Abstract
Stimulation of potassium uptake is the most rapid response to an osmotic upshock in bacteria. This cation accumulates by a number of different transport systems whose importance has not been previously addressed for rhizobia. In silico analyses reveal the presence of genes encoding four possible potassium uptake systems in the genome of Sinorhizobium meliloti 1021: Kup1, Kup2, Trk, and Kdp. The study of the relevance of these systems under a number of different growth conditions and in symbiosis showed that the integrity of Kup1 or Trk is essential for growth under laboratory conditions even in osmotically balanced media and the absence of both systems leads to a reduced infectivity and competitiveness of the bacteria in alfalfa roots. Trk is the main system involved in the accumulation of potassium after an osmotic upshift and the most important system for growth of S. meliloti under hyperosmotic conditions. The other three systems, especially Kup1, are also relevant during the osmotic adaptation of the cell, and the relative importance of the Kdp system increases at low potassium concentrations.
Rhizobia are gram-negative soil bacteria able to establish nitrogen-fixing symbiosis with leguminous plants under conditions of nitrogen deprivation. During this process, an exchange of molecular signals occurs between the two partners, leading to the formation of the root nodule, where biological nitrogen fixation takes place (12).
Nearly 40% of the world's land surface can be categorized as having potential salinity problems (38). Most crops are sensitive to relatively low levels of salinity (36), and in the case of legumes, there is an additional problem since not only the plant but also the symbiotic bacteria are sensitive to salinity both at the free-living stage and during the symbiotic process (22). The Rhizobium-legume symbiosis is highly sensitive to salt or osmotic stress, since these conditions may inhibit the initial steps of the symbiotic interaction (root colonization, nodule infection, and nodule development) and also have a depressive effect on nitrogen fixation (38). It has been observed that Rhizobium mutants affected in adaptation to high salinity present deficiencies in their symbiotic capacity (25). These results emphasize the importance of studying the mechanisms of rhizobial adaptation to changes in the osmotic conditions of the soil environment.
Response and adaptation to environmental stresses probably constitute a complex phenomenon involving many physiological and biochemical processes that likely reflect changes in gene expression and in the activities of enzymes and transport proteins (6, 7, 23). A rise in the external salinity and osmolarity triggers the outflow of water from the cell, resulting in a reduction in turgor and dehydration of the cytoplasm, causing a decrease in the cytoplasm volume and thus an increase in the ion concentration in the cytosol (33). A recent transcriptomic study of Sinorhizobium meliloti has revealed that the responses of this bacterium to ionic and nonionic compounds are very similar and include the induction of many genes of unknown function, suggesting the existence of still-unexplored osmoadaptation mechanisms (6). However, rhizobia are known to use distinct mechanisms for long-term adaptation to hyperosmotic stress, such as intracellular accumulation of low-molecular-weight organic solutes (amino acids, sugars, and polyamines), changes in cell morphology and size, or modifications in the pattern of extracellular polysaccharides (20, 22, 34, 38). Nevertheless, the most rapid response to an osmotic upshock, a sudden increase in external osmolarity, is the stimulation of potassium uptake (33).
Potassium (K+), being the most abundant intracellular cation, makes a major contribution to the turgor pressure of the cells, playing important roles in bacterial osmoadaptation, pH regulation, gene expression, and activation of cellular enzymes (8). The elevated K+ levels after an osmotic upshift act as a second messenger to turn on other osmotically activated responses (2).
K+ uptake after an osmotic upshift is achieved by bacteria through specialized transport systems, which have been well studied for certain bacteria (8, 17). Three different types of K+ transporters have been involved in this process: Trk, Kdp, and Kup. Trk is a multicomponent complex widespread in bacteria and archea. Trk systems have a moderate affinity for K+, are generally expressed constitutively, and are probably energized by taking up K+ in symport with a proton. They consist of a transmembrane protein named TrkH or TrkG, which is the actual K+-translocating subunit, and a cytoplasmic membrane surface protein, TrkA, which is a NAD+ binding protein required for the system's activity (33). Kdp is an inducible system with high affinity and specificity for K+, found in Escherichia coli and many other bacteria. Kdp is a P-type ATPase expressed only when the needs for K+ are not satisfied by other transport systems and thus plays a vital role when the ion is present at concentrations too low to be efficiently taken up by the other K+ transport systems and at higher K+ concentrations when other K+-transport systems are abolished by mutation (8). Kdp is the only bacterial K+ uptake system whose expression is strongly regulated at the transcriptional level, which is mediated by the KdpD sensor kinase and the KdpE response regulator (reviewed in reference 8). Finally, Kup is a constitutive K+ uptake system of modest affinity found in some bacteria. It is a single membrane protein that probably functions by proton symport. Kup activity in E. coli is more important at low pH, where its maximum rate exceeds that of Trk (35).
Although it has been described that the mutation of a K+ uptake system of Rhizobium tropici causes a decrease in the osmotic tolerance and the symbiotic competence of the strain (25), the role of K+ transport systems has not been comprehensively studied for rhizobia. In this work we have addressed a genetic and functional characterization of K+ uptake systems in S. meliloti. We obtained clear evidence that the presence of either Trk or Kup1 is required for bacterial growth even under osmotically balanced conditions. Furthermore, K+ accumulation after an osmotic upshift is triggered mainly by the Trk system. Nevertheless, Kup1, Kup2, and Kdp are also involved in the osmoadaptation process, and the relative importance of the Kdp system increases in media containing low K+ concentrations. The relevance of the different K+ uptake systems was also studied during establishment of symbiosis with alfalfa.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Sinorhizobium meliloti strains were grown at 30°C in TY complex medium (4) or in minimal medium (MM) containing glutamate (1.1 g/liter), mannitol (10 g/liter), and mineral salts (K2HPO4 [0.3 g/liter], KH2PO4 [0.3 g/liter], MgSO4·7H2O [0.15 g/liter], CaCl2·2H2O [0.05 g/liter], FeCl3 [0.006 g/liter], and NaCl [0.05 g/liter]) (29). The K+-free medium for S. meliloti was MM in which K+ phosphates were replaced with Na2HPO4-NaH2PO4 (0.5 mM). When a specific pH was required, the MM was buffered with Tris-morpolineethanesulfonic acid (5 mM). The K+ content of the MM used was adjusted when required by the addition of KCl from a 3 M stock solution. Escherichia coli strains were propagated on Luria-Bertani medium (30). When required, antibiotics were added at the following final concentrations: for E. coli, streptomycin (Sm), 50 μg/ml; spectinomycin (Sp), 50 μg/ml; kanamycin (Km), 50 μg/ml; gentamicin (Gm), 20 μg/ml; tetracycline (Tc), 10 μg/ml; and ampicillin (Ap), 200 μg/ml; for S. meliloti, Sm, 200 μg/ml; Sp, 100 μg/ml; Km, 200 μg/ml; Gm, 15 μg/ml; and Tc, 5 μg/ml. When osmotic stress conditions were imposed, the osmolarity of the medium was increased by the addition of appropriate amounts of NaCl from a 4 M stock solution (made in MM) or sucrose. Growth in the selected medium was followed, measuring the absorbance at 600 nm in cultures incubated in a gyratory shaker or by comparative analysis on solid medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| Sinorhizobium meliloti | ||
| 1021 | SU47 str-21 (wild type), Smr | 21 |
| 10K1K | 1021 (Δkup1::Km), Smr Kmr | This work |
| 10K2SS | 1021 (Δkup2::Sm/Sp), Smr Spr | This work |
| 10tAG | 1021 (ΔtrkA::Gm), Smr Gmr | This work |
| 10KdpSS | 1021 (ΔkdpA::Sm/Sp), Smr Spr | This work |
| 10K2K1 | 10K2SS (Δkup1::Km), Smr Spr Kmr | This work |
| 10tAK1 | 10tAG (Δkup1::Km), Smr Gmr Kmr | This work |
| 10tAK2 | 10tAG (Δkup2::Sm/Sp), Smr Gmr Spr | This work |
| 10tAKdp | 10tAG (ΔkdpA::Sm/Sp), Smr Gmr Spr | This work |
| 10tAK21 | 10tAK2 (Δkup1::Km), Smr Gmr Spr Kmr | This work |
| 10tAKK | 10tAKdp (Δkup1::Km), Smr Gmr Spr Kmr | This work |
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 relA1 hsdR17 | Bethesda Research Lab |
| S17.1 | thi pro recA hsdR hsdM; Rp4Tc::Mu; Km::Tn7; Tpr Smr Spr | 32 |
| Plasmids | ||
| pBluescript II KS | Cloning vector; Apr | Stratagene |
| pGEM-T easy | PCR cloning vector; Apr | Promega |
| pHP45Ω | pBR322 derivative with a streptomycin/spectinomycin resistance cassette; Apr Smr Spr | 9 |
| pHP45Ω-Km | pBR322 derivative with a kanamycin resistance cassette; Apr Kmr | 9 |
| pMS255 | pUC8 derivative with a gentamicin resistance cassette; Apr Gmr | 3 |
| pK18mobsacB | Suicide plasmid; Mob+, sacB; Kmr | 31 |
| pSUP202Pol4 | Suicide plasmid; Tcr | 11 |
| pGUS3 | pnfeD::gusA translational fusion in pBI101 (Clontech); Kmr | 14 |
| pJB3Tc19 | IncP cloning vector; Tcr Apr | 5 |
| pJB3K1 | pJB3Tc19 derivative containing a PCR-amplified fragment including the kup1 gene from S. meliloti 1021; Tcr Apr | This work |
| pJB3tA | pJB3Tc19 derivative containing a PCR-amplified fragment including the trkA gene from S. meliloti 1021; Tcr Apr | This work |
Smr, streptomycin resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance; Gmr, gentamicin resistance; Tpr, trimethoprim resistance; Apr, ampicillin resistance; Tcr, tetracycline resistance.
DNA sequence analysis and construction of S. meliloti mutant strains.
DNA sequence analysis was performed with the program BLAST from the network service at the NCBI (1). Standard techniques were used for DNA manipulations (30).
To construct plasmids for the disruption of the different genes, we first amplified the corresponding regions from the S. meliloti genome by PCR using custom-synthesized DNA primers (Table 2). After cloning the amplicons in appropriate vectors, we deleted part of the gene sequence by endonuclease digestion and inserted antibiotic resistance cassettes to obtain each mutant construct in vitro. The constructs were then subcloned into the appropriate shuttle vector, and the plasmids obtained were transferred by conjugation to the wild-type strain 1021. Transconjugants were selected on medium containing the appropriate antibiotics. In the case of pK18mobsacB, the use of sucrose addition during selection as described by Schäfer et al. (31) was avoided since its osmotic action could affect the obtention of osmosensitive mutants. Double and triple mutants were obtained by transduction with ϕM12 phage as described by Finan et al. (10). Disruption of the genes of interest was then confirmed by Southern hybridization with specific probes.
TABLE 2.
Primers used for gene cloning
| Gene | Forward/reverse primer (5′-3′)a |
|---|---|
| trkA (SMc01046) | TTATCTAGACGATCAGCATATCATGACC/XbaI |
| TAATCTAGACCACGATCCATACATTCGTCXbaI | |
| kdpA (SMa2333) | TAAGAATTCATACCTGCACTGACGAGAAG/EcoRI |
| TAAGAATTCACATCGAGAACGAAGAAGAGEcoRI | |
| kup1 (SMc00873) | TAAGAGCTCGCTACGACATCTTCGGTCAT/SacI |
| TAATCTAGAGCACTTCCAGGAAAAGTGTGXbaI | |
| kup2 (SMa1798) | TAATCTAGAGCCTGTTGACGACGATCAGT/XbaI |
| TAAGATATCGGGTCCAGATCTAGCTACGEcoRV |
Underlining denotes the action sites of the endonucleases used for cloning the mutated gene versions in the shuttle vector.
Determination of cell K+ content.
Cells were grown in MM to exponential phase (optical density at 600 nm = 0.6), and NaCl was added to a final concentration of 0.3 M to induce osmotic shock. Cells were then incubated under standard growth conditions, and aliquots were collected after 5, 30, and 60 min. An additional aliquot was collected right before NaCl addition to determine the cell K+ content just before the osmotic upshift.
For each time point, 10 ml of cultured cells was harvested by centrifugation at room temperature (12,000 rpm, 2 min), the medium was removed carefully, and the ionic content of the cells was extracted with 1 volume of HNO3 (0.1%) at room temperature for 20 h. The cellular debris was then removed by centrifugation and the K+ content determined in the supernatant obtained using an Iris Intrepid II XDL inductively coupled plasma-optical emission spectrometer.
Plant assays.
Alfalfa (Medicago sativa L. cv. Aragon) seeds were sterilized and germinated as described by Olivares et al. (26). Germinated seeds were then transferred to tubes or Leonard jars for growth.
To test the infectivity of the rhizobial strains, 24 individual plants grown in tubes were inoculated with each rhizobial suspension (106 CFU/plant). After inoculation, the number of nodulated plants and the number of nodules per plant were recorded daily.
To determine the competitive ability, 12 plants grown in tubes were inoculated with a mixture of S. meliloti 1021(pGUS3) and each strain tested, using a ratio of 1:1. The plasmid pGUS3 contains the marker gene coding for β-glucuronidase. Nodule occupancy was determined 12 days after inoculation. Roots were collected, briefly washed with water, and incubated overnight in the dark at 37°C in 1 mM 5-bromo-4-chloro-3 indolyl-β-d-glucuronide (Apollo Scientific) in 50 mM sodium phosphate buffer (pH 7.5) with 1% sodium dodecyl sulfate. Those nodules occupied by 1021(pGUS3) stain blue, so nodule occupancy could be determined by counting blue and white nodules. All the nodules from the 12 inoculated plants were screened (approximately 100 nodules). The results for any given strain were compared to those obtained with the wild-type strain, 1021, in the same experiment.
To study the symbiotic effectiveness of the strains, 32 individual plants grown in Leonard jars (8 plants per jar) were inoculated with each rhizobial suspension (109 CFU/plant). Seven weeks after inoculation, the plants were collected and the extent of nitrogen fixation was assessed by measurement of the nitrogen content of the aerial part (shoot) (24) and comparison of the shoot dry weight.
RESULTS
In silico analysis of putative K+ uptake systems genes in Sinorhizobium meliloti.
In silico analysis of the S. meliloti 1021 genome revealed the presence of genes coding for proteins homologous to the components of three K+ uptake systems: Trk, Kdp, and Kup (13). Genes homologous to trkA and trkH are present in the chromosome of S. meliloti 1021 (SMc01046 and SMc00936, respectively), together with a gene located in pSymA and described as trkH-like (SMa1691). All these genes code for proteins with high sequence identity (30 to 40%) to the corresponding components of the Escherichia coli Trk system. The presence of a Trk system seems to be an exception among rhizobia, since trk-like genes could be detected only in the S. meliloti, Sinorhizobium medicae, and Mesorhizobium sp. BCN1 genomes and appear to be missing in the Rhizobium etli CFN42, Rhizobium leguminosarum bv. viciae 3841, Mesorhizobium loti MAFF303099, Bradyrhizobium sp. strain BTAi1, Bradyrhizobium sp. strain ORS278, and Bradyrhizobium japonicum USDA110 genomes. The situation is very different regarding putative Kdp systems, since kdpABC genes are annotated in all sequenced rhizobial genomes, forming putative operons with at least two other genes homologous to those coding for the two-component system KdpDE in E. coli. The Kdp systems are conserved among rhizobia and show significant identities with other known Kdp P-type ATPases. In S. meliloti, the products of the kdpABC genes (SMa2333, SMa2331, and SMa2329) show 51%, 63%, and 47% sequence identity with the E. coli KdpA, KdpB, and KdpC subunits, respectively. Furthermore, the products of the S. meliloti genes SMa2327 and SMa2325, which form a putative operon with kdpABC, display 39% and 48% identity with the E. coli KdpD and KdpE subunits of the two-component regulator controlling kdp expression, respectively. On the other hand, we could not identify in the S. meliloti 1021 genome a gene homologous to kdpF, encoding the fourth component of the Kdp system in E. coli. Finally, there is a surprisingly high number of annotated kup-like genes in rhizobial genomes. Several species harbor three or even four genes coding for probable Kup systems. There are two annotated kup genes in M. loti MAFF303099 (18), three in R. etli CFN42 (16), R. leguminosarum bv. viciae 3841 (37), B. japonicum USDA110 (19), and Bradyrhizobium sp. strain ORS278 (15) and up to four in Bradyrhizobium sp. strain BTAi1 (15). S. meliloti 1021 harbors two putative kup genes: kup1 (SMc00873) and kup2 (SMa1798), whose products show 42% and 41% identity, respectively, with the Kup protein from E. coli.
Construction of S. meliloti mutants in K+ uptake systems.
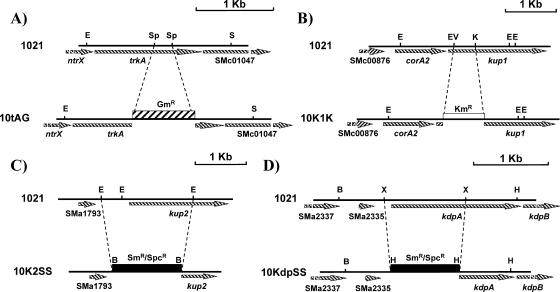
To study the importance of each of the four possible K+ uptake systems identified in S. meliloti, we constructed several 1021 derivative mutant strains: 10tAG, 10KdpSS, 10K1K, and 10K2SS, harboring interrupted versions of the genes trkA, kdpA, kup1, and kup2, respectively (Fig. 1). We also obtained a double mutant (10K2K1) lacking both Kup transporters, three double mutants (10tAK1, 10tAK2, and 10tAKdp) lacking TrkA and either Kup1, Kup2, or Kdp, respectively, and two triple mutants (10tAK21 and 10tAKK) lacking TrkA, Kup1, and either Kup2 or Kdp, respectively.
FIG. 1.
Construction of S. meliloti 1021 derivative mutants affected in the putative K+ uptake system Trk (A), Kup1 (B), Kup2 (C,) or Kdp (D). The wild-type version of the amplified region obtained by PCR is represented, and the mutated version is shown next. E, EcoRI; Sp, SphI; S, SmaI; EV, EcoRV; K, KpnI; B, BamHI; X, XbaI; H, HindIII. GmR, gentamicin resistance cassette; KmR, kanamycin resistance cassette; SmR/SpcR, streptomycin/spectinomycin resistance cassette.
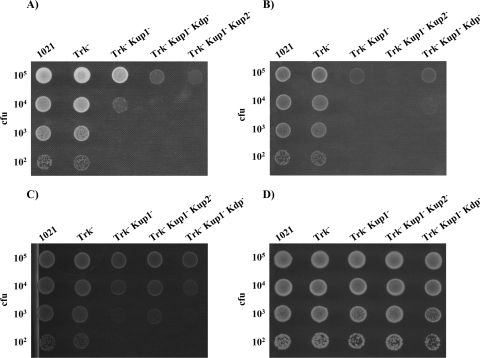
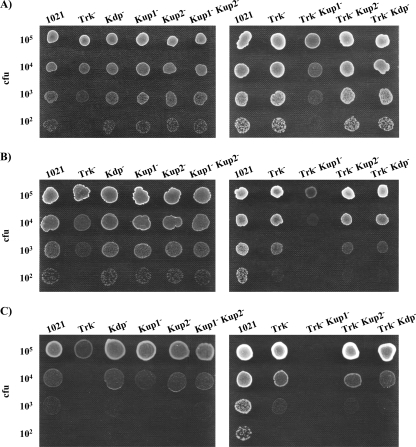
The growth of these 10 different mutants was tested in TY complex medium and in defined MM. In these media, all mutants grew as did the wild type except strains 10tAK1 (Trk− Kup1−), 10tAK21 (Trk− Kup2− Kup1−), and 10tAKK (Trk− Kdp− Kup1−), in which both the Trk and Kup1 systems were missing (Fig. 2A and B). The ability of the 10tAK1 (Trk− Kup1−) mutant to grow in TY or MM could be restored by complementation of the mutant with either the kup1 region (pJB3K1) or the trkA region (pJB3tA) (data not shown).
FIG. 2.
Ability of S. meliloti 1021-derivative mutants to grow in TY (A), MM (B), MM supplemented with 5 mM KCl (C), or MM supplemented with 10 mM KCl (D). In each row, drops contained approximately the number of CFU indicated on the left. A representative example of at least two experiments is shown. Pictures were taken after 2 to 4 days of incubation under the indicated conditions.
It has been described that growth rates of E. coli defective in K+ uptake systems depend on the K+ content of the medium. In fact, some of these mutants were isolated, taking advantage of their inability to grow at low K+ concentrations. An E. coli triple mutant lacking the Trk, Kdp, and Kup systems achieves a half-maximal growth rate at a K+ concentration of 20 mM (27). Therefore, we decided to explore the possibility that the growth defects of S. meliloti mutants lacking Trk and Kup1 could be relieved by externally added KCl. The growth abilities of these mutants were determined on solid MM amended with different concentrations of KCl (2 mM, 5 mM, 10 mM, and 20 mM), and we observed that 10 mM KCl was able to restore the growth of these mutants to a wild-type extent (Fig. 2). These results suggest that Trk and Kup1 are the main transport systems involved in K+ homeostasis during growth in osmotically balanced medium.
Role of K+ uptake systems in S. meliloti osmoadaptation.
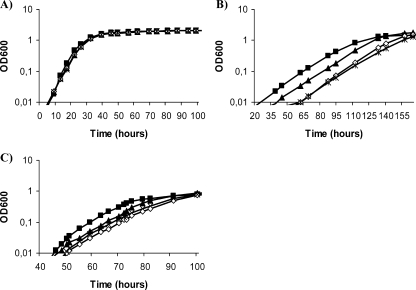
To study the involvement of the different transport systems in osmoadaptation, we first determined the growth ability of the wild-type strain in MM supplemented with NaCl (0.2 M, 0.3 M, 0.4 M, 0.5 M, and 0.6 M) or sucrose (0.3 M, 0.5 M, 0.7 M, and 0.9 M) (data not shown). We defined concentration ranges of 0.3 M to 0.5 M NaCl and 0.5 M to 0.7 M sucrose to be used for the determination of the osmoadaptive ability of the mutants. The growth curves of all mutants but 10tAK1 (Trk− Kup1−), 10tAKK (Trk− Kdp− Kup1−), and 10tAK21 (Trk− Kup2− Kup1−) were determined in liquid MM containing different concentrations of NaCl (0.3 M, 0.4 M, or 0.5 M) or sucrose (0.5 M or 0.7 M). Only results obtained with the highest concentrations of osmolytes are presented in Fig. 3. Among the single mutants, only the one lacking Trk displayed delayed growth in the presence of NaCl or sucrose. The effect of NaCl addition was stronger than that of sucrose addition. The delay in the mutant growth was clear with the three NaCl concentrations used; however, sucrose addition produced only a slight delay at the highest concentration tested. This behavior points out the importance of the Trk system for osmoadaptation in S. meliloti. However, the growth defects were stronger in trk kup2 and trk kdp double mutants, suggesting an involvement of both the Kup2 and Kdp systems in the osmoadaptation process which is revealed only in the absence of Trk (Fig. 3).
FIG. 3.
Growth curves of S. meliloti 1021 derivative strains in MM (A), MM supplemented with 0.5 M NaCl (B), or MM supplemented with 0.7 M sucrose (C). Only the growth curves of mutants significantly different from the wild type are shown. 1021 (wild type; black squares), 10tAG (Trk−; black triangles), 10tAK2 (Trk−, Kup2−; white diamonds), and 10tAKdp (Trk− Kdp−; asterisks) are analyzed. Data are representative of at least two replicate experiments. OD600, optical density at 600 nm.
Influence of pH in S. meliloti K+ uptake during osmoadaptation.
Among the conditions that determine the importance of different K+ uptake systems during osmoadaptation in bacteria, the pH and the K+ content of the medium are the most relevant ones (8). The Kup system has been reported to be the main system involved in osmoadaptation in E. coli at low pH, when the importance of Trk for K+ uptake is clearly reduced (35).
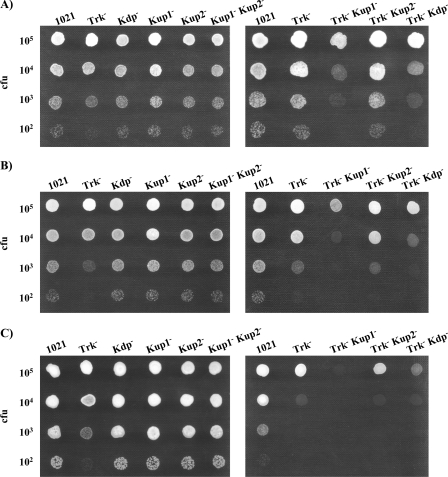
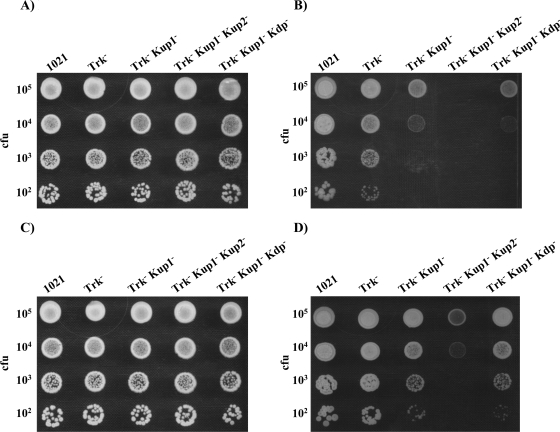
To establish the possible influence of pH on the relative importances of the different K+ transport systems of S. meliloti during osmotic adaptation, we used MM plates buffered at pH 6, pH 6.5, and pH 7 and supplemented with different concentrations of NaCl (0.3 M, 0.4 M, or 0.5 M) or sucrose (0.5 M, 0.6 M, or 0.7 M). The parental strain, 1021, was unable to grow on medium containing the highest concentrations of NaCl or sucrose at pH 6. Similar results were obtained under the remaining conditions (illustrative examples are presented in Fig. 4 and Fig. 5). The only single mutant affected by the addition of solutes was 10tAG (Trk−). Although NaCl addition caused a stronger effect than sucrose addition, we could observe the same trend in both cases and regardless of the pH or the osmolyte concentration. The ability of the 10tAG (Trk−) mutant to grow in NaCl-added medium could be restored to wild-type levels by complementation with the trkA region (pJB3tA) (data not shown). The growth ability of the 10tAK1 (Trk− Kup1−) mutant was strongly reduced by the addition of osmolytes to the medium (Fig. 4 and Fig. 5). This reduction could be relieved by complementation of the mutant with either the kup1 region (pJB3K1), which restored its growth ability to the levels of the single mutant 10tAG (Trk−), or the trkA region (pJB3tA), which restored its growth ability to the levels of the single mutant 10K1K (Kup1−) (data not shown). The double mutant 10tAKdp (Trk− Kdp−) also displayed a reduction in its growth ability with respect to 10tAG (Trk−) but only when challenged with high external NaCl (Fig. 4).
FIG. 4.
Ability of S. meliloti 1021 derivative mutants to grow in MM supplemented with 0.4 M NaCl at pH 7 (A), pH 6.5 (B) or pH 6 (C). In each row, drops contained approximately the number of CFU indicated on the left. A representative example of at least two experiments is shown. Pictures were taken after 4 to 6 days of incubation under the indicated conditions.
FIG. 5.
Ability of S. meliloti 1021-derivative mutants to grow in MM supplemented with 0.6 M sucrose at pH 7 (A), pH 6.5 (B), or pH 6 (C). In each row, drops contained approximately the number of CFU indicated on the left. A representative example of at least two experiments is shown. Pictures were taken after 4 to 6 days of incubation under the indicated conditions.
In contrast to the reported function of the Kup system at low pH in E. coli (35), we did not observe appreciable differences in the growth of the mutants 10K1K (Kup1−), 10K2SS (Kup2−), or 10K2K1 (Kup2− Kup1−) from that of the parental strain 1021 under any of the conditions tested.
Influence of external K+ in osmotolerance of S. meliloti K+ uptake mutants.
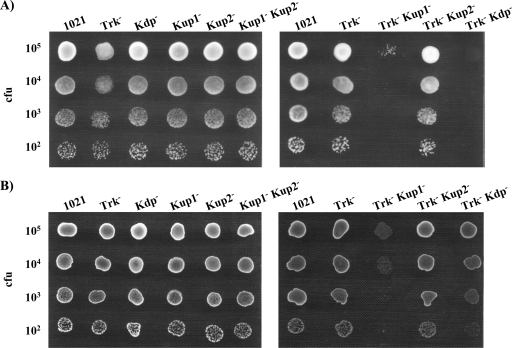
The deficient growth displayed by mutants lacking the Trk and Kup1 systems in osmotically balanced medium (Fig. 2) hampered the study of the possible involvement of the Kup1 system in osmoadaptation in the absence of Trk. This deficiency was more severe under abiotic stresses, such as low pH or high osmolyte concentrations (Fig. 4 and Fig. 5). Nevertheless, the actual involvement of Kup1 in the osmotolerance of strains lacking Trk could not be assessed, since the growth of these strains was not comparable to that of the wild type in nonstressing medium. As previously described, the addition of KCl to the medium releaved the growth defficiencies caused by the lack of Trk and Kup1 (Fig. 2). Therefore, we tested the osmoadaptation ability of these mutants in the presence of 10 and 20 mM KCl. The experiments were performed on solid MM to which were added different concentrations of NaCl (0.3 M or 0.4 M). The results for the different conditions tested were highly consistent, so we present only an illustrative example in Fig. 6. Even under these conditions, a reduction in the growth ability of 10tAG (Trk−) was observed upon NaCl addition, confirming a role of the Trk system in the osmoadaptation process. With these assays, we could also demonstrate a role of Kup1 during osmoadaptation in the absence of a functional Trk system. Furthermore, strain 10tAK21 (Trk− Kup2− Kup1−) presented even more difficulties in growth in the presence of NaCl than 10tAK1 (Trk− Kup1−), thus revealing an involvement of the Kup2 system during adaptation to hyperosmotic conditions, but only when Trk and Kup1 are absent. On the other hand, we did not notice any effect of the Kdp mutation on the growth of the 10tAKK (Trk− Kdp− Kup1−) mutant during osmoadaptation under these conditions. This result was not unexpected, since the Kdp system is known to be inhibited by high K+ concentrations (8). In fact, Kdp is believed to act in the K+ uptake process when the concentration of this cation is too low for it to be effectively accumulated by other systems. To test the functionality of the Kdp system of S. meliloti in response to the addition of NaCl or sucrose at low K+ concentrations, we used K+-free MM (described in Materials and Methods) (Fig. 7). The only single mutant affected by NaCl addition was 10tAG (Trk−), although the deffect was not as severe as that in K+-containing MM. On the other hand, the addition of NaCl to the K+-free medium almost abolished 10tAKdp (Trk− Kdp−) growth (Fig. 7). Even sucrose addition under these conditions resulted in decreased growth of 10tAKdp (Trk− Kdp−), not observed in normal K+-containing MM (Fig. 5). These results corroborate the higher importance of the Kdp system for osmoadaptation of S. meliloti at low K+ concentrations. Nevertheless, even under such conditions, Trk is still the main system involved in the osmoadaptation process, and the importance of Kdp can only be assessed in the absence of Trk.
FIG. 6.
Ability of S. meliloti 1021-derivative mutants to grow in MM supplemented with 10 mM KCl (A), 10 mM KCl and 0.3 M NaCl (B), 20 mM KCl (C), or 20 mM KCl and 0.3 M NaCl (D). In each row, drops contained approximately the number of CFU indicated on the left. A representative example of at least two experiments is shown. Pictures were taken after 4 to 6 days of incubation under the indicated conditions.
FIG. 7.
Ability of S. meliloti 1021 derivative mutants to grow in K+-free MM at pH 7 supplemented with 0.4 M NaCl (A) or 0.6 M sucrose (B). In each row, drops contained approximately the number of CFU indicated on the left. A representative example of at least two experiments is shown. Pictures were taken after 4 days of incubation under the indicated conditions.
Potassium accumulation.
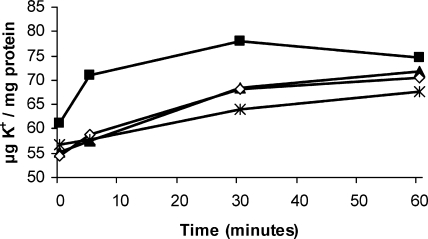
To test whether the growth deficiencies in hyperosmotic conditions observed in some K+ uptake mutants were related to defects in K+ accumulation, the levels of this cation present in the different mutants were determined after an osmotic upshift caused by addition of 0.3 M NaCl. In these assays, we used those mutants able to grow in liquid MM at a rate similar to that of the parental strain. Therefore, we could not test the mutants in which both Trk and Kup1 were absent. Among the strains tested, only those lacking the Trk system displayed accumulation kinetics different from that of the parental strain. We observed that the K+ accumulation process was significantly delayed in these mutants and seemed even slightly slower in 10tAKdp (Trk− Kdp−) than in 10tAG (Trk−) (Fig. 8). These results correlate well with the observed implication of these systems during adaptation to hyperosmotic conditions, suggesting that the osmoadaptation deficiencies are indeed due to the reduced rate of K+ accumulation in these mutants.
FIG. 8.
K+ accumulation kinetics of S. meliloti 1021 derivative strains after the addition of 0.3 M NaCl to midexponential cultures (optical density at 600 nm = 0.6) growing on MM. 1021 (wild type; black squares), 10tAG (Trk−; black triangles), 10tAK2 (Trk− Kup2−; white diamonds), and 10tAKdp (Trk− Kdp−; asterisks) are analyzed. Only the kinetics of mutants able to grow in MM at a rate similar to that of the parental strain and showing differences in K+ accumulation from that of the wild type are shown. Time zero corresponds to the point of application of the osmotic upshift. A representative example of two experiments is shown.
Importance of K+ transport systems for symbiosis.
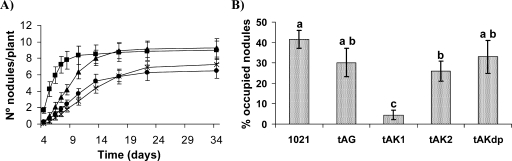
All K+ uptake mutants obtained were able to induce nitrogen-fixing root nodules on alfalfa plants, and most of them presented nodulation kinetics comparable to that of the parental strain, 1021 (data not shown). Nevertherless, the mutants lacking both the Trk and Kup1 systems (10tAK1, 10tAK21, and 10tAKK) exhibited delayed nodulation (Fig. 9A). This phenotype was not due to a loss of cell viability, since the numbers of CFU determined in the hydroponic solution were comparable for all strains (data not shown). Nevertheless, 10tAK1 (Trk− Kup1−) induced the development of a significantly lower number of nodules per plant than the parental strain (as determined by analysis-of-variance tests) until 11 days after inoculation. In the case of the triple mutants, the number of nodules per plant did not reach that of the parental strain until 22 days after inoculation.
FIG. 9.
Symbiotic phenotype of S. meliloti mutants in K+ uptake systems. (A) Nodulation kinetics of alfalfa plants inoculated with strain 1021 (wild type; black squares), 10tAK1 (Trk− Kup1−; black triangles), 10tAKK (Trk− Kdp− Kup1−; black dots), or 10tAK21 (Trk− Kup2− Kup1−; asterisks). The bars indicate standard errors. (B) Competition assays. Data represent the percentage of white nodules occupied by the wild type (1021), 10tAG (Trk−) (tAG), 10tAK1 (Trk− Kup1−) (tAK1), 10tAK2 (Trk− Kup2−) (tAK2), or 10tAKdp (Trk− Kdp−) (tAKdp) after inoculation with 1:1 mixtures of each strain and 1021(pGUS3). A representative example of two experiments is shown. Bars correspond to standard errors, and different letters indicate significative differences according to an analysis-of-variance test.
The lack of different K+ uptake systems had little effect on the competitiveness of most strains. However, a significant reduction in nodule occupancy was associated with the lack of Trk and Kup1 systems and, to a lesser extent, with the lack of Trk and Kup2 systems (Fig. 9B). These results strengthened again the importance of Trk and Kup1 in S. meliloti. We also studied the symbiotic effectiveness of the single mutants 10tAG (Trk−) and 10K1K (Kup1−). Nevertheless, there were no significant differences in the nitrogen content or the shoot weights of the plants inoculated with the mutants or wild type 1021 (data not shown).
DISCUSSION
In this work, we present the construction and functional characterization of Sinorhizobium meliloti 1021 mutants lacking each of the four possible K+ uptake systems identified in the genome sequence of this bacterium, as well as double and triple mutants in which several K+ transport systems were simultaneously inactivated. The lack of Trk and Kup1 caused a decrease in the viability of the cells during growth under osmotically balanced conditions (Fig. 2). Such a phenotype could be reverted by the addition of KCl to the culture medium, suggesting that the observed deficiencies were due to a reduction in the availability of K+ to these strains. Therefore, Trk and Kup1 would be the main systems involved in K+ homeostasis during S. meliloti growth. A similar phenotype has been described for Escherichia coli mutants in which all three Trk, Kup, and Kdp systems were inactivated by mutation (8).
We could determine the active implication of all four K+ transport systems of S. meliloti in osmoadaptation, studying the growth of the mutants in hyperosmotic medium. In contrast to the situation with other bacteria (35), the Trk system seems to be the most important K+ importer involved in the osmoadaptation of S. meliloti regardless of the pH, the osmolyte added, or the K+ content of the medium. Only in the absence of a functional Trk system could we establish the implication of the other K+ uptake systems in osmotolerance. The involvement of Kup1 in the adaptation to hyperosmotic conditions was revealed only after the behavior of the mutants lacking both Trk and Kup1 at high external K+ concentrations was assayed (Fig. 6). Furthermore, these experiments evidenced a role of the Kup2 system in osmoadaptation, although its implication in this process could be assessed only in the absence of both Trk and Kup1. It is well known that the presence of a K+ uptake system can mask a phenotype associated with the absence of other K+ transporters. For example, in E. coli the Trk system could be identified only after inactivation of the Kdp system (28), while in Bacillus subtilis the transcription of each of the two Ktr K+ uptake systems is elevated when the other is abolished by mutation (17).
Our results indicate that the Kdp system is inactive at high K+ concentrations in S. meliloti and its importance in osmoadaptation increases as the K+ content of the medium diminishes (Fig. 7), which agrees with observations with all Kdp systems studied so far. Nevertheless, we could detect an involvement of the Kdp system in osmoadaptation even in K+-containing MM, albeit only in the absence of a functional Trk system and only in response to NaCl addition and not to sucrose addition (Fig. 4 and 5). It has been previously reported that the Kdp response to an osmotic upshift depends on the nature of the osmolyte utilized in other bacteria. For example, the induction of the kdp operon in Salmonella is much more sensitive to ionic solutes than to nonpolar ones (2). This could explain the stronger effect of NaCl than of sucrose on the osmosensitivity of strains lacking a Kdp system in our experiments. Nevertheless, the differential effect caused by NaCl or sucrose on the osmoadaptive ability of the mutants lacking Trk (considered a constitutively expressed system in other bacteria [8]) suggests that a similar regulation might also control the transport activity of these systems.
Although all the 1021 derivatives obtained were able to nodulate alfalfa plants, the mutants unable to grow at rates comparable to that of the parental strain in standard media presented delayed nodulation kinetics (Fig. 9). This delay was not due to a loss of viability, and this fact, together with the observation that the absence of Trk and Kup1 caused a clear reduction of the competitiveness of the strain (Fig. 9), suggests that the growth defects of these mutants would affect their ability to colonize roots during symbiosis establishment. On the other hand, the study of the effectiveness of the symbioses established by 10tAG (Trk−) and 10K1K (Kup1−) with alfalfa plants did not reveal significant differences with that of the wild-type strain, 1021. This observation differs from the report by Nogales et al. (25), who observed that a mutation in a kup gene in R. tropici caused a reduction in the symbiotic effectiveness of the strain together with a deficiency in adaptation to hyperosmotic conditions. These results point out the differences in the involvement of K+ transport systems in the establishment of symbiosis by different rhizobial species. Furthermore, the genomic sequences of rhizobia reveal clear divergences with respect to the number and type of K+ uptake systems harbored by these bacteria. It would be necessary to widen our understanding of K+ transporter functionality in other rhizobia to draw conclusions on the importance of these systems for the symbiotic interaction.
Acknowledgments
This work was supported by grants AP2000-3118 and I3P-postgrado-2006 to A.D.-F. and BOS2002-04182-C02-01 and BIO2005-08089-CO2-01 to J.S.
Footnotes
Published ahead of print on 30 January 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaji, B., K. O'Connor, J. R. Lucas, J. M. Anderson, and L. N. Csonka. 2005. Timing of induction of osmotically controlled genes in Salmonella enterica serovar Typhimurium, determined with quantitative real-time reverse transcription-PCR. Appl. Environ. Microbiol. 718273-8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, A., M. Schmidt, W. Jäger, and A. Pühler. 1995. New gentamicin-resistance and promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 16237-39. [DOI] [PubMed] [Google Scholar]
- 4.Beringer, J. E. 1974. R factor fransfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84188-198. [DOI] [PubMed] [Google Scholar]
- 5.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djordjevic, M. A., H. C. Chen, S. Natera, G. Van Noorden, C. Menzel, S. Taylor, C. Renard, O. Geiger, the Sinorhizobium DNA Sequencing Consortium, and G. F. Weiller. 2003. A global analysis of protein expression profiles in Sinorhizobium meliloti: discovery of new genes for nodule occupancy and stress adaptation. Mol. Plant-Microbe Interact. 16508-524. [DOI] [PubMed] [Google Scholar]
- 7.Domínguez-Ferreras, A., R. Pérez-Arnedo, A. Becker, J. Olivares, M. J. Soto, and J. Sanjuán. 2006. Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti. J. Bacteriol. 1887617-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein, W. 2003. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 75293-320. [DOI] [PubMed] [Google Scholar]
- 9.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52147-154. [DOI] [PubMed] [Google Scholar]
- 10.Finan, T. M., E. Hartweig, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, H. M., M. Babst, T. Kaspar, G. Acuña, F. Arigoni, and H. Hennecke. 1993. One member of a groESL-like chaperonin multigene family of Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 122901-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dréano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Khan, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thébault, M. Vandenbol, F.-J. Vorhölter, S. Weidner, D. H. Wells, K. Wong, K.-C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293668-672. [DOI] [PubMed] [Google Scholar]
- 14.García-Rodríguez, F. M., and N. Toro. 2000. Sinorhizobium meliloti nfe (nodulation formation efficiency) genes exhibit temporal and spatial expression patterns similar to those of genes involved in symbiotic nitrogen fixation. Mol. Plant-Microbe Interact. 13583-591. [DOI] [PubMed] [Google Scholar]
- 15.Giraud, E., L. Moulin, D. Vallenet, V. Barbe, E. Cytryn, J. C. Avarre, M. Jaubert, D. Simon, F. Cartieaux, Y. Prin, G. Bena, L. Hannibal. J. Fardoux, M. Kojadinovic, L. Vuillet, A. Lajus, S. Cruveiller, Z. Rouy, S. Mangenot, B. Segurens, C. Dossat, W. L. Franck, W.-S. Chang, E. Saunders, D. Bruce, P. Richardson, P. Normand, B. Dreyfus, D. Pignol, G. Stacey, D. Emerich, A. Verméglio, C. Médigue, and M. Sadowsky. 2007. Legumes symbioses: absence of nod genes in photosynthetic bradyrhizobia. Science 3161307-1312. [DOI] [PubMed] [Google Scholar]
- 16.González, V., R. I. Santamaría, P. Bustos, I. Hernández-González, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramírez, V. Jiménez-Jacinto, J. Collado-Vides, and G. Davila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 1033834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holtmann, G., E. P. Bakker, N. Uozumi, and E. Bremer. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 1851289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7331-338. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9189-197. [DOI] [PubMed] [Google Scholar]
- 20.Lloret, J., B. B. H. Wulff, J. M. Rubio, J. A. Downie, I. Bonilla, and R. Rivilla. 1998. Exopolysaccharide II production is regulated by salt in the halotolerant strain Rhizobium meliloti EFB1. Appl. Environ. Microbiol. 641024-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meade, H. M., and E. R. Signer. 1977. Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 742076-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, K. J., and J. M. Wood. 1996. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50101-136. [DOI] [PubMed] [Google Scholar]
- 23.Miller-Williams, M., P. C. Loewen, and I. J. Oresnik. 2006. Isolation of salt-sensitive mutants of Sinorhizobium meliloti strain Rm1021. Microbiology 1522049-2059. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell, H. L. 1972. Microdetermination of nitrogen in plant tissues. J. Assoc. Off. Anal. Chem. 551-3. [Google Scholar]
- 25.Nogales, J., R. Campos, H. BenAbdelkhalek, J. Olivares, C. Lluch, and J. Sanjuan. 2002. Rhizobium tropici genes involved in free-living salt tolerance are required for the establishment of efficient nitrogen-fixing symbiosis with Phaseolus vulgaris. Mol. Plant-Microbe Interact. 15225-232. [DOI] [PubMed] [Google Scholar]
- 26.Olivares, J., J. Casadesús, and E. J. Bedmar. 1980. Method for testing degree of infectivity of Rhizobium meliloti strains. Appl. Environ. Microbiol. 39967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polarek, J. W., G. Williams, and W. Epstein. 1992. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J. Bacteriol. 1742145-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhoads, D. B., F. B. Waters, and W. Epstein. 1976. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J. Gen. Physiol. 67325-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertsen, B. K., P. Aiman, A. G. Darwill, M. McNeil, and P. Albersheim. 1981. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 67389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio-Technology 1784-791. [Google Scholar]
- 33.Sleator, R. D., and C. Hill. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 2649-71. [DOI] [PubMed] [Google Scholar]
- 34.Soussi, M., M. Santamaría, A. Ocaña, and C. Lluch. 2001. Effects of salinity on protein and lipopolysaccharide pattern in a salt-tolerant strain of Mesorhizobium ciceri. J. Appl. Microbiol. 90476-481. [DOI] [PubMed] [Google Scholar]
- 35.Trchounian, A., and H. Kobayashi. 1999. Kup is the major K+ uptake system in Escherichia coli upon hyper-osmotic stress at a low pH. FEBS Lett. 447144-148. [DOI] [PubMed] [Google Scholar]
- 36.Tuteja, N. 2007. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 428419-438. [DOI] [PubMed] [Google Scholar]
- 37.Young, J. P. W., L. C. Crossman, W. B. Johnston, N. R. Thomson, Z. F. Ghazoui, K. H. Hull, M. Wexler, A. R. J. Curson, J. D. Todd, P. S. Poole, T. H. Mauchline, A. K. East, M. A. Quail, C. Churcher, C. Arrowsmith, L. Cherevach, T. Chillingworth, K. Clarke, A. Cronin, P. Davis, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, S. Whitehead, and J. Parkhill. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahran, H. H. 1999. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 63968-989. [DOI] [PMC free article] [PubMed] [Google Scholar]