Abstract
The gene for the Thermotoga maritima Trk potassium transporter component TrkA was originally thought to be a frameshift mutation and not to encode a functional protein. However, expression from this gene yielded a complex consisting of two distinct proteins designated TM1088A and -B. Genetic complementation of Escherichia coli mutants unable to transport potassium suggests that TM1088A/B is part of a functional Trk potassium transporter complex with the membrane protein TM1089. The protein structure for TM1088A shows a characteristic Rossmann fold indicating an NAD+ binding site and has structural similarity to potassium channel-related proteins. Ligand binding studies indicated that ATP, ADP, and AMP stabilized TM1088A to a much greater degree than NADH and NAD, consistent with the crystal structure of TM1088A, which contains a bound AMP natural ligand at the characteristic GXGXXG nucleotide binding site. Mutation of single and all glycines at this nucleotide binding site eliminated in vitro protein stabilization by the ligand, yet these mutated proteins could still functionally complement the E. coli potassium uptake mutants. We predict that this new two-subunit class of TrkA proteins is present in a number of organisms. A further subclass of the predicted two-subunit TrkA proteins lack an identifiable membrane-spanning subunit of the Trk K+ transporter. This class, as exemplified by Mycobacterium tuberculosis, did not complement E. coli potassium transport with the native E. coli TrkH; thus, it may require a novel TrkH-like protein for activity or provide an alternate function in vivo.
Potassium is a major monovalent cellular cation that plays a key role in maintaining the cell turgor pressure (5), affects cytoplasmic pH (12), and even regulates the activity and expression of enzymes (26). The intracellular concentration of K+ (150 to 500 mM) (9, 13) is generally much higher than that in the extracellular environment. Thus, controlling intracellular K+ levels with channels and active transporters is vital for cell growth and survivability in the host and the natural environment. Bacteria often have multiple K+ uptake systems, and three of those systems (Kdp, Kup, and Trk) have been characterized in Escherichia coli (6). The Kdp-ATPase is expressed at very low K+ concentrations and in response to a decrease in turgor pressure (25). The Kup transporter is constitutive and transports K+ at a relatively low rate (39). The Trk potassium transporter is the major constitutive K+ transporter in E. coli, transporting K+ more than 10 times faster than the Kup transporter (39). Homologs of Trk can also be found in many bacterial genomes, illustrating its widespread role in bacterial physiology.
The Trk system, a proton symporter, is powered by the proton motive force and requires ATP (38), although the hydrolysis of ATP to ADP has not been shown (16, 23). In addition to K+ transport, it may also play a role in the resistance to antimicrobial peptides through K+ transport or by another means (35, 44). The Trk K+ transport system consists of a membrane-spanning protein and a peripheral membrane-associated nucleotide binding protein. E. coli contains homologous genes, trkH and trkG, only one of which is required, that encode the membrane-spanning protein (10). Both require trkA for function, and the trkH-dependent transport system requires an additional gene, trkE, encoding an ATP binding protein (10). In E. coli, these genes are dispersed on the chromosome and do not form an operon. TrkA, which contains a nucleotide binding domain, is a peripheral membrane protein located on the cytoplasmic side of the membrane (4). E. coli TrkA has been well studied and shown to bind NAD+/NADH but not ATP (41). It, like most TrkAs, is composed of two similar domains that have sequence similarity to dehydrogenases (41). In microbial genomes lacking a Trk transporter, such as Deinococcus radiodurans, Mycoplasma pneumoniae M129, and Shewanella oneidensis MR-1, an Na+/K+ symporter, KtrAB, is present. The Ktr transporter may also be present in addition to the Trk transporter. This transporter is also composed of a membrane-spanning protein, KtrB, and a peripheral membrane-associated nucleotide binding subunit, KtrA. KtrA has been shown to bind both ATP and NAD+/NADH (23). The peripheral membrane-associated cytoplasmic domains found in both TrkA and KtrA are present in K+ uptake and efflux transporters and channels and are thought to play a role in regulation by inducing a conformational change upon ligand binding (18). The approximately 140-amino-acid cytoplasmic domains are often referred to as RCK (regulating the conductance of K+) (19) or KTN (K+ transport, nucleotide binding) (15) domains. The KTN domains contain the characteristic GXGXXG nucleotide binding consensus, which may be absent in RCK domains.
We have studied the predicted potassium transport genes in Thermotoga maritima MSB8. This organism has been extensively studied as part of our NIGMS Protein Structure Initiative structural genomics program (28). In addition to structure determination, we have performed biochemical studies for hundreds of proteins across the T. maritima proteome in an attempt to validate functional annotations and understand structure/function relationships. This hyperthermophilic marine bacterium is a strict anaerobe that grows at an optimal temperature of 80°C by fermentation (17) and iron reduction (46). A unique feature of the Thermotogales is its deeply branching position on the 16S rRNA gene tree of life (1). The deeply branching phylogenetic position suggests a more ancient lineage with a close relationship to the last common ancestor and a closer relationship to archaea and eukarya than other bacteria.
In contrast to E. coli's three K+ transporters, only the Trk K+ transport system has been annotated in T. maritima. The proteins predicted to be involved in K+ uptake, based upon the annotation (32), are TM1088, annotated as the N-terminal half of TrkA; TM1089, the membrane-spanning protein of the Trk system; and TM1057, another TrkA-like annotated protein. Aspects of TM1088 are unusual for a putative K+ transporter. TM1088 has significant sequence divergence from an experimentally studied representative, and the protein is annotated as a suspected frameshift (32) in comparison with known Trk homologs. However, no other K+ transport systems are found in T. maritima, suggesting that TM1088 is part of a functional transporter. In this study, we show that TM1088, the T. maritima TrkA, represents a new class of two-subunit TrkA-like K+ transporters. This two-subunit TrkA complex can be identified in a number of sequenced bacterial genomes. However, a subclass of these two-subunit TrkA complexes, exemplified by Mycobacterium tuberculosis, lack a recognizable TrkH and may perform a unique function.
MATERIALS AND METHODS
Stains, plasmids, and recombinant techniques.
The strains and plasmids used in this study are listed in Table 1. Plasmids pTM1088A, pTM1088A/B-TM1089, pTM1088B-TM1089, pTM1088AB G14A G16A G19A TM1089, pTM1088B RCK, and pMT2765/6 were constructed as described previously (21). Primers were designed for amplification of the full-length proteins. The following primers sets were used to amplify the T. maritima genes for insertion into the pSpeedET expression plasmid: TM1088A/B to TM1089, forward 5′-CTGTACTTCCAGGGCATGTCAAAAAAACAAAAAAGC-3′ and reverse 5′-AATTAAGTCGCGTTATCAACTATTCACACCTCC-3′; TM1088A, forward 5′-CTGTACTTCCAGGGCATGTCAAAAAAACAAAAAAGC-3′ and reverse 5′-AATTAAGTCGCGTTATCAATCTTCTTCACTCCC-3′; TM1088B to TM1089, forward 5′-CTGTACTTCCAGGGCTTGAAAGTCATAATAATCGGTGG-3′ and reverse 5-AATTAAGTCGCGTTATCAACTATTCACACCTCC-3′; TM1089, forward 5′-CTGTACTTCCAGGGCATGACCAGTACCAAATACAGACTCAAGG-3′ and reverse 5′-AATTAAGTCGCGTTATCAACTATTCACACCTCC-3′, and TM1088B RCK domain (amino acids 1 to 136), forward 5′-CTGTACTTCCAGGGCATGAAAGTCATAATAATCGGTGG-3′ and reverse 5′-AATTAAGTCGCGTTATCAGAACTCATCTGGGAAGATGAGAGC-3′. Amplification of MT2765/6 was achieved with the primers forward 5′-CTGTACTTCCAGGGCATGCGGGTGGTTGTGATGGGGTGCG-3′ and reverse 5′-AATTAAGTCGCGTTATTACATGGACGGCAGCAGCAGCC-3′. The expression vector carries kanamycin resistance, and proteins are expressed by the arabinose induction system. Expressed proteins carry a cleavable N-terminal His6 tag followed by a TEV protease cleavage site for affinity purification. The PCR products were used to transform E. coli HK100. Expression plasmids containing TM1088A/B and TM1057 have been described previously as part of the whole-genome T. maritima clone collection (28). These plasmids are arabinose inducible and carry ampicillin resistance, and the expressed protein has an N-terminal His6 tag for affinity purification. Site-directed alanine substitution mutants of TM1088A were made as described previously (21). The triple-site directed mutant of the GXGXXG consensus sequence to G14A, G16A, and G19A was made sequentially.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype, gene locus present, or description | Source |
|---|---|---|
| E. coli strains | ||
| HK100 | Expression strain derived from GeneHogs (Invitrogen) | JCSG |
| TK2420 | F−thi rha lacZ nagA Δ(kdpFAB)5 Δ(trkA-mscL′) trkD1 | W. Epstein (6) |
| Plasmids | ||
| pTM1088A/B | TM1088A, TM1088B | JCSG |
| pTM1057 | TM1057 | JCSG |
| pTM1088A | TM1088A | This study |
| pTM1088A/B-TM1089 | TM1088A, TM1088B, TM1089 | This study |
| pTM1088B-TM1089 | TM1088B, TM1089 | This study |
| pTM1089 | TM1089 | This study |
| pTM1088A/B G14A G16A G19A TM1089 | TM1088A G14AG16AG19A, TM1088B, TM1089 | This study |
| pTM1088A G14A G16A G19A | TM1088A G14AG16AG19A | This study |
| pTM1088B RCK | Amino acids 1-136 of TM1088B | This study |
| pMT2765/6 | MT2756, MT2766 | This study |
| pSpeedET no insert | No insert added | JCSG |
| pSpeedET | Expression vector, arabinose inducible, kanamycin resistance | JCSG |
Expression and affinity purification.
TM1088A, TM1088A/B, TM1088B RCK, and MT2765/6 were expressed by E. coli HK100 cells harboring the respective plasmids and grown at 37°C in LB medium containing kanamycin or ampicillin as appropriate. Cultures were grown to an optical density at 600 nm of 0.5 and then induced with 0.02% arabinose. After 4 h, the cultures were harvested by centrifugation at 2,000 × g for 20 min. Cell pellets were resuspended in 20 mM Tris (pH 8), 0.1 M NaCl, and 0.5 mM TCEP [tris(2-carboxyethyl)phosphine] and disrupted with a Microfluidizer (Microfluidics). Unbroken cells and cellular debris were removed by centrifugation at 12,000 × g for 20 min. The supernatant was applied to a 1-ml Ni-nitrilotriacetic acid agarose (Qiagen) gravity column equilibrated with 50 mM Tris (pH 8), 0.05 M NaCl, 10 mM imidazole, and 0.5 mM TCEP. The column was washed with 50 mM Tris (pH 8), 0.3 M NaCl, 40 mM imidazole, 10% glycerol, and 0.5 mM TCEP. The His6-tagged proteins were then eluted with 20 mM Tris (pH 8), 0.3 M imidazole, 10% glycerol, and 0.5 mM TCEP. The protein concentration was determined by the Bradford method using a commercially available assay (Pierce) with bovine serum albumin as the standard. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (24) was performed as described previously.
Gel filtration.
The size of the TM1088A/B complex was determined by gel filtration on a Hiload 16/60 Superdex 75 preparative-grade column (GE Healthcare) with 20 mM HEPES (pH 7.9), 0.15 M NaCl, and 0.5 mM TCEP. Gel filtration standards were obtained from Bio-Rad.
Complementation analysis.
Complementation of Escherichia coli TK2420 [F− thi rha lacZ nagA Δ(kdpFAB)5 Δ(trkA mscL′) trkD1] was performed by standard methods. In brief, chemically competent cells of TK2420 were prepared and transformed as described previously (21) with the expression plasmids harboring TM1057, TM1088A/B, TM1088B plus TM1089, TM1088A/B plus TM1089, TM1089, TM1088A/B G14A G16A G19A TM1089, MT2765/6, and no insert. Transformed cells were plated on KML medium (4) containing either ampicillin (100 μg/ml) or kanamycin (50 μg/ml). Two to four individual colonies from each transformation were then picked and grown overnight on KML medium containing the antibiotic and then plated on modified Na-K minimal medium [1 mg/ml thiamine, 1 mg/ml nicotinic acid, 100 μM tryptophan, 4.6 mM Na2HPO4, 2.3 mM NaH2PO4, 8.0 mM (NH4)SO4, 6 μM FeSO4 0.4 mM MgCl2, 1.5% agar, 0.2% arabinose, 0.1 M NaCl, 0.5% glycerol, 10 mM sodium acetate,15 g agar, pH adjusted to 7.0] containing 0, 2, or 4 mM KCl. Growth was observed by colony formation after incubation at 25°C.
In vitro ligand binding.
Calorimetric studies were performed with an automated Microcal VP capillary differential scanning calorimeter. A 1.6-mg/ml stock solution of TM1088A was prepared in 20 mM Tris (pH 7.5), 40 mM NaCl, and 0.5 mM TCEP. TM1088A was diluted to 62.3 μM for each thermal scan. Data were collected at between 40 and 130°C at a scan rate of 90°C/h. Subsequent scans to assess protein-ligand or protein-protein interaction contained 500 μM ATP, ADP, AMP, NAD, NADH, NADP, and NADPH. Results were analyzed with Origin software provided by the manufacturer. A buffer-only reference scan was subtracted from each sample scan prior to data analysis. The melting temperatures were determined with an intrinsic software feature.
Thermofluor binding assay mixtures (8) contained 0.8 mg/ml purified TM1088A or 0.1 mg/ml TM1088B RCK, 1 mM ligand, and 17× Sypro Orange (Invitrogen) in 20 mM HEPES (pH 7.9) and 150 mM NaCl or in affinity purification elution buffer. The temperature was ramped on a Bio-Rad iCycler iQ5 real-time PCR detection system from 40 to 100°C at 1°C/min. Melting temperatures were determined by identifying the maximum derivative [d(RFU)/dt] of the curve.
Phylogenetic tree construction.
Identification of two-subunit TrkAs was performed using genome region comparison tools on JCVI-CMR (36). Only those TrkA proteins with both genes adjacent to each other were considered. The two TrkA proteins were concatenated for analysis. Single-component TrkAs were identified by BLAST of completed microbial genomes. The amino acid sequences were aligned using ClustalW with manual fine-tuning. Regions with poor alignment or regions that were not present in all proteins were removed from the analysis. The phylogenetic tree was constructed by maximum-likelihood analysis using Tree Puzzle (42) with 1,000 bootstraps. The tree was displayed using Fig Tree (http://tree.bio.ed.ac.uk/software/figtree/).
RESULTS
Sequence analysis of TM1088.
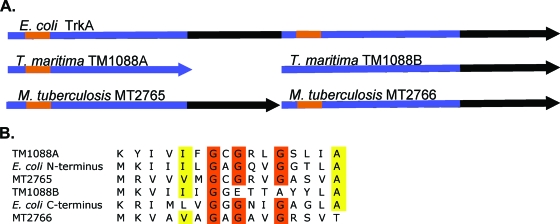
TM1088 was originally annotated as a frameshift mutation (32). As shown in Fig. 1A, TM1088 is missing 89 amino acids in the center of the protein compared with the E. coli TrkA, and the C-terminal half of the protein is in a different reading frame. If TM1088 encodes two separate proteins, TM1088A (the N-terminal half) and TM1088B (the C-terminal half), the reading frames would overlap by four nucleotides and the start codon for TM1088B would be TTG. TM1088A and TM1088B would have molecular masses of 16,033 Da and 24,036 Da, respectively, in contrast to the E. coli TrkA, which is 50,368 Da. TM1088A shares 26% sequence identity with the E. coli N-terminal TrkA, and TM1088B shares 24% sequence identity with the C-terminal TrkA. Upstream of TM1088 are two hypothetical proteins, TM1086 and TM1087. Downstream of TM1088A/B is TM1089, which shares 30% sequence identity to TrkH of E. coli. Unlike the E. coli trk genes, TM1088A/B and TM1089 are present in an operon (14) similar to that seen with Vibrio alginolyticus trkA and trkH, which are transcribed together (31). In addition to TM1088A/B, T. maritima possesses another gene annotated as TrkA-like, TM1057. The C-terminal half of this gene aligns with the N-terminal half of the E. coli TrkA with 22% sequence identity, but the N terminus possesses three predicted transmembrane helices which are not TrkA-like.
FIG. 1.
Schematic of aligned TrkA proteins. (A) The schematic shows the nature of the two-subunit TrkAs of T. maritima and M. tuberculosis CDC1551 in relation to the E. coli TrkA. The blue regions are the KTN/RCK domains. The characteristic nucleotide binding motifs GXGXXG are shown in orange. (B) An alignment of the nucleotide binding regions. Orange residues are the conserved glycines involved in nucleotide binding. The yellow residues are the extended conserved residues which indicate NAD(H) binding as described previously (20). TM1088B does not have the complete binding motif.
TM1088A and the E. coli TrkA both contain a characteristic GXGXXG nucleotide binding motif at the N terminus (Fig. 1B). The E. coli TrkA also contains a second nucleotide binding motif at the beginning of the second domain. In contrast, TM1088B does not contain this conserved sequence. TM1088A also contains an additional motif, V/IXGXGXXGXXXG/A, that builds upon the GXGXXG. This motif is suggested to further predict the binding of NAD(P)+ or an FAD+ cofactor (20). TM1057 also possesses a GXGXXG nucleotide binding domain in its C-terminal half.
The E. coli TrkA was recently shown to interact with dephosphorylated enzyme IIANtr product of the ptsN gene, which can serve to control K+ transport (26). No homolog of the ptsN gene was found in T. maritima, so whether K+ transport is controlled by TrkA interacting with another protein in T. maritima by a similar mechanism is not known.
Expression and copurification of TM1088A/B.
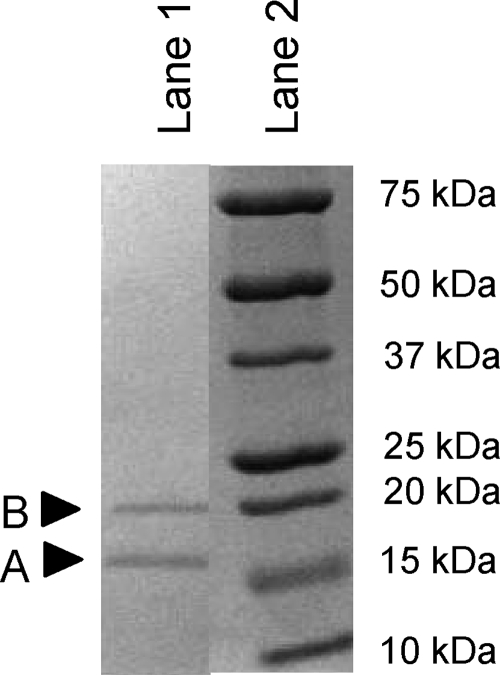
Expression of the putative TM1088 with the frameshift resulted in the expression and affinity purification of two proteins (Fig. 2). Therefore, TM1088 encodes two proteins, TM1088A and TM1088B, rather than being a frameshift mutation. The relative abundance of the proteins on an SDS-polyacrylamide gel following purification suggested a 1:1 to 2:1 (TM1088A to TM1088B) ratio of the two proteins, depending on the expression and purification run. The variable efficiency of TM1088B copurification may be a result of lower expression from the native ribosome binding site, dissociation of the untagged TM1088B subunit from the His-tagged TM1088A subunit, or partial assembly of the full complex with purification enrichment via the affinity tag. TM1088A can be expressed on its own, but aggregation becomes problematic, consistent with its normal state as a complex with TM1088B. Gel filtration was used to identify the native size of the complex, which was determined to be approximately 150 kDa. Assuming a ratio of 1:1, this would suggest that the TM1088A/B complex contains four subunits of both TM1088A and TM1088B. The TM1088A/B protein complex structure was recently determined (S. A. Lesley et al., unpublished data) and shows a tetramer of heterodimers.
FIG. 2.
SDS-polyacrylamide gel electrophoresis of copurification of TM1088A and TM1088B following Ni affinity chromatography, which interacts with the TM1088A His6 tag. Lane 1, copurified TM1088A and -B. TM1088B does not run true to size. Lane 2, protein size standard.
The three-dimensional crystal structure of TM1088A (PDB ID, 2g1u) was determined by the Joint Center for Structural Genomics (JCSG) (29), and coordinates were deposited in the Protein Data Bank (PDB). TM1088A forms a domain-swapped dimer upon crystallization, with the swapping of the C-terminal α helix. The structure is a classic Rossmann fold (3) consisting of a βαβαβ NAD+ binding motif with similarity to the nucleotide binding domains of K+ transporter-related proteins. This domain is one of the most represented folds in the PDB and encompasses proteins with over 50 functions (27), including oxidoreductases, K+ transporter-related proteins, and K+ channel-related proteins. A DALI search identified the closest structural similarity to the cytoplasmic nucleotide binding domains of Ktr K+/Na+ symporters: KTN MJA218 from Methanococcus jannaschii (PDB ID, 1lss; root mean square deviation, 1.9 Å) (40) and the KTN domain of KtrAB from Bacillus subtilis (PDB ID, 2hmt; root mean square deviation, 2.6 Å) (2). These structures were crystallized with NAD/NADH at the characteristic binding site. The B. subtilis KtrA has also been crystallized with ADP and ATP at the nucleotide binding site. The TM1088A structure shows an AMP molecule at this characteristic binding site. The structural homology to K+ transporters is consistent with TM1088A/B functioning in K+ transport.
Ligand binding to TM1088A, TM1088B RCK, and TM1088A G14A G16A G19A.
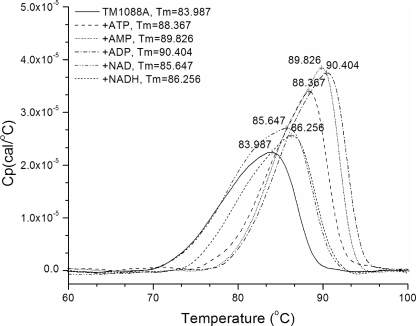
The crystal structure of TM1088A (PDB ID, 2g1u) showed an AMP molecule bound at the predicted site of NAD(H) binding (see below). This suggested that the thermophilic TrkA may function differently than the mesophilic E. coli protein, which had been shown to bind NADH but not ATP (41), and be more similar to the KtrA protein, which does bind ADP and ATP (2). Differential scanning calorimetry (DSC) was used to ascertain the nucleotide binding capabilities of TM1088A. As shown in Fig. 3, adenosine nucleotides stabilized TM1088A significantly (ATP, +4.4°C; AMP, +5.8°C; and ADP, +6.4°C). NAD+ (+1.7°C) and NADH (+2.3°C) did stabilize the protein but to a much lesser degree, which suggests that the required nucleotide may be AMP, ADP, or ATP. NADP+ and NADPH did not stabilize TM1088A (data not shown). In addition to DSC, ligand binding was evaluated by Thermofluor (34). This technique showed an identical melting temperature for TM1088A in the absence of ligand and an increase in protein stabilization by ATP (+4°C), AMP (+8°C), ADP (+8°C), and NADH (+4°C) but not by NAD+. A screen of TM1088A against a library of 192 small molecules (including nucleotides) revealed that TM1088A could also be stabilized by adenosine 5-phosphoribose but not by any additional nucleotides. Nucleotide binding to the RCK domain of TM1088B was also determined. ATP, ADP, AMP, IMP, NAD+, and NADH did not stabilize the protein. The TM1088A/B complex showed greatly increased thermal stability, with a melting temperature of greater than 130°C (the temperature limit of the DSC, as 100°C is the temperature limit for Thermofluor) versus 84°C and 87°C for TM1088A and TM1088B, respectively; thus, no ligand binding studies were performed on the complex.
FIG. 3.
DSC of TM1088A in the presence of no ligand, ATP, ADP, AMP, NAD+, or NADH. The melting temperature (Tm) of the protein is noted on the curve.
The nucleotide binding consensus sequence was disrupted in order to identify the role of nucleotide binding in protein function; the three glycines at the characteristic GXGXXG nucleotide binding site were replaced with alanine (TM1088A G14A G16A G19A). The Thermofluor ligand binding assay was used to ascertain the change, if any, in protein stabilization caused by ligand binding. Following replacement of any of the glycines with alanine or replacement of all three glycines with alanine, protein stabilization by ligand binding was completely disrupted. The melting temperature of the protein was not shifted in the presence of any of the ligands, i.e., ATP, ADP, AMP, or NADH, and was the same as that of the wild-type TM1088A.
Complementation and function.
In order to identify whether the two-subunit TM1088A/B complex actually functions in K+ transport, we complemented E. coli TK2420 (6), which is deficient in all forms of K+ transport, with the following plasmids: pTM1088A/B expressing TM1088A/B, pTM1088A/B-TM1089 expressing TM1088AB plus TM1089, pTM1088B-TM1089 expressing TM1088B plus TM1089, pTM1089 expressing TM1089, and pTM1057 expressing TM1057. As shown in Table 2, full complementation, which allowed E. coli TK2420 to grow in medium without added potassium, required TM1088A/B plus TM1089. This complementation strongly suggests that TM1088A/B and TM1089 function together as a K+ transporter. TM1088A/B alone could not complement K+ transport. TM1088A/B plus TM1089 could complement K+ transport but did not lead to an increased resistance to the antimicrobial peptide protamine. The slight complementation with TM1057 suggests that this protein may function as a K+ channel. The ability of TK2420 to grow at relatively low millimolar concentrations of K+ may be due to the presence of glycerol, acetate, and arabinose in the growth medium; the absence of glucose; and/or growth at 25°C. Growth was at 25°C for optimal expression. Growth in liquid medium to determine growth rates was not possible due to the high rate of suppressors of TK2420 to the wild-type phenotype under the growth and medium conditions used.
TABLE 2.
Complementation of K+ transport activity of TK2420 on modified NaK solid medium with added KCl
| Construct | Growtha with KCl addition (mM)
|
||
|---|---|---|---|
| 0 | 2 | 4 | |
| pTM1088A/B | − | (−) | + |
| pTM1057 | − | + | + |
| pTM1088A/B-TM1089 | + | + | + |
| pTM1088B-TM1089 | (−) | + | + |
| pTM1089 | − | (−) | + |
| pTM1088A/B G14A G16A G19A TM1089 | + | ND | ND |
| pSpeedET no insert | − | (−) | + |
+, good growth; (−), barely visible growth; −, no growth. ND, not determined.
In addition to plasmids bearing wild-type TM1088A (with TM1088B plus TM1089), constructs bearing TM1088A with all three glycines replaced with alanine (pTM1088A/B G14A G16A G19A TM1089) were introduced into E. coli TK2420. Growth of TK2420 pTM1088A/B G14A G16A G19A TM1089 on solid modified NaK medium without additional K+ was still observed, although it was at a lower rate. In vitro binding of nucleotide ligands to TM1088A G14A G16A G19A was no longer observed (data not shown), suggesting that nucleotide binding may not be essential for in vivo activity.
Two-subunit TrkAs shared with other organisms.
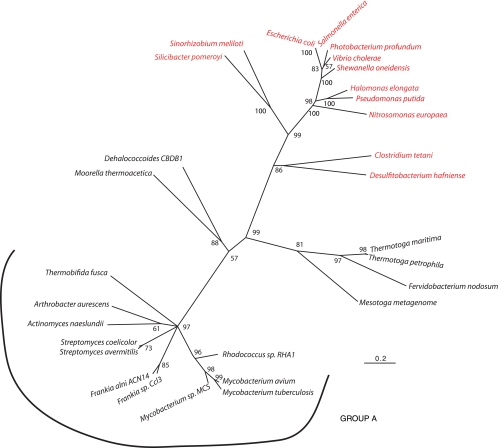
Bacterial genome sequencing has demonstrated similar two-subunit TrkAs in a number of organisms, including Mesotoga enrichment cultures (33), the Actinobacteria, and Dehalococcoides. A phylogenetic tree based on amino acid sequences of two- and one-component TrkAs was constructed. As shown in Fig. 4, the two-subunit TrkAs cluster as a monophyletic group, suggesting that the one- and two-subunit TrkAs evolved separately. An intriguing finding was that most of the organisms possessing a two-subunit TrkA do not have an identifiable TrkH (Fig. 4, group A). This would suggest that if these proteins are involved in K+ transport, they have a unique membrane-spanning partner. There are 21 genes shared by this group of organisms that encode hypothetical proteins with a minimum of one membrane-spanning region. One of these proteins may serve the functional role of TrkH. An alternative hypothesis is that these TrkA-like proteins have a unique function.
FIG. 4.
Maximum-likelihood phylogenetic tree of two- and one-component TrkAs. The tree shape and grouping of the one and two subunits were similar for both neighbor-joining and maximum-likelihood methods. Red and black indicate one- and two-subunit TrkAs, respectively. For the different branches, the organism name followed by the GenBank accession number and locus are as follows: Halomonas elongata, AAR91791, none; Pseudomonas putida KT2440, NP_742235, PP_0065; Escherichia coli, NP_417748, b3290; Salmonella enterica serovar Paratyphi ATCC 9150, YP_152404, SPA3276; Vibrio cholerae El Tor N16961, NP_229702, VC0043; Photobacterium profundum SS9, YP_131640, PBPRA3582; Shewanella oneidensis MR-1, AAN53116, SO 0029; Sinorhizobium meliloti 1021, ABR59944, SMc01046; Clostridium tetani E88, NP_780947, CTC00236; Fervidobacterium nodosum Rt17-B1, YP_001411178 + YP_001411177, Fnod1686 + Fnod1685; Thermotoga maritima MSB8, 2G1U_A + NP_228894, TM1087.1 + TM1088; Thermotoga petrophila, YP_001245237 + YP_001245236, Tpet_1656 + Tpet_1655; Mesotoga clone, CAJ75743 + CAJ75744, MES0053 + MES0054; Dehalococcoides sp. strain CBDB1, CAI82305 + CAI82304, cbdbA33 + cbdbA32; Moorella thermoacetic ATCC 39073, YP_430203 + YP_430204, moth_1347 + moth_1348; Thermobifida fusca YX, AAZ55961 + AAZ55962, Tfu_1928 + Tfu_1929; Frankia sp. strain CcI3, YP_480429 + YP_480428, Francci3_1323 + francci3_1322; Frankia alni ACN 14a, YP_712314 + YP_712313, FRAAL2085 + FRAAL2084; Mycobacterium avium 104, YP_882759 + YP_882760, Mav_3582 + Mav_3583; Mycobacterium tuberculosis CDC1551, NP_337266 + NP_337267, MT2765 + MT2766, Mycobacterium sp. strain MCS, YP_639361 + YP_639360, Mmcs_2197 + Mmcs_2196; Rhodococcus sp. strain RHA1, YP_706765 + YP_706764, RHA1_ro06835 + RHA1_ro06834; Streptomyces avermitilis MA-4680, NP_823567 + NP_823568, sav_2391 + sav2392; Streptomyces coelicolor A3(2), NP_629998 + NP_629997, SCO5876 + SCO5875; Actinomyces naeslundii MG1, none, ANA_0622 + ANA_0621; Arthrobacter aurescens TC1, YP_947545 + YP947544.
MT2765/6, a two-subunit TrkA?
As an example of a two-subunit TrkA that lacks a TrkH, the trkA genes (MT2765 and MT2766) were cloned from Mycobacterium tuberculosis CDC1551. These proteins were of interest because M. tuberculosis CDC1551 does not have an annotated TrkH, nor could one be identified by BLAST searches. Using a vector containing MT2765 and MT2766 constructed with MT2765 carrying the N-terminal His6 tag, MT2766 copurified with MT2765 by affinity purification. We screened MT2765/6 by Thermofluor against a 190-member ligand library of small molecules representing common cellular metabolites in the KEGG database. Interestingly, the ligands that increased the apparent melting temperature of MT2765/6 (50°C) were ATP (+10°C), ADP (+14°C), AMP (+11°C), NAD+ (+5°C), NADH (+5°C), and IMP (+11°C). These ligands are similar to the stabilizing ligands of TM1088A, except for IMP. IMP had no effect on the apparent melting temperature of TM1088A (data not shown). Attempts to complement K+ transport activity of E. coli TK2420 with pMT2765/6 were unsuccessful, suggesting that E. coli TrkH cannot interact with MT2765/6.
DISCUSSION
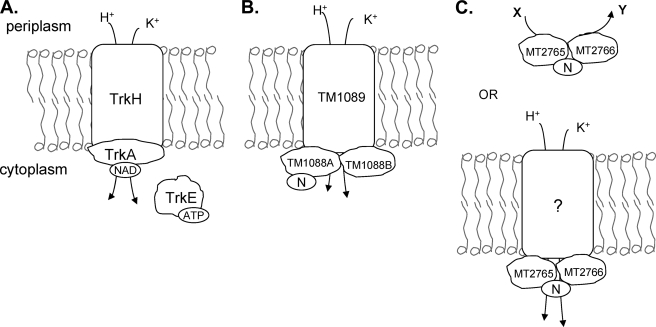
Here we describe T. maritima TrkA, a new two-subunit TrkA involved in K+ transport (Fig. 5). Our results show that the two-subunit T. maritima TrkA functions in vivo as a K+ transporter, binds adensosine nucleotides and NAD+/NADH yet may not require binding of these nucleotides for activity, and is phylogenetically related to other two-subunit TrkAs which may or may not function in K+ transport.
FIG. 5.
Schematic of the role of TrkA proteins. (A) TrkA serves as the peripheral NAD(H) binding protein of the Trk K+ transporter in E. coli. It is required in addition to TrkH or -G, which span the membrane. TrkE, which binds ATP, is required when TrkH serves as the pore. (B) Proposed model for the T. maritima Trk K+ transporter. Here the TrkA protein consists of two proteins, TM1088A and TM1088B. TM1088A binds nucleotides [N = adenosine nucleotides and NAD(H)]. (C) A third class of TrkA is found in M. tuberculosis. This two-subunit TrkA may interact with an unknown membrane protein to transport K+, or it may serve a novel function [N = adenosine nucleotides, NAD(H), and IMP].
The genetic arrangement of and the similarity between TM1088A and TM1088B suggest a likely gene duplication event, as has also been hypothesized for the E. coli TrkA, which contains homologous N- and C-terminal regions (41). If the T. maritima TrkA is a more ancient protein, then in the case of E. coli and other organisms with a one-component TrkA, the gene duplication event was followed by a gene fusion event. TM1088A/B is also an example of the limitations of annotation based on homology alone. Homology to the characteristic TrkAs suggested that TM1088 was an authentic frameshift (32) and that the Mesotoga A subunit (MES0053) is a pseudogene (33).
The ability to express and copurify TM1088A and -B strongly suggested that the protein was properly folded and functional. The similarity of the sequence and crystal structure of TM1088A to those of other K+ transport-related proteins also suggested that the protein was involved in K+ transport. However, the nucleotide binding domain, specifically the Rossmann fold, is found in many different proteins with many different functions. Thus, structural similarity to this fold may not be useful in deriving function (30). Therefore, complementation experiments were used to support the hypothesis that these proteins function in K+ transport. Complementation of TK2420 with TM1088A/B plus TM1089 supports the hypothesis that TM1088A/B is indeed a functional complex required for K+ transport. The lack of complementation of pTM1088A/B alone suggests that the differences between the one-component and two-subunit TrkAs and the approximately 25% sequence identity are significant enough to prevent a functional interaction with E. coli TrkH or TrkG. Previous studies showed that heterologously expressed TrkA of Vibrio alginolyticus and Halomonas elongata could complement an E. coli strain deficient in TrkA function (22, 31), yet in these cases, the complementing proteins are one-component TrkAs like the E. coli protein and the two TrkA proteins share 78% and 63% sequence identity, respectively. Complementation provides a powerful tool to determine protein function, but these experiments also demonstrate a limitation of this approach. When complementing for function with proteins possessing low sequence identity or existing in a different form (one component versus two components), such as the T. maritima TM1088A/B, interacting protein partners that work with the protein of interest must also be included to gain function. In this case the interacting protein, TM1089, was required. In the case of MT2765/6, where the interacting partner, if any, is unknown, little can be determined by lack of complementation.
Although the E. coli TrkA has been shown to bind NAD+ but not ATP (41), transport of K+ is dependent upon both the proton motive force and ATP (38). It has been hypothesized that in E. coli ATP binds the SapD/TrkE protein. SapD/TrkE then interacts with the Trk system, resulting in the transport dependence on ATP (16). In other organisms where the SapD/TrkE protein is absent, the Trk system is hypothesized to interact with an unknown ATP binding protein (16). There is no SapD homolog in T. maritima, and since the T. maritima TrkA interacts directly with the adenosine nucleotides, an additional ATP binding protein may not be required. It has also been shown that ATP and ADP bind the RCK domain of the KtrAB K+ transporter, an Na+ symporter, more tightly than NAD+ and NADH (2, 23). ATP binding to KtrA induces a conformational change that NAD+, NADH, ADP, or AMP does not, suggesting that ATP may be the physiological ligand. It is intriguing to suggest that K+ transport could be driven by ATP hydrolysis, but experimental data (16, 23) support only the role of ATP binding, not hydrolysis. NAD(H), ATP, ADP, or AMP may more likely serve a regulatory role, as the level of ATP correlates with transport activity (43). It should also be noted that the E. coli TrkA(H) may form a supercomplex with FoF1-ATPase (45). This notion has been suggested for E. coli when it is grown specifically under alkaline and anaerobic conditions. As T. maritima has an anaerobic lifestyle, it is intriguing to consider that binding of adenosine nucleotides may be a result of close interaction with FoF1-ATPase as a means to power K+ translocation.
TK2420 K+ uptake mutants complemented with pTM1088A/B G14A G16A G19A TM1089, which contain alanine substitutions in the GXGXXG nucleotide binding consensus sequence, were able to survive on unsupplemented K+ medium. This result is consistent with mutations of E. coli TrkA (44). Mutations at Gly-9 or Gly-241 affected the affinity and decreased the rate of K+ uptake but did not prevent K+ uptake. Whether any nucleotides still bound the E. coli TrkA Gly-9 and Gly-241 mutants in vitro was not determined. Nucleotide binding may act to fine-tune the rate of transport as opposed to acting as an on/off switch.
The structural similarity of TrkA and KtrA N-terminal nucleotide binding regions to the nucleotide binding regions of K+ channels supports the evolutionary relatedness of K+ symporters and channels (11). In fact, the similarity of T. maritima TrkA to KtrAB is quite intriguing. Durell et al. (11) suggested that the KtrB family (of K+ symporters) is most like the common ancestor of this group of proteins as opposed to the Trk family, which is thought to have diverged the most. Although the T. maritima TM1088A/B amino acid sequence is similar to those of TrkA proteins, the fact that the structure of TM1088A is very similar to the KtrA nucleotide binding domain may suggest a conservation of the more ancient structure despite the amino acid sequence divergence. It will be interesting to see if the structure of TM1089 will be more closely related to the models for TrkH/G or KtrB and how other TrkA-like proteins compare to KtrA.
As shown in Fig. 4, many two-subunit TrkA-like proteins do not have identifiable TrkH membrane components encoded in their genomes, suggesting that this group may represent a third class of TrkA proteins. One of these two-subunit TrkAs is present in M. tuberculosis CDC1551. These proteins are thought to be involved in K+ transport, yet K+ transport activity increases in knockout strains, and these strains show no difference in growth or K+ uptake in the presence of clofazimine (7), leaving open the possibility of another function for these two proteins. The function of these proteins may be integral for successful infection, as one of these proteins (Rv2691, the MT2765 homolog in M. tuberculosis H37Rv) is required for survival in macrophages (37).
These studies have identified a new class of two-subunit TrkAs exemplified by T. maritima TM1088A/B. Interestingly, a subclass of these two-subunit TrkAs may perform a unique function or interact with a novel membrane-spanning transporter. Understanding the role of the two-subunit TrkA in M. tuberculosis and other strains lacking a TrkH gene or identifying the interacting membrane protein if these TrkAs are involved in K+ transport should shed light upon the evolution and diversity of K+ transporters.
Acknowledgments
We thank Wolfgang Epstein for supplying strain TK2420 and the Joint Center for Structural Genomics (JCSG) and Heath Klock for HK100, pTM1088A/B, pTM1057, and pSpeedET. Marc Deller, Tina Trout, Julie Feuerhelm, and Anna Grzechnik provided helpful advice and assistance. Bernhard Geierstanger and Sebastian Sudek also developed the ligand library.
The project described was supported by award number U54GM074898 from the National Institute of General Medical Sciences.
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Achenbach-Richter, L., R. Gupta, K. O. Stetter, and C. R. Woese. 1987. Were the original eubacteria thermophiles? Syst. Appl. Microbiol. 934-39. [DOI] [PubMed] [Google Scholar]
- 2.Albright, R. A., J. L. Ibar, C. U. Kim, S. M. Gruner, and J. H. Morais-Cabral. 2006. The RCK domain of the KtrAB K+ transporter: multiple conformations of an octameric ring. Cell 1261147-1159. [DOI] [PubMed] [Google Scholar]
- 3.Bellamacina, C. R. 1996. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J. 101257-1269. [DOI] [PubMed] [Google Scholar]
- 4.Bossemeyer, D., A. Borchard, D. C. Dosch, G. C. Helmer, W. Epstein, I. R. Booth, and E. P. Bakker. 1989. K+-transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. J. Biol. Chem. 26416403-16410. [PubMed] [Google Scholar]
- 5.Brown, A. D. 1976. Microbial water stress. Bacteriol. Rev. 40803-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buurman, E. T., D. McLaggan, J. Naprstek, and W. Epstein. 2004. Multiple paths for nonphysiological transport of K+ in Escherichia coli. J. Bacteriol. 1864238-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cholo, M. C., H. I. Boshoff, H. C. Steel, R. Cockeran, N. M. Matlola, K. J. Downing, V. Mizrahi, and R. Anderson. 2006. Effects of clofazimine on potassium uptake by a Trk-deletion mutant of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 5779-84. [DOI] [PubMed] [Google Scholar]
- 8.Cummings, M. D., M. A. Farnum, and M. I. Nelen. 2006. Universal screening methods and applications of ThermoFluor. J. Biomol. Screen 11854-863. [DOI] [PubMed] [Google Scholar]
- 9.Dinnbier, U., E. Limpinsel, R. Schmid, and E. P. Bakker. 1988. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150348-357. [DOI] [PubMed] [Google Scholar]
- 10.Dosch, D. C., G. L. Helmer, S. H. Sutton, F. F. Salvacion, and W. Epstein. 1991. Genetic analysis of potassium transport loci in Escherichia coli: evidence for three constitutive systems mediating uptake of potassium. J. Bacteriol. 173687-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durell, S. R., Y. Hao, T. Nakamura, E. P. Bakker, and H. R. Guy. 1999. Evolutionary relationship between K(+) channels and symporters. Biophys. J. 77775-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein, W. 2003. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 75293-320. [DOI] [PubMed] [Google Scholar]
- 13.Epstein, W., and S. G. Schultz. 1966. Cation transport in Escherichia coli. V. Regulation of cation content. J. Gen. Physiol. 49221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ermolaeva, M. D., O. White, and S. L. Salzberg. 2001. Prediction of operons in microbial genomes. Nucleic Acids Res. 291216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harms, C., Y. Domoto, C. Celik, E. Rahe, S. Stumpe, R. Schmid, T. Nakamura, and E. P. Bakker. 2001. Identification of the ABC protein SapD as the subunit that confers ATP dependence to the K+-uptake systems Trk(H) and Trk(G) from Escherichia coli K-12. Microbiology 1472991-3003. [DOI] [PubMed] [Google Scholar]
- 17.Huber, R., T. A. Langworthy, H. Konig, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144324-333. [Google Scholar]
- 18.Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417515-522. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, Y., A. Pico, M. Cadene, B. T. Chait, and R. MacKinnon. 2001. Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron 29593-601. [DOI] [PubMed] [Google Scholar]
- 20.Kleiger, G., and D. Eisenberg. 2002. GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through C(alpha)-H… O hydrogen bonds and van der Waals interactions. J. Mol. Biol. 32369-76. [DOI] [PubMed] [Google Scholar]
- 21.Klock, H. E., E. J. Koesema, M. W. Knuth, and S. A. Lesley. 2008. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71982-994. [DOI] [PubMed] [Google Scholar]
- 22.Kraegeloh, A., B. Amendt, and H. J. Kunte. 2005. Potassium transport in a halophilic member of the Bacteria domain: identification and characterization of the K+ uptake systems TrkH and TrkI from Halomonas elongata DSM 2581T. J. Bacteriol. 1871036-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroning, N., M. Willenborg, N. Tholema, I. Hanelt, R. Schmid, and E. P. Bakker. 2007. ATP binding to the KTN/RCK subunit KtrA from the K+-uptake system KtrAB of Vibrio alginolyticus: its role in the formation of the KtrAB complex and its requirement in vivo. J. Biol. Chem. 28214018-14027. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 25.Laimins, L. A., D. B. Rhoads, and W. Epstein. 1981. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 78464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, C. R., S. H. Cho, M. J. Yoon, A. Peterkofsky, and Y. J. Seok. 2007. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc. Natl. Acad. Sci. USA 1044124-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, D., O. Redfern, and C. Orengo. 2007. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 8995-1005. [DOI] [PubMed] [Google Scholar]
- 28.Lesley, S. A., P. Kuhn, A. Godzik, A. M. Deacon, I. Mathews, A. Kreusch, G. Spraggon, H. E. Klock, D. McMullan, T. Shin, J. Vincent, A. Robb, L. S. Brinen, M. D. Miller, T. M. McPhillips, M. A. Miller, D. Scheibe, J. M. Canaves, C. Guda, L. Jaroszewski, T. L. Selby, M. A. Elsliger, J. Wooley, S. S. Taylor, K. O. Hodgson, I. A. Wilson, P. G. Schultz, and R. C. Stevens. 2002. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. USA 9911664-11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesley, S. A., and I. A. Wilson. 2005. Protein production and crystallization at the Joint Center for Structural Genomics. J. Struct. Funct Genomics. 671-79. [DOI] [PubMed] [Google Scholar]
- 30.Martin, A. C., C. A. Orengo, E. G. Hutchinson, S. Jones, M. Karmirantzou, R. A. Laskowski, J. B. Mitchell, C. Taroni, and J. M. Thornton. 1998. Protein folds and functions. Structure 6875-884. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura, T., N. Yamamuro, S. Stumpe, T. Unemoto, and E. P. Bakker. 1998. Cloning of the trkAH gene cluster and characterization of the Trk K(+)-uptake system of Vibrio alginolyticus. Microbiology 1442281-2289. [DOI] [PubMed] [Google Scholar]
- 32.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399323-329. [DOI] [PubMed] [Google Scholar]
- 33.Nesbo, C. L., M. Dlutek, O. Zhaxybayeva, and W. F. Doolittle. 2006. Evidence for existence of “mesotogas,” members of the order Thermotogales adapted to low-temperature environments. Appl. Environ. Microbiol. 725061-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantoliano, M. W., E. C. Petrella, J. D. Kwasnoski, V. S. Lobanov, J. Myslik, E. Graf, T. Carver, E. Asel, B. A. Springer, P. Lane, and F. R. Salemme. 2001. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6429-440. [DOI] [PubMed] [Google Scholar]
- 35.Parra-Lopez, C., R. Lin, A. Aspedon, and E. A. Groisman. 1994. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 133964-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 1028327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhoads, D. B., and W. Epstein. 1977. Energy coupling to net K+ transport in Escherichia coli K-12. J. Biol. Chem. 2521394-1401. [PubMed] [Google Scholar]
- 39.Rhoads, D. B., F. B. Waters, and W. Epstein. 1976. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J. Gen. Physiol. 67325-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roosild, T. P., S. Miller, I. R. Booth, and S. Choe. 2002. A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell 109781-791. [DOI] [PubMed] [Google Scholar]
- 41.Schlosser, A., A. Hamann, D. Bossemeyer, E. Schneider, and E. P. Bakker. 1993. NAD+ binding to the Escherichia coli K(+)-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Mol. Microbiol. 9533-543. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18502-504. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, L. M., E. P. Bakker, and I. R. Booth. 1985. Energy coupling to K+ uptake via the Trk system in Escherichia coli: the role of ATP. J. Gen. Microbiol. 13177-85. [DOI] [PubMed] [Google Scholar]
- 44.Stumpe, S., and E. P. Bakker. 1997. Requirement of a large K+-uptake capacity and of extracytoplasmic protease activity for protamine resistance of Escherichia coli. Arch. Microbiol. 167126-136. [PubMed] [Google Scholar]
- 45.Trchounian, A., K. Bagramyan, and A. Poladian. 1997. Formate hydrogenlyase is needed for proton-potassium exchange through the F0F1-ATPase and the TrkA system in anaerobically grown and glycolysing Escherichia coli. Curr. Microbiol. 35201-206. [DOI] [PubMed] [Google Scholar]
- 46.Vargas, M., K. Kashefi, E. L. Blunt-Harris, and D. R. Lovley. 1998. Microbiological evidence for Fe(III) reduction on early Earth. Nature 39565-67. [DOI] [PubMed] [Google Scholar]