In their seminal work, Stanier and van Niel (47) proposed that bacteria are distinguished from other forms of life, including viruses, protists, fungi, algae, plants, and animals, by their procaryotic cell structure. They defined the procaryotic cell by three major criteria: the absence of internal membranes that compartmentalize the nuclear material and the enzymatic machinery for respiration and photosynthesis, nuclear division which occurs by fission and not mitosis, and the presence of peptidoglycan in the cell wall. They also recognized that procaryotes possess enormous diversity, similar in scope to that of eucaryotic protists, and further proposed that the procaryotes represent a distinct mode of evolutionary diversification parallel to that found in the eucaryotes (47). Thus, the procaryotic-eucaryotic dichotomy was founded upon the recognition of two very different types of cellular organisms and not the phylogenetic relationships between them.
Recently, five major criticisms of the concept of the procaryote have been proposed (36, 55): it fundamentally contradicts the three-domain model of life, the procaryotes are not monophyletic, the procaryotes are defined by negative characteristics, the term procaryote “sustain[s] the concept that procaryotes evolved into eucaryotes,” and the term is imprecise. As shown below, these criticisms are misstatements of the original proposal and modern descriptions of the concept or otherwise erroneous (47, 27). In fact, since its original proposal in 1962, the experimental evidence for the procaryote concept has been enormously enriched.
Today, the concept of the procaryote includes a much greater understanding of the molecular basis for the differences between procaryotic and eucaryotic cells. In addition, it recognizes the antiquity, abundance, and diversity of procaryotes. Procaryotes likely dominated life on the early earth for over a billion years prior to the appearance of eucaryotes. Today, the biomass of the procaryotes is comparable to that of eucaryotes. The procaryotes are also extremely diverse, and representatives of two ancient domains, Bacteria and Archaea, are common today. Each domain includes organisms with many different metabolic and physiological capabilities, and the number of species is correspondingly so large that it has never been estimated accurately.
The procaryotic cell.
The cellular organization of procaryotes is of fundamental importance to their physiological and biochemical processes, and their differences from those of eucaryotes are well described (27). Three features are especially relevant. (i) Nuclear membranes are absent, which allows coupled transcription and translation (13, 33). Because the DNA is not segregated to the nucleus, it is also possible to regulate transcription with repressors and activators that bind metabolites. In this sense, transcriptional regulation is further coupled to metabolism. In the eucaryotes, the major metabolic processes occur in the mitochondria, chloroplast, and the cytoplasm and are isolated from transcription in the nucleus.
(ii) Procaryotic cells are usually smaller than eucaryotic cells. There are some notable exceptions. The sulfur-oxidizing bacterium Thiomargarita has a diameter up to 750 μm (44), which is larger than that of many protists. The eucaryotic marine picoalgae, which are 1 to 2 μm in diameter, are similar in size to many procaryotes (41). In spite of this diversity, size remains an important distinguishing characteristic (58). Size establishes the surface-to-volume ratio of the cell, which limits the rate and type of nutrient uptake. It also allows for rapid diffusion of small molecules and proteins throughout the entire cell, which provides a mechanism for coupling metabolism and regulation.
(iii) The cytoplasmic membrane is multifunctional in procaryotes and represents the defining structure of the cell. A proton motive force is generated on the cytoplasmic membrane by respiration, photosynthesis, or ATP hydrolysis to empower key cellular processes such as ATP biosynthesis, NAD+ reduction by reverse electron transport, nutrient uptake, motility, and secretion. Procaryotes utilize membrane transporters on the cell surface to assimilate nutrients dissolved in their environment. In many procaryotes, the cytoplasmic membrane possesses a complex topology composed of lamellae, tubules, or other cytoplasmic intrusions (27). In contrast, the cytoplasmic membrane of eucaryotes is very different in structure and function. Eucaryotes commonly take up particulate material by phagocytosis, a process that does not occur in procaryotes.
Evolution of procaryotes.
Geochemical and fossil evidence indicates that life on earth is at least 3.5 billion years old (1, 42, 43). While the form of ancient microfossils resembles that of modern procaryotes, there is little additional evidence in the fossil record for their molecular nature. However, by 2.5 billion years ago, there is evidence for abundant procaryotic life, including widespread microfossils and stromatolites or fossilized microbial mats, and major signatures of biological processes in the geochemical record, such as depletion of inorganic carbonates for 12C and deposits of complex organic carbon enriched in 12C (42). By this time, the oxygenation of the earth was also well under way (9), and it is likely that oxygenic photosynthesis was fully evolved within the domain Bacteria.
Molecular clocks based upon both rRNA and protein-encoding genes suggest that the domains Archaea and Bacteria had both diverged by this time (2, 45). Moreover, within the domain Bacteria, many of the deep groups or phyla had already formed, including the modern lineages of Cyanobacteria, Proteobacteria, and Firmicutes. Within the domain Archaea, the Crenarchaeota and Euryarchaeota had already diverged, as well as many of the major lineages of methanogens within the phylum Euryarchaeota. The presence of many diverse and highly specialized lineages, many of which share large numbers of complex biochemical pathways and molecular processes, suggests that the biochemical complexity of the procaryotes was fully evolved and that procaryotes very similar to modern organisms were abundant on earth 2.5 billion years ago. In contrast, the first fossils of clearly eucaryotic organisms appeared about 1.8 billion years ago (24). Analyses of the molecular diversity within the modern eucaryotes suggest that this group began to diversify about 1.1 to 2.0 billion years ago (8, 17). Thus, it is likely that the eucaryotes only evolved after the procaryotes had obtained their modern complexity.
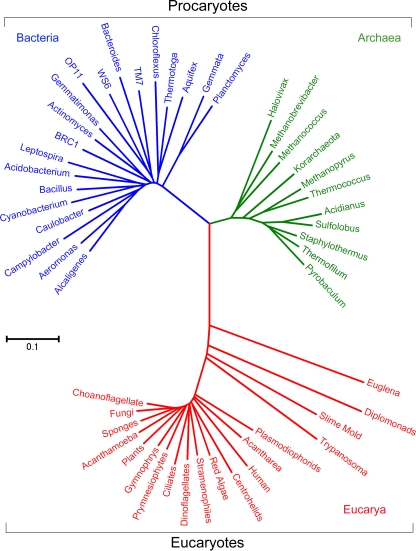
Although not stated explicitly, it is likely that Stanier and van Niel (47) believed that the procaryotes comprised more than one major phylogenetic lineage. They recognized that procaryotic diversity was comparable to that of the eucaryotes, as evidenced by various photosynthetic and chemotrophic metabolisms; unicellular, multicellular, and coenocytic lifestyles; and cellular multiplication by binary fission, budding, or the formation of conidia. Moreover, they proposed that the procaryotes represented one of two parallel forms of evolutionary diversification, with the second form being the eucaryotes. At the time, the eucaryotes were believed to comprise multiple lineages, so this dichotomy implied that the procaryotes also comprised more than a single lineage. Originally discovered based upon comparisons of 16S rRNA sequences, the presence of the two ancient lineages or domains Bacteria and Archaea within the procaryotes is now well established (Fig. 1) (56). Fundamental differences in the molecules required for information-processing systems, such as translation, transcription, and replication, further support the early emergence of these domains (28, 32).
FIG. 1.
Phylogenetic tree of the 16S rRNA genes in the three domains Archaea, Bacteria, and Eucarya. The procaryotes comprise the archaeal and bacterial domains. The eucaryotes contain solely the domain Eucarya. This unrooted tree was calculated by the minimum evolutionary algorithm in the software MEGA4 (48). The tree is drawn to scale, with branch lengths in evolutionary distance calculated by the maximum composite likelihood method and units of numbers of base substitutions per site.
Procaryotic domains.
In addition to the information-processing systems, fundamental differences exist in the cellular lipids of archaea and bacteria. Bacterial lipids are generally composed of fatty acids linked to glycerol by ester bonds. The stereochemistry of the glycerol moiety is the 1,2-sn configuration. Archaeal lipids are composed of isoprenoid side chains linked to glycerol by ether bonds, where the glycerol has the 2,3-sn configuration (25, 50). The isoprenoid side chains may contain linear or cyclopentane units. While these lipids commonly form bilayers as in bacteria, some archaeal membranes are monolayers containing glycerol tetraethers that span the membrane.
Few other features are as distinctive of either the archaea or the bacteria. For instance, the presence of peptidoglycan in the cell wall was one of the original characteristics used to define the procaryotes (47). It has since been recognized that the cell walls of both bacteria and archaea are very diverse. Both groups contain wall-less organisms, such as Mycoplasma within the domain Bacteria and the genus Thermoplasma within the domain Archaea. Most of the described bacterial cell walls contain a peptidoglycan composed of muramic acid. Only a few archaea possess a cell wall composed of peptidoglycan, but muramic acid is absent, and the polymer differs profoundly from bacterial peptidoglycan (29). Instead, archaeal cell walls are most often composed solely of S-layers (26). These are monolayers of a single protein that provide the cell with structural integrity and shape. S-layers are components of many bacterial cell walls as well, but they only rarely serve as the sole wall structure as in archaea.
Both groups are also metabolically diverse, and their members have adapted to a wide range of physiological conditions and life-styles (27). They share many of the major types of transporters of the common organic and inorganic nutrients (37). For sugar catabolism, variations of the Embden-Meyerhof and Entner-Doudoroff pathways have been described in both archaeal and bacterial heterotrophs (5). While chlorophyll-based photosynthesis is unknown within the domain Archaea, both archaea and bacteria possess the retinal-based bacteriorhodopsin systems (14). Similarly, the ability to utilize inorganic substrates for lithotrophic growth, such as H2, reduced sulfur compounds, or ammonium, is shared by members of both lineages. The diversity of anabolic pathways is comparable. While only bacteria are known to utilize the Calvin cycle, archaeal and bacterial autotrophs use the Ljungdahl-Wood, reverse trichloroacetic acid, or hydroxypropionate pathway of CO2 fixation (3). Interestingly, while some archaea possess ribulose 1,5-bisphosphate carboxylase/oxygenase, the key enzyme of the Calvin cycle, this enzyme is involved in AMP metabolism and not autotrophic CO2 fixation (40, 52). Complex variations of intermediary metabolism also occur in both lineages (5). Many major pathways of monomer biosynthesis are the same in both groups, and some common variations, such as the citramalate pathway of isoleucine biosynthesis, are also found in both lineages (10).
Thus, in contrast to the genes for the information-processing systems, there are no clear distinctions between archaeal and bacterial metabolic genes (51). As an example, the genes of the tryptophan biosynthetic operon are widely distributed in both groups (57). While the genes within each domain are generally more similar to the genes from the same domain, they are clearly homologous across both domains. There are also no special features that are characteristic of either domain. Instead, the genes are distributed according to the physiological adaptations of the specific organisms and complex histories of gene fusions, insertions, horizontal transfers, and other evolutionary events. There are a few exceptions. Chlorophyll-based photosynthesis appears to be entirely bacterial and eucaryotic. Methanogenesis appears to be limited to the domain Archaea.
Genomic comparisons are consistent with these conclusions. Within the domain Archaea, 7,538 clusters of orthologous groups of proteins (arCOGs) were identified (32). Of these, 53% possess a high affinity for bacterial genes and 42% appear to be uniquely archaeal. A small number of arCOGs are common to all archaeal genomes. This “core” of 166 arCOGs largely includes genes which encode information-processing functions. Of these arCOGs, 77% possess high similarities to eucaryotic genes. Archaeal genomes also possess a much larger “shell” of about 2,200 arCOGs which are widely distributed throughout archaeal genomes but are not universal. Many of these genes encode metabolic functions and are shared with bacteria. The conserved core distinguishes the domain Archaea from the domain Bacteria, but the shell encodes the metabolic features shared among the procaryotes.
In summary, examination of the distinct and shared characters between the archaea and bacteria provide compelling evidence for two domains. However, the nature of the evolutionary processes that led to their formation is much less clear (7). Because of the complexity of the shared features, a model that encompasses only “vertical” evolution would suggest that the common ancestor was a sophisticated organism capable of accurate translation and transcription and possessing the metabolic capability of making all the small molecules in the cell, including complex vitamins such as cobamides (21). However, the physiological characters shared by both lineages exceed those found in any single modern organism. For instance, multiple pathways of glycolysis, autotrophy, and respiration are common to both the archaea and the bacteria. This observation suggests that the modern domains are the products of a more complex evolutionary history which must have included horizontal gene transfers and gene creations and losses as common events (7). This conclusion is supported by the observation of fairly recent horizontal gene transfers between organisms from each domain (35). Remarkably, in at least one case, the number of genes involved was comparable to the number acquired by the ancestor of the eucaryotes during endosymbiosis (6). Although the consequences may not have been as profound for biology, this massive horizontal gene transfer illustrates the potential scale of this process in procaryotic evolution.
Distribution of procaryotes.
Procaryotes are found nearly everywhere in the modern world, and their presence defines the biosphere (54). They have been detected at altitudes of 77 km in the atmosphere and depths of 2 km in the subsurface. Soil, water, sea ice, leaves and roots of trees, guts of invertebrate and vertebrate animals, and subsurface aquifers are all fully colonized by highly specialized populations of procaryotes. The number of individual cells is probably on the order of 5 × 1030, and their biomass is comparable to that of plants (54).
Their abundance enables them to play key roles in the geochemical cycles that process the major elements of life, including C, N, and S. For instance, except for the noble gases, procaryotes contribute to the production of all of the abundant gases within the earth's atmosphere. For some, such as methane and nitrous oxide, procaryotes represent the major sources. Likewise, for some of the cycles, key steps are catalyzed nearly exclusively by procaryotes. Within the nitrogen cycle, biological nitrogen fixation, denitrification, and nitrification are exclusively procaryotic processes. Within the sulfur cycle, dissimilatory sulfate reduction and anaerobic sulfide oxidation are exclusively procaryotic.
Diversity of procaryotes.
Microbiologists have long been impressed with the extreme diversity of procaryotes (47). For instance, procaryotes in the order Planctomycetales possess true intracytoplasmic membranes, which is one of the defining characteristics of eucaryotes (30). Within the genus Gemmata, these membranes surround the nucleoid. This structure superficially resembles that of the eucaryotic nucleus, but the presence of ribosomes inside the compartment is a fundamental difference. Procaryotes also possess numerous fundamentally different strategies for motility, including flagellation, gliding, twitching, and gas vesicles. Remarkably, even archaeal and bacterial flagella are not homologous, demonstrating that microorganisms have acquired similar mechanisms by different means (20). Similar diversity is found in many other features, including respiration, photosynthesis, cell structure, and cell division.
Recently, it has been possible to investigate procaryotic diversity quantitatively. For instance, surveys of procaryotic 16S rRNA genes in environmental samples have detected greater than 50 bacterial “phyla,” of which only half have cultivated representatives (19, 38). Procaryotic phyla represent the deepest classification within the domain Bacteria or Archaea. Molecular clocks and correlations with the biogeochemical record indicate that these phyla probably formed greater than 2.5 billion years ago (2, 45). The antiquity of these lineages is consistent with their enormous diversity. Importantly, procaryotic phyla are much more diverse than eucaryotic phyla, which formed much later. For instance, the mammals, reptiles, and amphibians probably formed within the last 450 million years (8, 17). If they were classified by the same criteria used for many procaryotes, they would be placed in separate genera within the same family.
Given the diversity of ancient groups, it is not surprising that the number of modern groups is enormous. A procaryotic species is much deeper than common in eucaryotic biology and includes strains with >70% DNA-DNA hybridization and a change in the melting temperature of the DNA hybrids of <5°C (53). By this same criterion, most of the primates would be considered a single species (54). At present, there are no certain estimates of the total number of procaryotic species on earth. Within soil, which contains a relatively diverse population, various methods have detected 103 to 104 different molecular species or operational taxonomic units (OTUs) per sample (39, 49). Theoretical estimates suggest that soil could contain well over 106 OTUs (4). Similar observations have been made in the deep sea (46). In the most extensive study to date, partial sequencing of 900,000 16S rRNA procaryotic genes from two deep-sea sites encountered 36,087 unique sequences representing 20,468 OTUs (18). The OTUs detected in these experiments are defined at 97% sequence similarity of the 16S rRNA and are deeper taxonomic groups than a conventional procaryotic species as defined above (23). Importantly, only a small fraction of the total number of species known to exist have ever been characterized.
Classification of procaryotes.
For more than 20 years, a large community of microbiologists have sought a phylogenetic classification of procaryotic life (12, 15, 31). A major goal of this effort has been to produce a classification that summarizes our understanding of procaryotes and provides an explanatory, predictive, and conceptual framework for further investigations. Even though the procaryotes are not monophyletic and the evolutionary processes they have experienced are extremely complex (34), this classification strategy remains useful and knowledge of the evolutionary processes which formed modern organisms provides a great deal of insight into their biological properties. However, phylogenetic classifications must extend beyond the simple recognition of monophyletic groups. They must recognize the wide variety of evolutionary processes known to occur and the realization that not all evolutionary processes have the same biological consequences. Thus, one important goal of classification is to distinguish groups of organisms which share transformative evolutionary events.
In the case of the procaryote-eucaryote dichotomy, the evolution of the eucaryotes was such a transformative event. Even though the origin of the eucaryotes is not certain, the consequences were profound enough to produce an organism fundamentally different from those not descendant from this event (11). It could be argued that the procaryotes are then defined by a negative, i.e., the failure to undergo this transformative event (36). This criticism neglects the large number of similarities within the procaryotes that are part of the basis of this classification. The procaryotic classification could also be criticized because it is paraphyletic, but this objection is not substantive if the identification of monophyletic groups is not a major goal of the classification.
Phylogenetic analyses provide additional insights by identifying relationships among organisms that would not otherwise be obvious. This feature is important because, while the general properties of procaryotes are sufficient to unite them into a single group, the diversity is so large that no single phenotypic or functional property is likely to be universal. However, even groups with exceptional properties can be related to more conventional procaryotes by identifying the evolutionary processes responsible for their formation. This principle was established prior to the development of molecular phylogenetic methods in the classification of the cyanobacteria within the procaryotes (47). Although their photosynthetic apparatus and aspects of cellular differentiation are similar to those of eucaryotes, these microorganisms possess many features typical of other procaryotes, such as peptidoglycan in their cell walls, the capacity for nitrogen fixation, and the absence of intracytoplasmic organelles. In fact, this classification has since withstood the test of molecular phylogenetic methods, and the cyanobacterial genome is distinctively procaryotic (22). Similarly, even though the planctomycete Pirellula possesses a true intracytoplasmic membrane, most of its genes are bacterial, consistent with its classification (16). Genome reduction is also a common process observed in many independent lineages of symbiotic procaryotes that produces highly distinctive organisms. In spite of these differences, these organisms are reasonably classified as procaryotes. From this perspective, even if a modern descendant of the eucaryotes was phenotypically indistinguishable from the procaryotes, it would remain a eucaryote based upon its phylogeny.
A phylogenetic classification that considers many types of evolutionary processes, as well as their biological consequences, easily accommodates chimeric organisms that are very distinctive by creating a new category. Arguably, the eucaryotes are precisely such a classification. While the details of their formation remain to be elucidated, most evidence suggests that they formed from both archaeal and bacterial lineages fairly late in the history of life (11). The classification of groups of chimeric organisms where the consequences are less dramatic is not necessarily problematic. However, it requires a more complete understanding of horizontal gene transfer and its role in the properties of the specific organism.
Conclusions.
The last 50 years has produced a wealth of new information that greatly enriches our understanding of procaryotes. These organisms have proven to be of enormous abundance and diversity, the product of complex evolutionary processes over billions of years. They dominated life on the earth prior to the appearance of eucaryotes. Their antiquity implies that many of the most salient features of modern life evolved in an entirely procaryotic world, including most of the organizing principles of the cell; the basic mechanisms of replication, transcription, and translation; the major catabolic and anabolic pathways; and the biogeochemical cycles which maintain the biosphere. While the term procaryote was precisely defined by Stanier and van Niel (47), it has occasionally been misused, usually as synonymous with bacteria. The solution to this problem is education about its true meaning and not to discard a valuable and central concept in biology.
Acknowledgments
I am grateful to my colleagues Dave Coleman, Ford Doolittle, Matt Kane, Anna Karls, Noel Krieg, John Leigh, Jan Mrazek, Mary Ann Moran, Larry Shimkets, Jim Staley, and Juergen Wiegel, whose constructive criticisms greatly improved this work. Thanks also to Kamlesh Jangid for preparation of the figure.
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Altermann, W., and J. Kazmierczak. 2003. Archaean microfossils: a reappraisal of early life on Earth. Res. Microbiol. 154611-617. [DOI] [PubMed] [Google Scholar]
- 2.Battistuzzi, F. U., A. Feijao, and S. B. Hedges. 2004. A genomic timescale of prokaryotic evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, I. A., D. Kockelkorn, W. Buckel, and G. Fuchs. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 3181782-1786. [DOI] [PubMed] [Google Scholar]
- 4.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 9910494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danson, M. J., H. J. Lamble, and D. W. Hough. 2007. Central metabolism, p. 260-287. In R. Cavicchioli (ed.), Archaea: molecular and cellular biology. ASM Press, Washington, DC.
- 6.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, et al. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4453-461. [PubMed] [Google Scholar]
- 7.Doolittle, W. F., and W. Bapteste. 2007. Pattern pluralism and the tree of life hypothesis. Proc. Natl. Acad. Sci. USA 1042043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douzery, E. J. P., E. A. Snell, E. Bapteste, F. Delsuc, and H. Philippe. 2004. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl. Acad. Sci. USA 10115386-15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eigenbrode, J. L., and K. H. Freeman. 2006. Late archean rise of aerobic microbial ecosystems. Proc. Natl. Acad. Sci. USA 10315759-15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekiel, I., I. C. P. Smith, and G. D. Sprott. 1984. Biosynthesis of isoleucine in methanogenic bacteria: a 13C NMR study. Biochemistry 231683-1687. [Google Scholar]
- 11.Embley, T. M., and W. Martin. 2006. Eukaryotic evolution, changes and challenges. Nature 440623-630. [DOI] [PubMed] [Google Scholar]
- 12.Fox, G. E., E. Stackebrandt, R. B. Hespell, J. Gibson, J. Maniloff, T. A. Dyer, R. S. Wolfe, W. E. Balch, R. S. Tanner, L. J. Magrum, L. B. Zablen, R. Blakemore, R. Gupta, L. Bonen, B. J. Lewis, D. A. Stahl, K. R. Luehrsen, K. N. Chen, and C. R. Woese. 1980. The phylogeny of prokaryotes. Science 209457-463. [DOI] [PubMed] [Google Scholar]
- 13.French, S. L., T. J. Santangelo, A. L. Beyer, and J. N. Reeve. 2007. Transcription and translation are coupled in Archaea. Mol. Biol. Evol. 24893-895. [DOI] [PubMed] [Google Scholar]
- 14.Frigaard, N.-U., A. Martinez, T. J. Mincer, and E. F. DeLong. 2006. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439847-850. [DOI] [PubMed] [Google Scholar]
- 15.Garrity, G. M., and J. G. Holt. 2001. The road map to the manual, p. 119-141. In D. R. Boone, and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, NY. [Google Scholar]
- 16.Glöckner, F. O., M. Kube, M. Bauer, H. Teeling, T. Lombardot, W. Ludwig, D. Gade, A. Beck, K. Borzym, K. Heitmann, R. Rabus, H. Schlesner, R. Amann, and R. Reinhardt. 2003. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 1008298-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedges, S. B., J. E. Blair, M. L. Venturi, and J. L. Shoe. 2004. A molecular timescale of eukaryotic evolution and the rise of complex multicellular life. BMC Evol. Biol. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber, J. A., D. B. M. Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 31897-100. [DOI] [PubMed] [Google Scholar]
- 19.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1804765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarrell, K. F., D. P. Bayley, and A. S. Kostyukova. 1996. The archaeal flagellum: a unique motility structure. J. Bacteriol. 1785057-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, W. J., D. P. Nagle, Jr., and W. B. Whitman. 1987. Methanogens and the diversity of archaebacteria. Microbiol. Rev. 51135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, et al. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3109-136. [DOI] [PubMed] [Google Scholar]
- 23.Keswani, J., and W. B. Whitman. 2001. Relationships of 16S rRNA sequence similarity to DNA hybridization in prokaryotes. Int. J. Syst. Evol. Microbiol. 51667-678. [DOI] [PubMed] [Google Scholar]
- 24.Knoll, A. H., E. J. Javaux, D. Hewitt, and P. Cohen. 2006. Eukaryotic organisms in Proterozoic oceans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 3611023-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koga, Y., and H. Morii. 2007. Biosynthesis of ether-type polar lipids in Archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 7197-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.König, H., R. Rachel, and H. Claus. 2007. Proteinaceous surface layers of Archaea: ultrastructure and biochemistry, p. 315-340. In R. Cavicchioli (ed.), Archaea: molecular and cellular biology. ASM Press, Washington, DC.
- 27.Krieg, N. R. 2005. Procaryotic domains, p. 21-25. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2A. Springer, New York, NY. [Google Scholar]
- 28.Lao-Sirieix, S.-H., V. L. Marsh, and S. D. Bell. 2007. DNA replication and cell cycle, p. 93-109. In R. Cavicchioli (ed.), Archaea: molecular and cellular biology. ASM Press, Washington, DC.
- 29.Leps, B., H. Labischinski, G. Barnickel, H. Bradaczek, and P. Giesbrecht. 1984. A new proposal for the primary and secondary structure of the glycan moiety of pseudomurien. Eur. J. Biochem. 144279-286. [DOI] [PubMed] [Google Scholar]
- 30.Lindsay, M. R., R. I. Webb, M. Strous, M. S. M. Jetten, M. K. Butler, R. J. Forde, and J. A. Fuerst. 2001. Cell compartmentalisation in planctomycetes: novel types of structural organization for the bacterial cell. Arch. Microbiol. 175413-429. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig, W., and H.-P. Klenk. 2005. Overview: a phylogenetic backbone and taxonomic framework for procaryotic systematics, p. 49-65. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2A. Springer, New York, NY. [Google Scholar]
- 32.Makarova, K. S., A. V. Sorokin, P. S. Novichkov, Y. I. Wolf, and E. V. Koonin. 2007. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol. Direct 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, W., and E. V. Koonin. 2006. A positive definition of prokaryotes. Nature 442868. [DOI] [PubMed] [Google Scholar]
- 34.McInerney, J. O., J. A. Cotton, and D. Pisani. 2008. The prokaryotic tree of life: past, present…future? Trends Ecol. Evol. 23276-281. [DOI] [PubMed] [Google Scholar]
- 35.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, et al. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399323-329. [DOI] [PubMed] [Google Scholar]
- 36.Pace, N. R. 2006. Time for a change. Nature 441289. [DOI] [PubMed] [Google Scholar]
- 37.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 30175-100. [DOI] [PubMed] [Google Scholar]
- 38.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57369-394. [DOI] [PubMed] [Google Scholar]
- 39.Roesch, L. F. W., R. R. Fulthorpe, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. H. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, T., H. Atomi, and T. Imanaka. 2007. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 3151003-1006. [DOI] [PubMed] [Google Scholar]
- 41.Scherr, E., and B. Scherr. 2000. Marine microbes: an overview, p. 13-46. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, NY.
- 42.Schidlowski, M., J. M. Hayes, and I. R. Kaplan. 1983. Isotopic inferences of ancient biochemistries: carbon, sulfur, hydrogen, and nitrogen, p. 149-186. In J. W. Schopf (ed.), Earth's earliest biosphere. Princeton University Press, Princeton, NJ.
- 43.Schopf, J. W. 2006. Fossil evidence of archaean life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361869-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulz, H. N., T. Brinkhoff, T. G. Ferdelman, M. Hernández Mariné, A. Teske, and B. B. Jorgenson. 1999. Dense populations of a giant sulfur bacterium in Namibian Shelf sediments. Science 284493-495. [DOI] [PubMed] [Google Scholar]
- 45.Sheridan, P. P., K. H. Freeman, and J. E. Brenchley. 2003. Estimated minimal divergence times of the major bacterial and archaeal phyla. Geomicrobiol. J. 201-14. [Google Scholar]
- 46.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the unexplored “rare biosphere.” Proc. Natl. Acad. Sci. USA 10312115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanier, R. Y., and C. B. van Niel. 1962. The concept of a bacterium. Arch. Microbiol. 4217-35. [DOI] [PubMed] [Google Scholar]
- 48.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 49.Torsvik, V., L. Øvreås, and T. T. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 2961064-1066. [DOI] [PubMed] [Google Scholar]
- 50.Valentine, D. L. 2007. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 5316-323. [DOI] [PubMed] [Google Scholar]
- 51.Walsh, D. A., and W. F. Doolittle. 2005. The real ‘domains’ of life. Curr. Biol. 15R237-R240. [DOI] [PubMed] [Google Scholar]
- 52.Watson, G. M. F., J.-P. Yu, and F. R. Tabita. 1999. Unusual ribulose 1,5-bisphosphate carboxylase/oxygenase of anoxic Archaea. J. Bacteriol. 1811569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37463-464. [Google Scholar]
- 54.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 956578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woese, C. R. 2007. The Archaea: an invitation to evolution, p. 1-13. In R. Cavicchioli (ed.), Archaea: molecular and cellular biology. ASM Press, Washington, DC.
- 56.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 874576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie, G., N. O. Keyhani, C. A. Bonner, and R. A. Jensen. 2003. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 67303-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young, K. D. 2006. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70660-703. [DOI] [PMC free article] [PubMed] [Google Scholar]