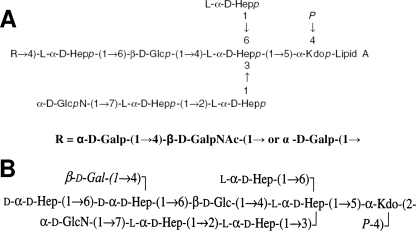Abstract
Comparison between the lipopolysaccharide (LPS) core structures of Aeromonas salmonicida subsp. salmonicida A450 and Aeromonas hydrophila AH-3 shows great similarity in the inner LPS core and part of the outer LPS core but some differences in the distal part of the outer LPS core (residues ld-Hep, d-Gal, and d-GalNAc). The three genomic regions encoding LPS core biosynthetic genes in A. salmonicida A450, of which regions 2 and 3 have genes identical to those of A. hydrophila AH-3, were fully sequenced. A. salmonicida A450 region 1 showed seven genes: three identical to those of A. hydrophila AH-3, three similar but not identical to those of A. hydrophila AH-3, and one without any homology to any well-characterized gene. A. salmonicida A450 mutants with alterations in the genes that were not identical to those of A. hydrophila AH-3 were constructed, and their LPS core structures were fully elucidated. At the same time, all the A. salmonicida A450 genes identical to those of A. hydrophila AH-3 were used to complement the previously obtained A. hydrophila AH-3 mutants for each of these genes. Combining the gene sequence and complementation test data with the structural data and phenotypic characterization of the mutant LPSs enabled a presumptive assignment of all LPS core biosynthesis gene functions in A. salmonicida A450. Furthermore, hybridization studies with internal probes for the A. salmonicida-specific genes using different A. salmonicida strains (strains of different subspecies or atypical strains) showed a unique or prevalent LPS core type, which is the one fully characterized for A. salmonicida A450.
Aeromonas salmonicida subsp. salmonicida is an important pathogen of fish, producing the systemic disease furunculosis (30). Since the annual worldwide losses of farmed fish due to diseases involve millions of dollars, this pathogen has been subjected to considerable investigation. One of the principal virulence factors of this pathogen is an S-layer (named the A-layer) that consists principally of a 2-dimensional crystalline tetragonal protein (A-protein, with a molecular mass of 49 kDa) array (15), which is tethered to the cell by lipopolysaccharide (LPS) (5). Labeling studies have shown that the A-layer appears to cover most of the surface of virulent A. salmonicida (10), although some LPS may also be exposed (23). This structure has been shown to protect this bacterium from killing by serum in a manner that somehow requires both LPS and the A-layer (22). Although the A-layer is not completely necessary for the bacterium to be resistant to serum killing, it is an important barrier against opsonophagocytosis (19). In gram-negative bacteria, the LPS is one of the major structural and immunodominant molecules of the outer membrane. It consists of three domains: lipid A, core oligosaccharide, and O-specific polysaccharide, or O-antigen. Some studies have characterized the structures of the O-antigen polysaccharide and the core oligosaccharide region of LPS from A. salmonicida strain SJ-15 (32, 33), and a recent study described the structural elucidation of the core oligosaccharide region of the A. salmonicida subsp. salmonicida LPS from strains A449 and 80204-1 (34).
Genes involved in LPS core biosynthesis in several members of the Enterobacteriaceae, such as Escherichia coli, Salmonella enterica, and Klebsiella pneumoniae, are usually found clustered in a region of the chromosome, the wa gene cluster (13, 25). On the other hand, a careful analysis of several fully sequenced genomes of nonenteric bacteria suggested that the genes for LPS core biosynthesis may not be clustered and may be distributed among several regions. Recently, we reported the identification and characterization of three genomic regions in Aeromonas hydrophila AH-3 (serotype O34) involved in LPS core biosynthesis (16). These data, together with the elucidation of the structure of the LPS core in mutants for each gene from the three gene clusters, enabled the assignment of all LPS core biosynthesis gene functions.
Comparison of the LPS core structures of A. salmonicida subsp. salmonicida A449 (34) and A. hydrophila AH-3 (16, 17) shows great similarities in the inner LPS core and part of the outer LPS core but some differences in the distal part of the outer LPS core (residues l-glycero-d-manno-heptose [ld-Hep], d-Gal, and d-GalNAc) (Fig. 1). For this reason, we characterized the genes and their functions in A. salmonicida subsp. salmonicida A450 so as to proceed with the complete assignment of all LPS core biosynthesis gene functions. Furthermore, the genome of strain A449 has recently been published (26).
FIG. 1.
Chemical structures of the LPS cores of A. salmonicida A449 (A) (33) and A. hydrophila strain AH-3 (B) (16, 17).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Aeromonas strains were routinely grown on tryptic soy broth (TSB) or tryptic soy agar (TSA) at 20°C (A. salmonicida) and 30°C (A. hydrophila). E. coli strains were grown in Luria-Bertani (LB) Miller broth and on the same medium with agar at 37°C. Kanamycin (50 μg ml−1), ampicillin (100 μg ml−1), tetracycline (20 μg ml−1), rifampin (rifampicin) (100 μg ml−1), nalidixic acid (20 μg ml−1), chloramphenicol (25 μg ml−1), or gentamicin (20 μg ml−1) was added to the different media.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−endA hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA96 φ80lacZM15 | 12 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F−proAB lacIqZΔM15 Tn10) | Stratagene |
| S17-1 | hsdR pro recA; RP4-2 in chromosome; Km::Tn7 (Tc::Mu) | 8 |
| A. hydrophila | ||
| AH-3 | O34; wild type | 18 |
| AH-405 | AH-3; spontaneous Rifr | 18 |
| AH-3005 | AH-3 ΔWahD; Kmr | 16 |
| AH-3006 | AH-3 ΔWaaE; Kmr | 16 |
| AH-3007 | AH-3 ΔWaaC; Kmr | 16 |
| AH-3Δ2.1 | AH-3 ΔWahA | 16 |
| AH-3Δ3.1 | AH-3 ΔWaaL | 16 |
| AH-3Δ4.1 | AH-3 ΔWahB | 16 |
| AH-3Δ5.1 | AH-3 ΔWahC | 16 |
| AH-3Δ2.2 | AH-3 ΔWahF | 16 |
| AH-3Δ7.1 | AH-3 ΔWahE; Kmr | 16 |
| AH-3Δ4.2 | AH-3 ΔWaaF; Kmr | 16 |
| A. salmonicida | ||
| A450 | Wild type | 19 |
| A450 Nalr | Spontaneously nalidixic acid-resistant A450 | 20 |
| A450ΔWaaL | A450 WaaL mutant with pDM4; Kmr | This study |
| A450ΔORF2 | A450 ORF2 mutant with pDM4; Kmr | This study |
| A450ΔWahBlike | A450 WahBlike mutant with pDM4; Kmr | This study |
| A450ΔWahDlike | A450 WahDlike insertion mutant with pFS100; Kmr | This study |
| Plasmids | ||
| pRK2073 | Helper plasmid; Spcr | 8 |
| pGEM-T | PCR cloning vector; Apr | Promega |
| pGEMT-WaaA | pGEM-T with A450 WaaA; Apr | This study |
| pGEMT-WaaAKdkA | pGEM-T with A450 WaaA and KdkA; Apr | This study |
| pDM4 | pir dependent with sacAB genes; oriR6K; Cmr | 21 |
| pDM4ΔORF2 | pDM4 with A450:ORF2:Km; Cmr Kmr | This study |
| pDM4ΔWaaL | pDM4 with A450:WaaL:Km; Cmr Kmr | This study |
| pDM4ΔWahBlike | pDM4 with A450:WahBlike:Km; Cmr Kmr | This study |
| pFS100 | pGP704 suicide plasmid, pir-dependent; Kmr | 27 |
| pFS-WahDlike | pFS100 with an internal fragment of WahBlike; Kmr | This study |
| pBAD33 | Arabinose-inducible expression vector; Cmr | ATCC |
| pBAD33-Gm | pBAD33 vector with Gmr | This study |
| pBAD33-Gm-ORF | pBAD33-Gm with the corresponding A. salmonicida ORF; Gmr | This study |
General DNA methods.
General DNA manipulations were carried out essentially as described previously (28). DNA restriction endonucleases (Promega), T4 DNA ligase (Invitrogen), E. coli DNA polymerase (Klenow fragment; Promega), and alkaline phosphatase (GE Healthcare) were used as recommended by the suppliers.
DNA sequencing and computer analysis of sequence data.
Double-stranded DNA sequencing was performed by using the dideoxy chain termination method (29) with the ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer). Oligonucleotides used for genomic DNA amplification experiments and for DNA sequencing were purchased from Pharmacia LKB Biotechnology. The DNA sequence was translated in all six frames, and all open reading frames (ORFs) were inspected. Deduced amino acid sequences were compared with those of DNA translated in all six frames from nonredundant GenBank and EMBL databases by using the BLAST (2, 4) network service at the National Center for Biotechnology Information and the European Biotechnology Information, respectively. Clustal W was used for multiple sequence alignments.
Dot blot hybridizations.
For dot blot hybridizations, DNA was denatured by boiling for 5 min, chilled on ice for another 5 min, and spotted onto a Hybond N1 nylon membrane (Amersham). The DNA was fixed onto the membrane, which had been prewetted in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), by UV irradiation; then it was prehybridized in a solution of 5× SSC, 0.1% N-lauroyl sarcosine, 0.02% sodium dodecyl sulfate (SDS), 5% blocking reagent (Roche), and 50% formamide for 2 h at 42°C. A digoxigenin-labeled probe (20 ng/ml) was added to the prehybridization solution, and the blots were hybridized for 18 h at 42°C. Alkaline phosphatase detection was finally carried out using the enhanced chemiluminescence detection system (Amersham) according to the manufacturer's instructions.
Mutant construction.
To produce nonpolar mutants A450ΔORF2, A450ΔWaaL, and A450ΔWahBlike, the corresponding genes were amplified by PCR using A450 chromosomal DNA with the primers shown in Table SA in the supplemental material, ligated into vector pGEM-T Easy (Promega), and transformed into E. coli XL1-Blue. The Tn5-derived kanamycin resistance cartridge (nptll) from pUC4-KIXX was inserted into each of these genes. The cartridge contains an outward-reading promoter that ensures the expression of downstream genes when it is inserted in the correct orientation (7). The SmaI-digested cassette was inserted into a restriction site internal to each gene: Klenow fragment-treated BglII, Klenow fragment-treated NarI, and BalI sites, respectively. Constructs containing the mutated genes were recovered by BamHI digestion, ligated into a BglII-digested and phosphatase-treated pDM4 suicide vector, which contains the counterselectable marker sacB (21), transformed into E. coli MC1061 (λpir), and selected on chloramphenicol and kanamycin plates. Triparental mating with the mobilizing strain HB101/pRK2073 was used to transfer the plasmids containing the engineered mutated genes (pDM4ΔORF2, pDM4ΔWaaL, and pDM4ΔWahBlike) into an A. salmonicida A450 nalidixic acid-resistant strain. The A. salmonicida A450 nalidixic acid-resistant strain was previously obtained as a spontaneous mutant. Transconjugants were selected on plates containing chloramphenicol, kanamycin, and nalidixic acid at 20°C. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA by homologous recombination. To complete the allelic exchange, the integrated suicide plasmid was forced to excise from the chromosome by growth on agar plates containing 10% sucrose. The mutants were confirmed by sequencing of the whole constructs in amplified PCR products.
Mutant A450ΔWahDlike was produced as a defined insertion mutant by using the suicide plasmid pFS100 (27). Briefly, an internal fragment of the selected gene was amplified by PCR using A450 chromosomal DNA with the primers shown in Table SA in the supplemental material, ligated into the pGEM-T Easy vector (Promega), and transformed into E. coli XL1-Blue. The DNA insert was recovered by EcoRI digestion and ligated into EcoRI-digested, phosphatase-treated pFS100 (28). The ligation product was transformed into E. coli MC1061 (λpir) and selected for kanamycin resistance. Triparental mating with the mobilizing strain HB101/pRK2073 was used to transfer the recombinant plasmid into the A. salmonicida A450 nalidixic acid-resistant strain, and transconjugants were selected on plates containing nalidixic acid and kanamycin.
Construction of plasmids for gene overexpression and mutant complementation studies.
For gene overexpression studies and for complementation of the constructed A450 core mutants and some A. hydrophila AH-3 core mutants, the corresponding genes from A. salmonicida strain A450 were PCR amplified from chromosomal DNA by using specific primer pairs designed with the XbaI (for the forward primer) and HindIII (for the reverse primer) restriction sites, as shown in Table SB in the supplemental material. The DNA fragment was digested with XbaI and HindIII and was ligated to the XbaI and HindIII sites in the previously digested pBAD33-Gm vector. To generate plasmid pBAD33-Gm, the gentamicin resistance gene (aacC1) of the pUCGmlox vector (24) was used. pUCGmlox was digested with SmaI, and the fragment containing the lox-flanked aacC1 gene was purified and cloned into the PvuII and ScaI sites in previously digested pBAD33, replacing its chloramphenicol resistance gene (cat). The plasmid pBAD33-Gm constructs were transformed into E. coli LMG194 by electroporation, plated onto gentamicin LB agar plates, and incubated at 30°C. Plasmid constructs were then transferred into the different mutants by triparental mating using the mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing gentamicin, kanamycin, and nalidixic acid (for A450 mutants) or gentamicin and rifampin (for AH-3 mutants) and were confirmed by PCR. Each gene was expressed from the arabinose-inducible and glucose-repressible pBAD33-Gm promoter. Repression from the PBAD promoter was achieved by growth in a medium containing 0.2% (wt/vol) d-glucose, and induction was achieved by the addition of l-arabinose to a final concentration of 0.2% (wt/vol). The cultures were grown for 18 h at 20°C in TSB medium supplemented with gentamicin and kanamycin (for A450 mutants) or at 30°C with gentamicin alone (for AH-3 mutants) and 0.2% glucose. These cultures were diluted 1:100 in fresh medium (without glucose) and grown until they reached an A600 of about 0.2. l-Arabinose was then added, and the cultures were grown for another 2 h. Repressed controls were maintained in glucose-containing medium.
To produce the pGEMT-WaaAKdkA plasmid (carrying A. salmonicida waaA and kdkA together), the A. salmonicida A450 kdkA gene and its putative promoter site were amplified by PCR using chromosomal DNA with primers 2.3-F1 (5′-GAGCCGCAGTATGCAGATA-3′) and 2.3-R1 (5′-CCTCGAAACCGAATCTGA-3′). The amplified DNA fragment was ligated into the pGEM-T vector (Promega) to produce the pGEMT-KdkA plasmid, which was transformed into E. coli XL1-Blue. Transformants were selected on LB plates containing ampicillin. The A. salmonicida A450 waaA gene was PCR amplified from chromosomal DNA with primers 1.2-F1 (5′-ACGCGTCGACCGAGTCGCCAGATCAACC-3′) and 1.2-R1 (5′-ACGCGTCGACGATAATGCGTTGACCGATG-3′). The PCR product was digested with SalI (underlined) and ligated to the SalI-digested, phosphatase-treated pGEMT-KdkA plasmid to generate plasmid pGEMT-WaaAKdkA, which was transformed into E. coli XL1-Blue.
LPS isolation and SDS-PAGE.
Cultures for analysis of LPS were grown in TSB at 20°C. LPS was purified by the method of Galanos et al. (11), resulting in a 2.1% yield. For screening purposes, LPS was obtained after proteinase K digestion of whole cells (9). LPS samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) or SDS-Tricine-PAGE and were visualized by silver staining as previously described (9, 14).
Large-scale isolation and mild-acid degradation of LPS.
Dry bacterial cells of each mutant in 25 mM Tris-HCl buffer containing 2 mM CaCl2, pH 7.63 (10 ml g−1), were treated at 37°C with RNase and DNase (24 h; 1 mg g−1 each) and then with proteinase K (36 h; 1 mg g−1). The suspension was dialyzed and lyophilized, and the LPS was extracted by the phenol-water procedure (35).
A portion of the LPS (∼50 mg) from each strain was heated with aqueous 2% acetic acid (6 ml) at 100°C for 45 min. The precipitate was removed by centrifugation (13,000 × g, 20 min), and the supernatant was fractionated on a column (56 by 2.6 cm) of Sephadex G-50 in 0.05 M pyridinium acetate buffer (pH 4.5) with monitoring using a differential refractometer. An oligosaccharide fraction was obtained at a yield of 9 to 20% depending on the strain.
Methylation analysis.
A sample of each oligosaccharide (0.5 mg) was dissolved in 1 ml dimethyl sulfoxide. An excess of powdered NaOH was added, and the reaction glass was flushed with dry N2 and sealed. After stirring at 20°C for 1 h, 0.5 ml of cold CH3I was added, and the mixture was stirred at 20°C for 1 h. Then water was added, the methylated product was extracted with CHCl3, and the extract was washed with water, which was evaporated with a stream of dry nitrogen. The methylated oligosaccharide was hydrolyzed with 2 M CF3CO2H (120°C, 2 h), and acid was removed with a stream of nitrogen. The methylated monosaccharides were conventionally reduced with NaBH4, acetylated with acetic anhydride in pyridine, and analyzed by gas-liquid chromatography (GLC) on a Hewlett-Packard 5880 chromatograph and by GLC-mass spectrometry on a Hewlett-Packard HP 5989A instrument using an HP-5ms capillary column and a temperature gradient of 150°C (3 min) to 320°C at 5°C min−1.
Mass spectrometry.
Positive-ion reflectron time-of-flight (matrix-assisted laser desorption ionization—time-of-flight [MALDI-TOF]) mass spectra were acquired on a Voyager DE-PR instrument (Applied Biosystems) equipped with a delayed-extraction ion source. The ion acceleration voltage was 20 kV, the grid voltage was 14 kV, the mirror voltage ratio was 1.12, and the delay time was 100 ns. Samples were irradiated at a frequency of 5 Hz by 337-nm photons from a pulsed nitrogen laser. Post-source decay was performed using an acceleration voltage of 20 kV. The reflectron voltage was decreased in 10 successive 25% steps. Mass calibration was obtained with a maltooligosaccharide mixture from corn syrup (Sigma). A solution of 2,5-dihydroxybenzoic acid in 20% CH3CN in water at a concentration of 25 mg/ml was used as the MALDI matrix. One microliter of matrix solution and 1 μl of the sample were premixed and then deposited on the target. The droplet was allowed to dry at the ambient temperature. Spectra were calibrated and processed under computer control using Applied Biosystems Data Explorer software.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the three A. salmonicida A450 chromosomal regions containing the LPS core biosynthetic genes described here have been assigned GenBank accession numbers FJ238464, FJ238465, and FJ238466, respectively.
RESULTS
Sequencing of three genomic regions of A. salmonicida A450 coding for LPS core genes.
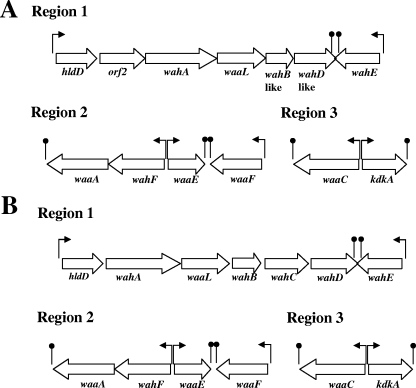
We generated three internal DNA probes from A. hydrophila AH-3 waaC (probe 1, region 3), waaA (probe 2, region 2), and wahA (probe 3, region 1) (16). The reason for choosing these DNA probes is that A. salmonicida A449 showed the same structural motifs encoded by these genes in the LPS core as A. hydrophila AH-3 (16, 34) (Fig. 1). All of these DNA probes showed high hybridization with the A. salmonicida A450 genome. For this reason, we used several of the previously described oligonucleotides (16) obtained when we sequenced the three regions of A. hydrophila AH-3 containing LPS core genes in PCRs with A. salmonicida A450 genomic DNA. Furthermore, in order to complete the DNA sequence of the three regions containing LPS core genes in A. salmonicida A450, the nucleotide sequence was spanned, either downstream or upstream of the region, by genome-walking PCR (36) when needed. Analysis of the sequenced region in A. salmonicida A450 named region 1 showed seven genes involved in LPS core biosynthesis (Fig. 2), and the corresponding analysis of the proteins encoded by these ORFs, with their homology, is presented in Table 2. All the A. salmonicida A450 ORFs showed >99% similarity with their corresponding ORFs from the A. salmonicida A449 genome (26). Three of the A. salmonicida A450 genes (hldD [orf1], wahA [orf3], and wahE [orf7]) are clearly the same genes as those in A. hydrophila AH-3, and three showed some similarity but probably are not the same genes (waaL [orf4], wahB-like [orf5], and wahD-like [orf6]). Finally, A. salmonicida A450 orf2 showed no similarity to A. hydrophila AH-3 LPS core genes or other well-characterized genes. Analysis of the sequenced regions named regions 2 and 3 showed the same genes involved in LPS core biosynthesis in both regions as in A. hydrophila AH-3 (Fig. 2), and the corresponding analysis of the proteins encoded by these ORFs in both regions, with their homology, is presented in Table 2. With these results obtained, we decided to study the complementation of the A. hydrophila AH-3 LPS core mutants previously obtained (16) with the sequenced genes of A. salmonicida A450.
FIG. 2.
Genetic organization of the three A. salmonicida A450 (A) and A. hydrophila AH-3 (B) chromosomal regions containing genes for LPS core biosynthesis. The direction of transcription and transcription stops (lollipops) are indicated.
TABLE 2.
Characteristics of the three regions of A. salmonicida A450 containing genes for the biosynthesis of LPS core
| ORF no. | Protein | Nucleotide position | Protein size (aa) | Homologous protein, organism (accession no.) | % Identity, similaritya |
|---|---|---|---|---|---|
| Region 1 | |||||
| 1 | HldD | 210-1169 | 319 | HldD, Aeromonas hydrophila AH-3 (ACA14473) | 98, 99 |
| ADP-l-glycero-d-manno-heptose 6-epimerase, Aeromonas hydrophila ATCC 7966 (YP_858654) | 97, 99 | ||||
| 2 | ORF2 | 1255-2376 | 373 | Glycosyltransferase, Clostridium perfringens (YP_697915) | 34, 55 |
| 3 | WahA | 2467-4227 | 586 | WahA, Aeromonas hydrophila AH-3 (ACA14474) | 93, 96 |
| Glycosyltransferase, Aeromonas hydrophila ATCC 7966 (YP_858653) | 91, 94 | ||||
| 4 | WaaL | 4234-5445 | 403 | WaaL, Aeromonas hydrophila AH-3 (ACA14475) | 73, 85 |
| O-antigen polymerase, Aeromonas hydrophila ATCC 7966 (YP_858652) | 72, 84 | ||||
| 5 | WahBlike | 5442-6125 | 227 | WahB, Aeromonas hydrophila AH-3 (ACA14476) | 69, 82 |
| Putative LPS biosynthesis protein, Aeromonas hydrophila (AAR27965) | 71, 84 | ||||
| 6 | WahDlike | 6122-7174 | 350 | WahD, Aeromonas hydrophila AH-3 (ACA14478) | 34, 56 |
| Putative d-glycero-d-manno-heptosyltransferase, Aeromonas hydrophila (AAR27967) | 35, 54 | ||||
| 7 | WahE | 8213-7149 | 354 | WaaE, Aeromonas hydrophila AH-3 (ACA14479) | 87, 91 |
| Putative ADP-heptose:LPS, Aeromonas hydrophila (AAR27968) | 86, 91 | ||||
| Region 2 | |||||
| 1 | WaaA | 1364-99 | 421 | WaaA, Aeromonas hydrophila AH-3 (ACA14480) | 91, 94 |
| 3-Deoxy-d-manno-octulosonic acid transferase, Aeromonas hydrophila ATCC 7966 (YP_854703) | 89, 93 | ||||
| 2 | WahF | 2481-1366 | 371 | WahF, Aeromonas hydrophila AH-3 (ACA14481) | 95, 97 |
| Lipopolysaccharide heptosyltransferase II, Aeromonas hydrophila ATCC 7966 (YP_854702) | 92, 95 | ||||
| 3 | WaaE | 2723-3511 | 262 | WaaE, Aeromonas hydrophila AH-3 (ACA14482) | 96, 97 |
| Glycosyltransferase, Aeromonas hydrophila ATCC 7966 (YP_854701) | 93, 96 | ||||
| 4 | WaaF | 4616-3573 | 347 | WaaF, Aeromonas hydrophila AH-3 (ACA14483) | 98, 99 |
| Heptosyltransferase, Aeromonas hydrophila ATCC 7966 (YP_854700) | 95, 97 | ||||
| Region 3 | |||||
| 1 | WaaC | 1372-317 | 351 | WaaC, Aeromonas hydrophila AH-3(ACA14486) | 96, 96 |
| Lipopolysaccharide heptosyltransferase, Aeromonas hydrophila ATCC 7966 (YP_854567) | 90, 94 | ||||
| 2 | KdkA | 1558-2271 | 237 | KdkA, Aeromonas hydrophila AH-3(ACA14485) | 97, 98 |
| 3-Deoxy-d-manno-octulosonic-acid kinase, Aeromonas hydrophila ATCC 7966 (YP_854568) | 94, 97 |
All the A. salmonicida A450 ORFs showed similarities greater than 99% with the corresponding ORFs from the A. salmonicida A449 genome (26).
Complementation studies with A. salmonicida A450 LPS core biosynthesis genes.
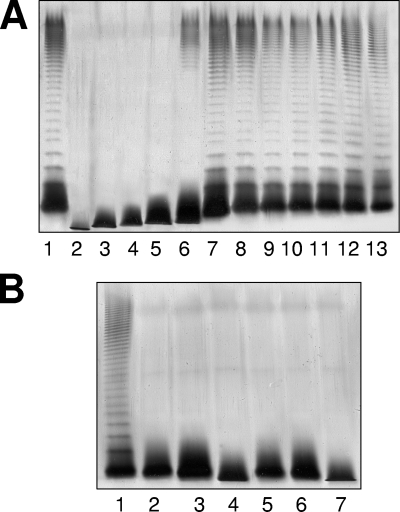
Plasmids pBAD33-Gm-WaaC, pBAD33-Gm-WaaF, pBAD33-Gm-WaaE, pBAD33-Gm-WahF, pBAD33-Gm-WahE, and pBAD33-Gm-WahA, containing the A. salmonicida A450 LPS core biosynthesis genes waaC, waaF, waaE, wahF, wahE, and wahA, respectively, were able to fully restore the wild-type LPS profile in gels (Fig. 3A) when introduced into the corresponding A. hydrophila AH-3 LPS core mutants (AH-3007, AH-3Δ4.2, AH-3006, AH-3Δ2.2, AH-3Δ7.1, and AH-3Δ2.1, respectively). However, this was not the case for plasmids pBAD33-Gm-WahBlike and pBAD33-Gm-WahDlike, containing the A. salmonicida A450 LPS core biosynthesis genes wahB-like (orf5) and wahD-like (orf6), respectively, when these genes were introduced into A. hydrophila AH-3 LPS core mutants AH-3Δ4.1 and AH-3005, as judged by the LPS profile in gels (Fig. 3B). The putative A. salmonicida A450 waaL (orf4) gene was also unable to complement the A. hydrophila AH-3Δ3.1 (WaaL) mutant (Fig. 3B).
FIG. 3.
LPS samples were extracted and analyzed by SDS-PAGE (12%) according to the method of Darveau and Hancock (9). (A) SDS-PAGE analysis of LPS from A. hydrophila AH-3 (wild type) (lane 1) and mutants AH-3007, AH-3Δ4.2, AH-3006, AH-3Δ2.2, AH-3Δ2.1, and AH-3Δ7.1 (ΔwaaC, ΔwaaF, ΔwaaE, ΔwahF, ΔwahA, and ΔwahE, respectively [16]) (lanes 2 to 7, respectively). These mutants were independently complemented with plasmid pBAD33-Gm-WaaC, pBAD33-Gm-WaaF, pBAD33-Gm-WaaE, pBAD33-Gm-WahF, pBAD33-Gm-WahA, or pBAD33-Gm-WahE, respectively, carrying the corresponding single gene from A. salmonicida A450 (lanes 8 to 13, respectively). (B) SDS-PAGE analysis of LPS from A. hydrophila AH-3 (wild-type) (lane 1); from mutants AH-3Δ3.1, AH-3Δ4.1, and AH-3005 (ΔwaaL, ΔwahB, and ΔwahD, respectively [16]) (lanes 2 to 4, respectively); and from mutants AH-3Δ3.1, AH-3Δ4.1, and AH-3005 independently carrying plasmid pBAD33-Gm-WaaL, pBAD33-Gm-WahBlike, or pBAD33-Gm-WahDlike, respectively, with the corresponding single gene from A. salmonicida A450 (lanes 5 to 7, respectively). The strains with pBAD33-Gm plasmids were grown under induced conditions.
E. coli strain CJB26 harbors a kanamycin determinant inserted into the waaA gene and carries its wild-type waaA gene in a temperature-sensitive plasmid (pJSC2), leading to a growth temperature-sensitive phenotype (6). Plasmid pGEMT-WaaA (carrying A. salmonicida waaA alone) was unable to complement E. coli CJB26, whereas plasmid pGEMT-WaaAKdkA (carrying A. salmonicida waaA and kdkA together) was able to abolish the E. coli CJB26 growth temperature-sensitive phenotype. This experiment strongly suggests that these two genes encode 3-deoxy-d-manno-oct-2-ulosonic (ketodeoxyoctonic) acid (Kdo) transferase and Kdo kinase, respectively, of the A. salmonicida LPS inner core biosynthetic pathway. Both of these A. salmonicida A450 genes are extremely similar to those of A. hydrophila AH-3 (16).
Mutant isolation and phenotypic characterization.
Due to the results obtained, we isolated four A. salmonicida A450 mutants. The mutants had alterations in ORFs 2 (no clear homology), 4 (WaaL), 5 (WahB-like), and 6 (WahD-like), and all of them were devoid of the O-antigen LPS by SDS-PAGE except for the ORF2 mutant (A450ΔORF2) (Fig. 4). They also showed faster migration in the LPS core on a Tricine-SDS-PAGE gel, except for the WaaL mutant (A450ΔWaaL), than the corresponding wild-type LPS core (Fig. 4).
FIG. 4.
SDS-Tricine-PAGE gels of the wild-type nalidixic acid-resistant A450 strain (lane 1); LPS core mutants A450ΔWaaL, A450ΔWahBlike, A450ΔORF2, and A450ΔWahDlike (lanes 2 to 5, respectively); and these LPS core mutants complemented with pBAD33-Gm-WaaL, pBAD33-Gm-WahBlike, pBAD33-Gm-ORF2, or pBAD33-Gm-WahDlike, respectively (lanes 6 to 9, respectively). The strains with pBAD33-Gm plasmids were grown under induced conditions.
Complementation with the corresponding A. salmonicida A450 wild-type gene (orf2, waaL, wahB-like, or wahD-like) expressed in the pBAD33-Gm vector plasmid restored the complete LPS from the wild-type strain (either by the presence of the O antigen or by the relative electrophoretic moiety of the LPS core) in the respective mutant (A450ΔORF2, A450ΔWaaL, A450ΔWahBlike, or A450ΔWahDlike), as revealed by SDS-PAGE (Fig. 4). We used vector plasmid pBAD33-Gm to complement A. salmonicida A450 mutants due to the intrinsically low chloramphenicol resistance of strain A450. No such complementation was achieved with the plasmid vector alone (data not shown).
Furthermore, by using the same oligonucleotides described in Table SB in the supplemental material for A450, we amplified the homologous genes waaL, orf2, wahB-like, and wahD-like from A449 genomic DNA and cloned them into pBAD33-Gm. These A. salmonicida A449 genes were able to fully complement their corresponding A. salmonicida A450 mutants (data not shown). No such complementation was achieved with the plasmid vector alone.
Elucidation of the structures of the mutant LPSs.
LPSs were isolated by phenol-water extraction; LPS samples were degraded by mild-acid hydrolysis; and the released core oligosaccharides were isolated by gel permeation chromatography on a Sephadex G-50 column. Mass spectrometry studies and methylation analysis, when necessary, were performed. Methylation studies enabled not only determination of the linkage positions in most sugar residues but also differentiation between dd-Hep and ld-Hep residues by their different retention times in GLC.
(i) Mutant A450ΔWaaL.
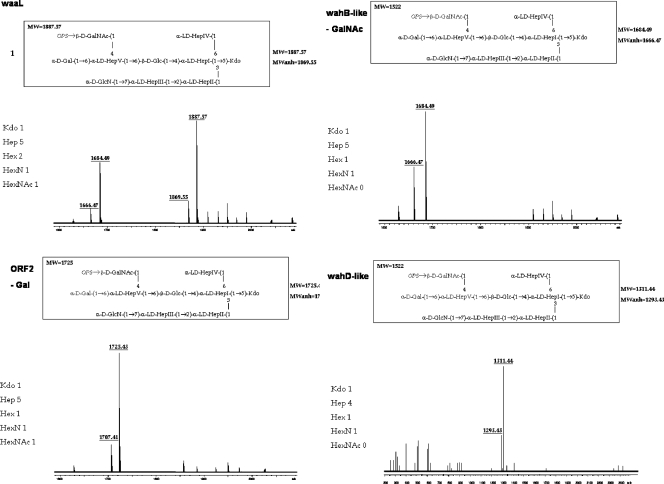
Composition analysis of the oligosaccharide from mutant A450ΔWaaL showed the presence of Glc, Gal, GlcN, GalNAc, ld-Hep, and Kdo. The mass spectrum from this oligosaccharide sample showed a major molecular ion peak at m/z 1,888.60 (Fig. 5), corresponding to the full core (calculated molecular mass, 1,887.60 atomic mass units). This molecular mass is essentially similar to those reported for both wild-type A. salmonicida subsp. salmonicida strains A449 and 80204-1 and an in vivo-derived rough phenotype of these strains (34). As in other reported cases (16, 17, 34), structural heterogeneity was observed, which was associated with the existence of Kdo in both normal and anhydro forms. Methylation analysis showed that the core oligosaccharide from this mutant was characterized by containing similar molar ratios of terminal Gal, GlcN, GalNAc, and ld-Hep. In addition, 6-substituted Glc, 2-substituted Hep, 7-substituted Hep, 4,6-bisubstituted Hep, and 3,4,6-trisubstituted Hep were found. The complete presumptive structure of the LPS from A450ΔWaaL mutant is shown in Fig. SA in the supplemental material.
FIG. 5.
Positive-ion MALDI-TOF spectra of acid-released core LPSs from A. salmonicida A450 mutants A450ΔWaaL (WaaL), A450ΔORF2 (ΔORF2), A450ΔWahBlike (WahB-like), and A450ΔWahDlike (WahD-like). Schematic structures of the most representative compounds are shown above the spectra.
(ii) Mutant A450ΔWahBlike.
The oligosaccharide from mutant A450ΔWahBlike showed a major molecular ion peak at m/z 1,685.82 (Fig. 5), in agreement with a structure lacking a GalNAc residue. Methylation analysis showed the presence of terminal Gal, GlcN, Hep, and Kdo residues. In addition, 6-substituted Hep was found instead of 4,6-bisubstituted Hep.
(iii) Mutant A450ΔORF2.
Due to the presence of O-antigen LPS in mutant A450ΔORF2 (Fig. 4), we isolated and studied the core oligosaccharide fraction. This core fraction was found to lack Gal (molecular ion peak at m/z 1,726.70) (Fig. 5). Methylation analysis showed the presence of similar molar ratios of terminal GlcN and GalNAc, and the presence of 4-substituted Hep instead of 4,6-bisubstituted Hep.
(iv) Mutant A450ΔWahDlike.
The mass spectrum of the LPS from mutant A450ΔWahDlike (Fig. 5) showed the lack of the outer core trisaccharide fragment consisting of Gal, ld-HepV, and GalNAc, whereas the rest of the core, with the molecular ion peak at m/z 1,331.54, was unaffected. Methylation analysis confirmed the conservation of the terminal ld-HepIV, the absence of Gal and GalNAc, and the appearance of the terminal Glc.
The complete LPS structures of the mutants are shown in Fig. SA in the supplemental material.
Gene distribution among various typical and atypical strains of A. salmonicida.
We generated internal DNA probes of the A. salmonicida A450 genes orf2, wahB-like, and wahD-like, obtained by PCR amplification of the A. salmonicida A450 genome with primers derived from the DNA sequence. The oligonucleotides used were 2.1-F3 (5′-TTATTCCCACCTGGA TGGTC-3′) and 2.1-R3 (5′-CCATCAGCTAACGCTTGTG-3′) for orf2 amplification, 5.1-F3 (5′-TTCTCGAAGTCAA GCTCGG-3′) and 5.1-R3 (5′-CTACAACTTGGGGTGCGAT-3′) for wahB-like, and 6.1-F and 6.1-R for wahD-like (see Table SA in the supplemental material), giving DNA bands of 658 bp, 416 bp, and 545 bp, respectively. The three probes hybridized completely with purified DNA from several A. salmonicida strains belonging to different subspecies: A. salmonicida subsp. salmonicida (n = 10), Aeromonas salmonicida subsp. achromogenes (n = 4), Aeromonas salmonicida subsp. pectinolytica (n = 4), Aeromonas salmonicida subsp. masoucida (n = 4), and Aeromonas salmonicida subsp. smithia (n = 2). Also, these DNA probes hybridized fully with the purified DNA from five atypical A. salmonicida strains (AS19, AS30, AS42, AS203, and AS224 [3)]).
DISCUSSION
A. salmonicida subsp. salmonicida strain A450 showed three different genomic regions containing LPS core genes, a situation similar to that reported for A. hydrophila strain AH-3 (16). Region 1 contained seven genes flanked by genes encoding a putative transcriptional regulator from the TetR family and a hypothetical protein; region 2 contained four genes flanked by genes encoding an integral membrane protein of unknown function and CoaD; and region 3 contained two genes flanked by genes encoding a guanylate kinase and a protein with a GGDEF domain. The three regions and the genes contained are in complete agreement with the recently obtained DNA sequence of the A. salmonicida strain A449 genome (26). Region 1 (ASA_0093 to ASA_0099), region 2 (ASA_4219 to ASA_4222), and region 3 (ASA_0037 and ASA_0038) are flanked by the same putative genes and contain identical genes in the genomes of A. salmonicida A449 and strain A450. A. salmonicida A450 mutants were fully complemented by the corresponding A. salmonicida A449 genes, and >99% homology was observed for all the wa genes (the three genomic regions) between the two strains. Furthermore, the positions and the genes for regions 2 and 3 are in agreement with those of A. hydrophila AH-3 and ATCC 7966T (16, 31). The presence of the structural motifs in the LPS cores of A. salmonicida A450 and A449 and A. hydrophila AH-3 (16) encoded by these genes (waaC, kdkA, waaA, waaF, waaE, and wahF), as well as the complete complementation of the A. hydrophila AH-3 mutants by the corresponding A. salmonicida A450 genes, led us to assign the latter the same functions previously described (16).
The positions for the A. salmonicida A450 region 1 genes are the same as those in the other Aeromonas strains previously studied (16, 31). However, the genes of this region are identical to those of A. salmonicida A449, but four of them are different from the A. hydrophila AH-3 and ATCC7966T genes from this region. Nevertheless, three of these genes (hldD, wahA and wahE), either by sequence homology alone (hldD) or also by complete complementation of the corresponding A. hydrophila AH-3 mutants and the presence of the structural motif in the LPS core structure of strain A450, could be directly assigned the same function performed in A. hydrophila AH-3 (16, 17, 34). One of the four different genes corresponds to waaL, because mutant A450ΔWaaL produced an LPS with the full core but was completely devoid of the O antigen. A. salmonicida A450 WaaL also showed 11 transmembrane helices between amino acid residues 12 to 31, 36 to 55, 68 to 85, 90 to 107, 112 to 134, 144 to 166, 173 to 192, 197 to 216, 222 to 239, 357 to 374, and 379 to 396, characteristic for lipid A core O-antigen ligases (1).
The mass spectra and methylation results obtained with the four A. salmonicida A450 mutants suggest that the core oligosaccharide structure is slightly different from that previously reported (34) (Fig. 1A), where the ld-HepV was proposed to be substituted at position 4 by either an α-d-Gal residue or the disaccharide α-d-Gal-(1→4)-β-d-GalNAc (Fig. 1A and 6). The presence of 4,6-bisubstituted ld-Hep instead of 4-substituted Hep and the absence of 4-substituted GalNAc strongly suggest that in strain A450 the GalNAc is a terminal residue most probably linked to the 4 position of ld-HepV (Fig. 6). This was confirmed by the appearance of 4-substituted and 6-substituted Hep residues in mutants A450ΔORF2 and A450ΔWahBlike, respectively. In addition, no 4,6-bisubstituted Hep was found in these two mutants. The absence of O antigen in mutants A450ΔWahDlike and A450ΔWahBlike and its presence in the A450ΔORF2 mutant strongly suggest that the O antigen is linked to the GalNAc residue in a manner similar to that in A. hydrophila AH-3 (16).
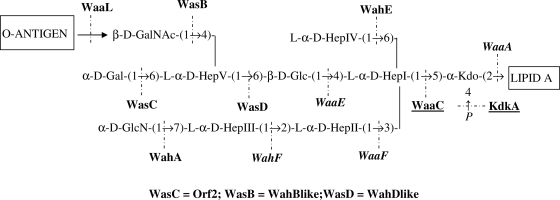
FIG. 6.
Complete presumptive assignment of the A. salmonicida A450 genes involved in LPS core biosynthesis. Proteins encoded by genes from different regions are shown in roman type (region 1), in italics (region 2), or underlined (region 3).
We also proposed to rename the three A. salmonicida A450 genes and proteins to distinguish them from the A. hydrophila AH-3 genes. To wa (for LPS core genes) we added s (for A. salmonicida) instead of h (for A. hydrophila). B, C, and D were kept as the designations for their positions in Aeromonas LPS structures. Thus, wahB-like, orf2, and wahD-like were renamed wasB, wasC, and wasD, respectively (Fig. 6). Now that the functions of all genes involved with the A. salmonicida A450 LPS core biosynthetic pathway have been assigned and some corresponding single-gene mutants have been characterized, experimental work on the role of the LPS core in the virulence of this pathogenic bacterium can proceed. Furthermore, the hybridization results obtained with internal probes of the A. salmonicida-specific genes indicated that all the A. salmonicida strains tested, including those of different subspecies as well as atypical strains, possess these genes. This fact strongly suggested that A. salmonicida strains showed a unique or highly predominant LPS core type, which is the one characterized for A. salmonicida A450 and shown in Fig. 6.
Supplementary Material
Acknowledgments
This work was supported by Plan Nacional de I + D and FIS grants (Ministerio de Educación, Ciencia y Deporte and Ministerio de Sanidad, Spain) and from the Generalitat de Catalunya (Centre de Referència en Biotecnologia). N.J. and A.L. hold predoctoral fellowships from the Generalitat de Catalunya and the Ministerio de Educación, Ciencia y Deporte, respectively.
We also thank Maite Polo for technical assistance and the Servicios Científico-Técnicos of the University of Barcelona.
Footnotes
Published ahead of print on 16 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abeyrathne, P. D., C. Daniels, K. K. H. Poon, M. J. Matewish, and J. S. Lam. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 1873002-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin, B., D. A. Austin, I. Dalsgaard, B. K. Gudmundsdöttir, S. Høie, J. M. Thornton, J. L. Larsen, B. O'Hici, and R. Colwell. 1998. Characterization of atypical Aeromonas salmonicida by different methods. Syst. Appl. Microbiol. 2150-64. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belland, R. J., and T. J. Trust. 1985. Synthesis, export, and assembly of Aeromonas salmonicida A-layer analyzed by transposon mutagenesis. J. Bacteriol. 163877-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belunis, C. J., T. Clementz, S. M. Carty, and C. R. Raetz. 1995. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J. Biol. Chem. 27027646-27652. [DOI] [PubMed] [Google Scholar]
- 7.Bott, M., M. Meyer, and P. Dimroth. 1995. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol. Microbiol. 18533-546. [DOI] [PubMed] [Google Scholar]
- 8.Canals, R., N. Jiménez, S. Vilches, M. Regué, S. Merino, and J. M. Tomás. 2006. The UDP N-acetylgalactosamine 4-epimerase gene is essential for mesophilic Aeromonas hydrophila serotype O34 virulence. Infect. Immun. 74537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darveau, R. P., and R. E. Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dooley, J. S. G., H. Engelhardt, W. Baumeister, W. W. Kay, and T. J. Trust. 1989. Three-dimensional structure of an open form of the surface layer from the fish pathogen Aeromonas salmonicida. J. Bacteriol. 171190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galanos, C., O. Lüderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9245-249. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 13.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30221-232. [DOI] [PubMed] [Google Scholar]
- 14.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishiguro, E. E., W. W. Kay, T. Ainsworth, J. B. Chamberlain, J. T. Buckley, and T. J. Trust. 1981. Loss of virulence during culture of Aeromonas salmonicida at high temperature. J. Bacteriol. 148333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiménez, N., R. Canals, A. Lacasta, A. N. Kondakova, B. Lindner, Y. A. Knirel, S. Merino, M. Regué, and J. M. Tomás. 2008. Molecular analysis of three Aeromonas hydrophila AH-3 (serotype O34) lipopolysaccharide core biosynthesis gene clusters. J. Bacteriol. 1903176-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knirel, Y. A., E. Vinogradov, N. Jimenez, S. Merino, and J. M. Tomás. 2004. Structural studies on the R-type lipopolysaccharide of Aeromonas hydrophila. Carbohydr. Res. 339787-793. [DOI] [PubMed] [Google Scholar]
- 18.Merino, S., S. Camprubí, and J. M. Tomás. 1992. Effect of growth temperature on outer-membrane components and virulence of Aeromonas hydrophila strains of serotype O:34. Infect. Immun. 604343-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merino, S., S. Albertí, and J. M. Tomás. 1994. Aeromonas salmonicida resistance to complement-mediated killing. Infect. Immun. 625483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino, S., S. Vilches, R. Canals, S. Ramirez, and J. M. Tomás. 2005. A C1q-binding 40 kDa porin from Aeromonas salmonicida: cloning, sequencing, role in serum susceptibility and fish immunoprotection. Microb. Pathog. 38227-237. [DOI] [PubMed] [Google Scholar]
- 21.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1781310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munn, C. B., E. E. Ishiguro, W. W. Kay, and T. J. Trust. 1982. Role of the surface components in serum resistance of virulent Aeromonas salmonicida. Infect. Immun. 361069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phipps, B. M., and W. W. Kay. 1988. Immunoglobulin binding by the regular surface array of Aeromonas salmonicida. J. Biol. Chem. 2639298-9303. [PubMed] [Google Scholar]
- 24.Quénée, L., D. Lamotte, and B. Polack. 2005. Combined sacB-based negative selection and cre-lox antibiotic marker recycling for efficient gene deletion in Pseudomonas aeruginosa. BioTechniques 3863-67. [DOI] [PubMed] [Google Scholar]
- 25.Regué, M., N. Climent, N. Abitiu, N. Coderch, S. Merino, L. Izquierdo, M. Altarriba, and J. M. Tomás. 2001. Genetic characterization of the Klebsiella pneumoniae waa gene cluster, involved in core lipopolysaccharide biosynthesis. J. Bacteriol. 1833564-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reith, M. E., R. K. Singh, B. Curtis, J. M. Boyd, A. Bouevitch, J. Kimball, J. Munholland, C. Murphy, D. Sarty, J. Williams, J. H. E. Nash, S. C. Johnson, and L. L. Brown. 2008. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubirés, X., F. Saigí, N. Piqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 1797581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott, M. 1968. The pathogenicity of Aeromonas salmonicida (Griffin) in sea and brackish waters. J. Gen. Microbiol. 50321-327. [DOI] [PubMed] [Google Scholar]
- 31.Seshadri, R., S. W. Joseph, A. K. Chopra, J. Sha, J. Shaw, J. Graf, D. Haft, M. Wu, Q. Ren, M. J. Rosovitz, R. Madupu, L. Tallon, M. Kim, S. Jin, H. Vuong, O. C. Stine, A. Ali, A. J. Horneman, and J. F. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J. Bacteriol. 1888272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw, D. H., Y. Z. Lee, M. J. Squires, and O. Lüderitz. 1983. Structural studies on the O-antigen of Aeromonas salmonicida. Eur. J. Biochem. 131633-638. [DOI] [PubMed] [Google Scholar]
- 33.Shaw, D. H., M. J. Hart, and O. Lüderitz. 1992. Structure of the core oligosaccharide in the lipopolysaccharide isolated from Aeromonas salmonicida ssp. salmonicida. Carbohydr. Res. 23183-91. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Z., J. Li, E. Vinogradov, and E. Altman. 2006. Structural studies of the core region of Aeromonas salmonicida subsp. salmonicida lipopolysaccharide. Carbohydr. Res. 341109-117. [DOI] [PubMed] [Google Scholar]
- 35.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharide extraction with phenol-water and further application of the procedure. Methods Carbohydr. Chem. 583-89. [Google Scholar]
- 36.Yu, H. B., Y. L. Zhang, Y. L. Lau, F. Yao, S. Vilches, S. Merino, J. M. Tomas, S. P. Howard, and K. Y. Leung. 2005. Identification and characterization of putative virulence genes and gene clusters in Aeromonas hydrophila PPD134/91. Appl. Environ. Microbiol. 714469-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.