulls consist primarily of fish and terrestrial food, such as insects (7, 18), whereas human food is a major component of the diets of urban ring-billed gulls (2, 3). Moreover, gulls in urban areas like the Duluth-Superior harbor are also likely to be exposed to E. coli strains that are discharged from wastewater treatment facilities, while gull colonies in more remote areas are not exposed to such strains. Another possible explanation, which may be specific to this study, is the way in which gull fecal materials were collected at the different sites. Adult gulls in Duluth were baited over plastic sheets on which they voided fecal material, whereas gull fecal materials from the other sites were collected directly on the ground and therefore might have been contaminated with other E. coli strains. Alternatively, more culturable E. coli strains and possibly a greater diversity of strains might have been obtained from freshly voided fecal material collected in the Duluth region from plastic sheeting than from older fecal materials collected on beaches. While all of these scenarios seem plausible, our research was not designed to identify which of these factors, if any, was responsible for the difference between the E. coli populations in ring-billed gulls from the Duluth site and those from other regions of Lake Superior and Lake Michigan.
E. coli strains from juvenile and adult ring-billed gulls.
The E. coli strains collected from juvenile ring-billed gulls in Duluth were compared to those obtained from adult ring-billed gulls in Duluth and the other Great Lakes regions (Table 1). Comparisons were made for E. coli strains collected during the same year, so only E. coli strains collected from adult gulls at Duluth in 2006 (n = 91) and from Lake Michigan sites in Wisconsin in 2005 (n = 35) and in Illinois in 2003 (n = 11) could be compared to strains collected from juvenile birds at the Duluth location in 2006 (n = 31), 2005 (n = 65), and 2003 (n = 11). In most cases, adult ring-billed gulls harbored E. coli populations different from those harbored by juvenile gulls.
The strongest comparisons can be made between E. coli strains from adult and juvenile gulls living at the same site. The E. coli populations from adult and juvenile gulls collected in 2006 in only the Duluth area were different when they were compared by using MANOVA (P < 0.05). Only 18% of the subclusters in dendrograms contained E. coli strains from both adult and juvenile ring-billed gulls (data not shown), and only three strains were found in both adult and juvenile birds. E. coli strains isolated from adult and juvenile gulls in different regions of the Great Lakes were also different. For example, E. coli strains collected from adult gulls in the Door County, WI, region of Lake Michigan during 2005 were different from strains collected from juvenile gulls in Duluth during the same year (P < 0.05). No E. coli strains were found in both adult gulls from Door County, WI, and juvenile gulls at the Duluth location. Similarly, E. coli strains obtained from adult gulls in Illinois were different (P < 0.05) from strains collected during the same year (2003) from juvenile gulls in the Duluth region of Lake Superior. Since adult birds bring food to nonfledged gulls, these results indicated that the rookery habitat might be responsible for the different E. coli strains found in juvenile gulls. It is clear from these results that juvenile (“flightless”) birds should not be the sole source of E. coli strains for representing waterfowl species in MST databases.
E. coli strains from juvenile gulls and terns that inhabit the same island.
The population structure of E. coli strains collected from juvenile ring-billed gulls was not statistically different (P > 0.05) from that of strains obtained from juvenile common terns that lived on Interstate Island concurrently during 2002, 2003, and 2006 (Table 3). Cluster analysis did not discretely group the E. coli strains from juvenile gulls and juvenile terns for any year (2002, 2003, or 2006), and about 58, 44, and 57% of the subclusters, respectively, contained E. coli strain fingerprints from juvenile birds of both species. These results indicate that the intestinal tracts of juvenile birds of different waterfowl species that live close to each other can have very similar E. coli strain compositions.
TABLE 3.
Comparison of E. coli strains collected from juvenile (“flightless”) common terns and ring-billed gulls at the Duluth, MN, locationa
| Comparison
|
No. of E. coli isolatesc | P value | |
|---|---|---|---|
| Speciesb | Yr | ||
| CT vs RBG | 2002 | 25, 57 | 0.78 |
| CT vs RBG | 2003 | 56, 11 | 0.93 |
| CT vs RBG | 2006 | 11, 31 | 0.95 |
| RBG | 2002 vs 2003 | 57, 11 | 0.08 |
| RBG | 2003 vs 2005 | 11, 65 | <0.01 |
| RBG | 2005 vs 2006 | 65, 31 | 0.01 |
| CT | 2002 vs 2003 | 25, 56 | <0.01 |
| CT | 2002 vs 2006 | 25, 11 | <0.01 |
| CT | 2003 vs 2006 | 56, 11 | <0.01 |
HFERP fingerprint patterns from different avian species and from the same bird species in different years were compared by MANOVA.
RBG, ring-billed gull; CT, common tern.
Numbers of isolates in the groups compared. All E. coli strains were isolated from juvenile birds on Interstate Island in the Duluth-Superior harbor.
Considering that the diets of juvenile common terns and ring-billed gulls are different (3, 31), the confined rookery area that juvenile birds share may influence the E. coli strains that they harbor as much as their food sources. This possibility implies that the composition of a host's E. coli is influenced not only by diet (16, 27, 33) but also by the habitat in which the host animal resides. While previous studies have shown that different species of domestic animals may share the same E. coli strains (1), to our knowledge this is the first time that this phenomenon has been demonstrated for wild bird species.
Temporal changes in waterfowl E. coli populations.
The genetic relatedness of E. coli strains isolated from juvenile ring-billed gulls, juvenile common terns, and adult Canada geese during different years in the Duluth-Superior harbor was examined to determine if temporal changes occur in E. coli populations in waterfowl. The HFERP patterns of E. coli strains in juvenile ring-billed gulls were different in different years, except for 2002 and 2003 (P > 0.05) (Table 3). Only one E. coli strain was found in both the 2002 and 2003 samples. A dendrogram comparing juvenile gull E. coli strains obtained in all 4 years showed that 81% of the subclusters contained isolates from each year. The maximum level of similarity of any two E. coli strains between 2002 and 2006 was 89.7%. The HFREP patterns of E. coli strains isolated from juvenile common terns were different in 2002, 2003, and 2006 (P < 0.05) (Table 3). Only one to three strains were found in all these years. While the levels of similarity between strains ranged from 8.3 to 96.6% when all 3 years were considered, the maximum level of similarity was lower (less than 86%) when pairs of years were compared. The majority (59%) of subclusters in the dendrogram included strains from at least two different years.
Similarly, the HFERP patterns of E. coli strains isolated from adult Canada geese were different in 2005 (48 isolates) and 2006 (74 isolates) (P < 0.05). Although cluster analyses revealed that E. coli strains isolated in each year were present throughout the dendrogram, eight strains were present in both years, demonstrating that some E. coli strains can persist from year to year in waterfowl populations. These results agree with those of other studies (1, 21) that found that a few E. coli strains persist in warm-blooded animals over time, but the majority of E. coli strains may not be part of a permanent intestinal microflora.
Taken together, the results of these studies demonstrate that the variety of E. coli strains can change annually in waterfowl species, that juvenile and adult birds of the same species harbor different populations of E. coli, and that adult gulls from three regions of Lake Michigan and the Wisconsin shore of Lake Superior may contain similar populations of E. coli that differ from E. coli populations in adult gulls from the Duluth region of Lake Superior. Thus, while some portions of source tracking databases might be successfully used in other geographic regions, it is clear that juvenile birds should not be the sole source of E. coli strains used for source tracking databases and that comprehensive libraries should be constructed with E. coli host strains from animal hosts that are collected over several years in order to identify the potential sources of E. coli in the environment.
Acknowledgments
We thank Greg Kleinheinz (University of Wisconsin, Oshkosh), W. T. Evert Ting, Charles C. Tseng, and Deborah Johnson (Purdue University, Calumet) for providing E. coli isolates from gulls. We also thank Fred Strand (Wisconsin Department of Natural Resources) and Rich Staffon (Minnesota Department of Natural Resources) for help with obtaining bird fecal swabs on Interstate Island and at Park Point, MN, and Jenna Bergin, Jason Kish, John Gralewski, and Ryan Oster for technical assistance in the field and the lab.
This work was supported in part by a grant from the Minnesota Sea Grant College Program, NOAA Office of Sea Grant, United States Department of Commerce (grant NA03-OAR4170048 to R.E.H. and M.J.S.).
Footnotes
Published ahead of print on 9 January 2009.
This paper is journal reprint no. JR560 of the Minnesota Sea Grant College Program.
REFERENCES
- 1.Anderson, M. A., J. E. Whitlock, and V. J. Harwood. 2006. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 72:6914-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belant, J. L., S. K. Ickes, and T. W. Seamans. 1998. Importance of landfills to urban-nesting herring and ring-billed gulls. Landscape Urban Planning 43:11-19. [Google Scholar]
- 3.Brousseau, P., J. Lefebvre, and J.-F. Giroux. 1996. Diet of ring-billed gull chicks in urban and non-urban colonies in Quebec. Colonial Waterbirds 19:22-30. [Google Scholar]
- 4.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 5.Caugant, D. A., B. R. Levin, and R. K. Selander. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98:467-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caugant, D. A., B. R. Levin, and R. K. Selander. 1984. Distribution of multilocus genotypes of Escherichia coli within and between host families. J. Hyg. 92:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chudzik, J. M., K. D. Graham, and R. D. Morris. 1994. Comparative breeding success and diet of ring-billed and herring gulls on South Limestone Island, Georgian Bay. Colonial Waterbirds 17:18-27. [Google Scholar]
- 8.Dick, L. K., and K. G. Field. 2004. Rapid estimation of numbers of fecal Bacteriodetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufour, A. P. 1984. EPA health effects criteria for fresh recreational waters. Office of Research and Development, United States Environmental Protection Agency, Research Triangle Park, NC.
- 10.Gabrey, S. W. 1996. Migration and dispersal in Great Lakes ring-billed and herring gulls. J. Field Ornithol. 67:327-339. [Google Scholar]
- 11.Gordon, D. M. 1997. The genetic structure of Escherichia coli populations in feral house mice. Microbiology 143:2039-2046. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, D. M., and J. Lee. 1999. The genetic structure of enteric bacteria from Australian mammals. Microbiology 145:2673-2682. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton, M. J., T. Yan, and M. J. Sadowsky. 2006. Development of goose- and duck-specific DNA markers to determine sources of Escherichia coli in waterways. Appl. Environ. Microbiol. 72:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartel, P. G., J. D. Summer, J. L. Hill, J. V. Collins, J. A. Entry, and W. I. Segars. 2002. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed] [Google Scholar]
- 16.Hartel, P. G., J. D. Summer, and W. I. Segars. 2003. Deer diet affects ribotype diversity of Escherichia coli for bacterial source tracking. Water Res. 37:3263-3268. [DOI] [PubMed] [Google Scholar]
- 17.Harwood, V. J. 2007. Assumptions and limitations associated with microbial source tracking, p 33-64. In J. W. Santo Domingo and M. J. Sadowsky (ed.), Microbial source tracking. ASM Press, Washington, DC.
- 18.Hebert, C. E., J. L. Shutt, K. A. Hobson, and D. V. C. Weseloh. 1999. Spatial and temporal differences in the diet of Great Lakes herring gulls (Larus argentatus): evidence from stable isotope analysis. Can. J. Fish. Aquat. Sci. 56:323-338. [Google Scholar]
- 19.Ishii, S., D. L. Hansen, R. E. Hicks, and M. J. Sadowsky. 2007. Beach sand and sediments are temporal sinks and sources of Escherichia coli in Lake Superior. Environ. Sci. Technol. 41:2203-2209. [DOI] [PubMed] [Google Scholar]
- 20.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkins, M. B., P. G. Hartel, T. J. Olexa, and J. A. Stuedemann. 2003. Putative temporal variability of Escherichia coli ribotypes from yearling steers. J. Environ. Qual. 32:305-309. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatib, L. A., Y. L. Tsai, and B. H. Olson. 2003. A biomarker for the identification of swine fecal pollution in water, using the STII toxin gene from enterotoxigenic Escherichia coli. Appl. Microbiol. Biotechnol. 63:231-238. [DOI] [PubMed] [Google Scholar]
- 24.Kirs, M., and D. C. Smith. 2007. Multiplex quantitative real-time reverse transcriptase PCR for F+-specific RNA coliphages: a method for use in microbial source tracking. Appl. Environ. Microbiol. 73:808-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ksoll, W. B., S. Ishii, M. J. Sadowsky, and R. E. Hicks. 2007. Presence and sources of fecal coliform bacteria in epilithic periphyton communities of Lake Superior. Appl. Environ. Microbiol. 73:3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Layton, A., L. McKay, D. Williams, V. Garrett, R. Gentry, and G. Saylor. 2006. Development of Bacteriodes 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72:4214-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leser, T. D., R. H. Lindecrona, T. K. Jensen, B. B. Jensen, and K. Moller. 2000. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl. Environ. Microbiol. 66:3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molina, M. 2005. Temporal and spatial variability of fecal indicator bacteria: implications for the application of MST methodologies to differentiate sources of fecal contamination. Ecosystems Research Division, National Exposure Research Laboratory Athens, GA.
- 31.Nisbet, I. C. T. 2002. Common tern (Sterna hirundo), p. 1-40. In A. Poole and F. Gill (ed.), The birds of North America, no. 618. The Academy of Natural Sciences, Philadelphia, PA.
- 32.Ochman, H., T. S. Whittam, D. A. Caugant, and R. K. Selander. 1983. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J. Gen. Microbiol. 129:2715-2726. [DOI] [PubMed] [Google Scholar]
- 33.Russell, J. B., F. Diez-Gonzalez, and G. N. Jarvis. 2000. Invited review: effects of diet shifts on Escherichia coli in cattle. J. Dairy Sci. 83:863-873. [DOI] [PubMed] [Google Scholar]
- 34.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, T. M., J. Caren, G. R. Nelson, T. M. Jenkins, and J. Lukasik. 2004. Tracking sources of fecal pollution in a South Carolina watershed by ribotyping Escherichia coli: a case study. Environ. Forensics 5:15-19. [Google Scholar]
- 36.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking—state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 37.Souza, V., M. Travisano, P. E. Turner, and L. E. Eguiarte. 2002. Does experimental evolution reflect patterns in natural populations? E. coli strains from long-term studies compared with wild isolates. Antonie van Leeuwenhoek 81:143-153. [DOI] [PubMed] [Google Scholar]
- 38.Stewart, J., J. W. Santo Domingo, and T. J. Wade. 2007. Fecal pollution, public health, and microbial source tracking, p. 1-32. In J. W. Santo Domingo and M. J. Sadowsky (ed.), Microbial source tracking. ASM Press, Washington, DC.
- 39.Stoeckel, D. M., and V. J. Harwood. 2007. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73:2405-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, Y. L., J. Y. Le, and B. H. Olson. 2003. Magnetic bead hybridization to detect enterotoxigenic Escherichia coli strains associated with cattle in environmental water sources. Can. J. Microbiol. 49:391-398. [DOI] [PubMed] [Google Scholar]
- 41.U.S. Environmental Protection Agency. 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). EPA-821-R-02-023. Office of Water (4303T), Washington, DC.
- 42.U.S. Environmental Protection Agency. 2005. Microbial source tracking guide. Document EPA/600/R-05/064. U.S. Environmental Protection Agency, Washington, DC.
- 43.Vogel, J. R., D. M. Stoeckel, R. Lamendella, R. B. Zelt, J. W. Santo Domingo, S. R. Walker, and D. B. Oerther. 2007. Identifying fecal sources in a selected catchment reach using multiple source-tracking tools. J. Environ. Qual. 36:718-729. [DOI] [PubMed] [Google Scholar]
- 44.Whittam, T. S. 1989. Clonal dynamics of Escherichia coli in its natural habitat. Antonie van Leeuwenhoek 55:23-32. [DOI] [PubMed] [Google Scholar]
- 45.Whittam, T. S., H. Ochman, and R. K. Selander. 1983. Geographic components of linkage disequilibrium in natural populations of Escherichia coli. Mol. Biol. Evol. 1:67-83. [DOI] [PubMed] [Google Scholar]
- 46.Yan, T., and M. J. Sadowsky. 2007. Determining sources of microorganisms in waterways. Environ. Monit. Assess. 129:97-106. [DOI] [PubMed] [Google Scholar]