Abstract
Peptide inhibitors of phage lambda site-specific recombination were previously isolated by screening synthetic combinatorial peptide libraries. These inhibitors cause the accumulation of complexes between the recombinase and the Holliday junction intermediate of several highly divergent tyrosine recombinases. Peptide WRWYCR and its d-amino acid derivative bind to the center of protein-free junctions and prevent their resolution either by site-specific recombinases or by junction resolvases or helicases. With lesser affinity, the peptides also bind to branched DNA molecules that mimic replication forks. The peptides are bactericidal to both gram-positive and gram-negative bacteria, presumably because they can interfere with DNA repair and with chromosome dimer resolution by the XerC and XerD tyrosine recombinases. In order to test the correspondence between their mechanism in vivo and in vitro, we have tested and shown peptide wrwycr's ability to inhibit the excision of several prophages (lambda, P22, Gifsy-1, Gifsy-2, Fels-1, Fels-2) and to trap Holliday junction intermediates of phage lambda site-specific recombination in vivo. In addition, we found that the peptide inhibits replication of the Salmonella prophage Fels-1 while integrated in the chromosome. These findings further support the proposed mechanistic basis for the antimicrobial activity of the peptide and its use as a tool to dissect strand exchange-dependent DNA repair within cells.
Bacteriophage lambda uses a phage-encoded integrase (Int) to catalyze the site-specific recombination reaction that integrates its chromosome into and excises it out of the Escherichia coli chromosome (e.g., see references 2 and 34). We have previously identified and characterized hexapeptides that inhibit site-specific recombination by the phage lambda Int in vitro by binding to the Holliday junction (HJ) intermediates of the reaction and preventing their resolution (4, 7, 13, 22). The most potent of these peptides (WRWYCR, KWWCRW, and related peptides) were subsequently found to be bactericidal, very likely due to their causing the accumulation of DNA breaks and inhibiting chromosome segregation (18; C. Gunderson and A. Segall, unpublished data). In vivo, however, the d-amino acid forms of the peptides (wrwycr and kwwcrw) were more potent than their l-form counterparts, presumably because they resist peptidases (18).
The question remains whether these peptides block site-specific recombination and accumulate HJ inside bacterial cells. Int is the archetype of a large family of site-specific recombinases that use a tyrosine nucleophile for sequential transesterification reactions. The LT2 strain of Salmonella enterica serovar Typhimurium has four naturally occurring prophages (bacteriophages integrated in its chromosome): Gifsy-1, Gifsy-2, Fels-1, and Fels-2 (11, 15, 36). Each of these prophages encodes an Int-like tyrosine recombinase and can be induced to excise and replicate in a manner very similar to that of phage lambda. DNA damage is the predominant signal that leads to activation of these (and many other) prophages. The prophages respond to DNA damage either because the repressors that maintain the lysogenic state are similar to the SOS regulon repressor LexA (8, 25, 28) and, like it, are sensitive to autocleavage stimulated by RecA's coprotease activity (25) or because the phage antirepressor proteins are part of the SOS regulon, as is the case for phages 186 and N15 of E. coli and phage Fels-2 of Salmonella (5, 6, 23, 26).
Since the peptides cause DNA damage and at least moderately induce the SOS response (18), it was reasonable to expect that peptide treatment might also activate the prophages. However, the peptides were isolated as inhibitors of site-specific recombination, and they may concomitantly inhibit prophage excision. We found that peptide wrwycr inhibits phage lambda excision and/or replication in vivo as well as the excision of the Salmonella phages P22, Gifsy-1, Gifsy-2, Fels-1, and Fels-2. In one case, Fels-1, the peptide also inhibits in situ replication. We also demonstrated that the peptide causes accumulation of the HJ intermediate of phage lambda excision in vivo.
MATERIALS AND METHODS
Strains and bacterial culture methods.
Bacterial strains used in this work and the sources from which they came are listed in Table 1. Strains were maintained on LB agar plates, and cultures were grown in LB broth, except during peptide experiments, for which we used Mueller-Hinton broth (MHB; Becton-Dickinson), a rich medium lacking NaCl. All chemicals were purchased from Sigma (St. Louis, MO), unless otherwise indicated.
TABLE 1.
Bacterial strains used in this study
| Strain designationa | Species and strain | Genotype/description | Source |
|---|---|---|---|
| G255 | S. enterica serovar Typhimurium LT2 | Wild type | Lab collection |
| RW138/G478 | E. coli K-12 | recA galK lacZ(ochre) Spcr (λ cIts857 λ dgal-138 cIts857) | R. Weisberg |
| MA8507/G754 | S. enterica serovar Typhimurium LT2 | Gifsy-1− Gifsy-2− Fels-2− | L. Bossi |
| MA8508/G755 | S. enterica serovar Typhimurium LT2 | Gifsy-1− Gifsy-2− Fels-2− Fels-1Δ(int-attR)::Camr | L. Bossi |
| G889 | E. coli K-12 N99 | λ cIts857 virr | J. Gardner |
| SDT2679 | S. enterica serovar Typhimurium LT2 | ΔFels-1::(frt::Kanr::frt) | Lab collection |
| SDT2739 | S. enterica serovar Typhimurium LT2 | MA8507 (Gifsy-1− Gifsy-2− Fels-2−) ΔFels-1::frt | Lab collection |
| SDT2704 | S. enterica serovar Typhimurium LT2 | P22 lysogen | Lab collection |
| N6377/G158 | E. coli K-12 | C600 thr leu thi pro gal+ (chlD-pgl)Δ8/λ int+xis+ Δ(SalI-XhoI) cI857 (cro-chl(A))ΔH1 | A. Oppenheim via H. Nash |
| EDT1084 | E. coli K-12 | N6377/pLR110 | This study |
The Segall lab strain designation is given after the strain designation from the lab of origin.
Peptides.
All peptides were synthesized with an amidated C terminus and purified to a >95% purity at Sigma-Genosys (St. Louis, MO). According to convention, peptides made of l amino acids are designated in uppercase letters, while those made of d amino acids are designated in lowercase letters. Peptide stock solutions (10 mM) were maintained in 100% dimethyl sulfoxide (DMSO). Final DMSO concentrations in experimental procedures were, at most, 0.32%, and DMSO at the appropriate concentration was added in the absence of peptide to control for DMSO effects. Typically, the d-form peptides are more potent in vivo over the course of 24-h experiments because they are resistant to peptidases (4, 18), and therefore, we used d peptides when available.
Lambda prophage excision.
A lambda lysogen, E. coli N99 λ cIts857 virr (G889), was used to test both survival of the bacteria and phage production. This strain was isolated in Jeff Gardner's lab by first lysogenizing E. coli N99 with λ cIts857, then selecting a λ-resistant derivative by plating the lysogen onto a lawn of λvir phage. Although the allele wasn't mapped, the mutations that generally answer this selection are those affecting the lamB phage receptor (J. Gardner, personal communication). The temperature-sensitive repressor allele, cI857, allows production of plaque-forming phages at 42°C, while no phages are produced at 30°C. Three independent colonies were grown overnight at 30°C, diluted 1:100 in fresh MHB, and grown at 30°C for 2 h. Aliquots (0.5 ml each) were treated with DMSO or peptides (24 μM wrwycr or wrwyar, 12 μM wrwyrggrywrw) and immediately incubated at 42°C for 5 min to induce the prophage. Subsequently, the cultures were returned to 30°C to permit outgrowth of the cured lysogens. Aliquots were taken for plating of the dilutions to determine the percentage of survival of bacteria, and chloroform was added to the remaining cultures to obtain phage lysates. The lysates were diluted in 10 mM Tris-Cl (pH 8) and 10 mM MgCl2, and 2.5-μl spots were spotted onto a lawn of E. coli strain MG1655 grown in 0.2% maltose overnight and supplemented with 10 mM MgSO4 to allow efficient adsorption of the phage. After overnight incubation, plaques were counted to determine phage titer. The experiment was independently repeated on 3 to 5 days, with three independent colonies each day for most peptide or DMSO treatments, and was performed on 2 days with three independent colonies each for peptide wrwyrggrywrw.
Salmonella phage plaquing assays.
A Salmonella P22 lysogen (SDT2704) was used for the P22 induction assays and the wild-type Salmonella strain LT2 (G255) for the Gifsy and Fels induction assays. The P22 lysogen was isolated in the lab by infecting wild-type LT2 with ΦP22 and isolating phage-resistant colonies that produced phage when treated with 1 μg/ml mitomycin C (MMC). Overnight cultures of each of these strains were diluted 1:100 in fresh MHB and grown for 2 hours at 37°C. The cultures were then treated with MMC, peptide, or both and incubated for an additional 3 h at 37°C. As mentioned above, the peptides were stored at 10 mM in DMSO, while MMC stocks were kept at a concentration of 0.5 mg/ml in water; they were added at final concentrations of 32 μM and 1 μg/ml, respectively. After treatment, chloroform was added to the cultures to liberate the phage particles, and the cultures were centrifuged to remove cell debris. The resulting lysates were diluted and spotted onto lawns of recipient strains in the top agar. The recipient strain for the P22 lysate was wild-type LT2. The recipient strain for the Gifsy/Fels lysates was Salmonella strain MA8508 (a gift from L. Bossi and N. Figueroa-Bossi), in which three of the prophages are excised and most of Fels-1, including the immunity region, is deleted and replaced by a cat gene.
qPCR.
Salmonella strain LT2 (G255) was subcultured at a 1:100 dilution from an overnight culture into 1 ml fresh MHB and grown for 2 hours at 37°C. Treatments (DMSO, peptide, and/or MMC) were added, and the cultures were returned to the 37°C shaker for 3 hours. Genomic DNA from the treated cultures was isolated using cetyltrimethylammonium bromide and chloroform extractions (31). Five micrograms of this genomic DNA was used per quantitative PCR (qPCR) as the template. Primer sets used to detect the attL and attB sites of Fels-1, the attR and attB sites of Fels-2, Gifsy-1, and Gifsy-2, and the host rpe and sulA loci are listed in Table 2. The attB sites were amplified using the appropriate attL up and attR down primers specific for that phage. Each primer set was tested for specificity prior to use (data not shown). Bio-Rad iQ SYBR green supermix was used to provide the polymerase, deoxynucleoside triphosphates, and detection dye (SYBR green). The qPCR mixtures were heated at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 56°C for 20 s, followed by a plate reading. After the 40 cycles, a melt curve was done to check the specificity of the amplified products. An MJ Research Chromo4 instrument was used.
TABLE 2.
Primers used to detect Salmonella prophage attachment sites via qPCRa
| Oligonucleotide | Sequence (5′ to 3′) | Product |
|---|---|---|
| Fels-1 attL up | GCAGATTGAGTACACGCAGC | Fels-1 attL |
| Fels-1 attL down | TTCTGCGAAAGGTACTATCTGCG | |
| Fels-1 attR up | ATTGGCCGGAACAACCAG | Fels-1 attR |
| Fels-1 attR down-2 | GTGTAGCTTCACGCTGGC | |
| Gifsy-1 attL up | TCCATCGAATCAAGTACCTGAGC | Gifsy-1 attL |
| Gifsy-1 attL down | GTGGAATGTCCACCGCTG | |
| Gifsy-1 attR up | TTGCGGGGATCTTGAGAGG | Gifsy-1 attR |
| Gifsy-1 attR down | GTAGTGTAGAATGCGGCGTTTC | |
| Gifsy-2 attL up | CCGCTGGAGTATACCTTGTTTAGC | Gifsy-2 attL |
| Gifsy-2 attL down | GTGGGATGTCAGAGAAGAGCG | |
| Gifsy-2 attR up | GGTCTGTGAAGTTCGTTAAAGTTCG | Gifsy-2 attR |
| Gifsy-2 attR down | CAAATCGCGCTACGCAGAATG | |
| sulA up | CCTACCTTCGCTGCTTCAAC | sulA |
| sulA down | CAGTCTTCAGGTTTGCCATT | |
| rpe for | CCAGCTCACTGCGCATTTCA | rpe |
| rpe rev | CCGCATTGATGCGTCCGGTT |
The attB sites of any of the phages can be amplified using the appropriate combination of attL up and attR down primers specific for that phage (up signifies upstream and down signifies downstream). Similarly, the attP sites can be amplified by using the appropriate combination of attR up and attL down primers.
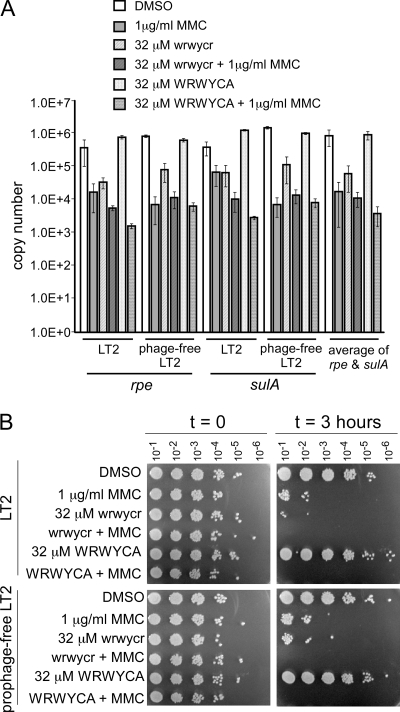
The MMC and peptide treatments, even at sublethal concentrations, were expected to reduce the amount of usable template for qPCR due to both the modification of the DNA by MMC (3) and the decreased cell number in the treated cultures (18; Gunderson and Segall, unpublished). To correct for any treatment-related template changes, the qPCR data were first normalized to two non-prophage-associated loci, rpe and sulA. These two genes showed very similar copy number fluctuations with the different treatments, which also correlated with the decrease in bacterial viability (Fig. 1). However, because a constant amount of DNA was used for each PCR, viability should not influence the results. To test whether induction of the prophages and host cell lysis might also affect the results, the rpe and sulA copy numbers were also measured in a strain deleted for all four prophages, SDT2739. SDT2739 was derived by transduction of the Kanr marker from strain SDT2679, in which the Fels-1 prophage was deleted using linear lambda Red-mediated transformation into strain MA8507, and subsequently, the drug resistance cassette was removed using Flp-mediated recombination, as described previously (9). Strain 2739 showed the same pattern of chromosomal locus copy numbers as did LT2, and thus, prophage induction and replication do not directly reduce PCR efficiency. Thus, the lower apparent copy numbers of rpe and sulA show that the treatments affected the ability of DNA to act as a PCR template. This has been noted previously with DNA-active antibiotics (19).
FIG. 1.
Effect of peptide and/or MMC on chromosome copy numbers of rpe and sulA (A) and viability (B) of treated Salmonella cultures. The wild-type LT2 and LT2 lacking resident prophages were compared to assess the potential contribution of prophage induction to the observed copy number and viability changes. As described in the text, qPCRs were done with a constant 5 μg of isolated genomic DNA.
Excisive site-specific recombination between plasmid-borne att sites in vivo.
E. coli EDT1084, containing plasmid pLR110 (1), was grown overnight in MHB with 100 μg/ml ampicillin (Amp100) at 30°C and subcultured 1:20 in the same medium until an optical density at 600 nm of 0.6 to 0.7 was reached. DMSO or peptide was added to 2.5-ml culture aliquots, and all except one of the aliquots were diluted 1:1 with 2.5 ml of MHB-Amp100 preheated at 65°C to achieve 42°C in order to induce Int and Xis production from the defective prophage. One 2.5-ml culture was not diluted and served as the uninduced control. The diluted cultures were incubated for 30 min at 42°C and then downshifted to 37°C and incubated for another 30 min, since Int does not perform as well at 42°C. Plasmid DNA was isolated from all cultures by alkaline lysis extraction according to published protocols (31), and 20 μg plasmid from each culture was restricted with EcoRI overnight. Parallel in vitro recombination reactions, treated with wrwyrggrywrw or not, were performed as described previously for use as markers (4). Digests and markers were separated by electrophoresis on a 0.8% agarose-1× Tris-borate-EDTA gel. The DNA was depurinated, denatured, and transferred by capillary action to a ZetaProbe nylon membrane (Bio-Rad). A 496-bp fragment of DNA containing the lambda attL site (centered around position 0 of the overlap region) (32) was nick-translated with the Klenow fragment of DNA polymerase I and [α32P]dATP and was used to probe the transferred DNA. Note that at higher concentrations (5 μM and above) of peptide wrwyrggrywrw, recovery of plasmid DNA is less efficient (worse the higher the concentration), potentially because the peptide interferes with plasmid replication. Gels were exposed to a phosphorimager screen and read using the Storm 860 phosphorimager (GE Healthcare) and then quantitated using the ImageQuant v.1.1 program.
RESULTS
Peptide wrwycr inhibits strand exchange by Int and many other related recombinases, including the Cre protein of phage P1, the Flp protein of the Saccharomyces cerevisiae 2μm plasmid, and the integrase of the conjugative element NBU1 (4, 16, 30; P. Rice, personal communication). These proteins are all tyrosine recombinases, site-specific recombination enzymes that generate HJ intermediates during their catalytic cycle. If this peptide has the same mechanism of action in vivo as it does in vitro, it should inhibit the excision of prophages that use tyrosine recombinases.
Phage lambda induction and excision.
In order to test whether wrwycr inhibits the activity of lambda Int in vivo, we used an E. coli strain lysogenic for λ cI857 virR (G889). Bacteria grown at 30°C and subcultured in log phase were treated with the DMSO solvent or one of three peptides and immediately subjected to a 5-min pulse at 42°C. The cells were then returned to 30°C; phages that excised and replicated were prevented from reinfecting by the virR mutation, which affects expression of the lamB (malB) gene (J. Gardner, personal communication). The results of this assay are presented in Table 3. Peptides wrwycr and wrwyrggrywrw (the latter is a single-chain peptide designed to mimic a dimer of wrwycr [30; J. Boldt, A. Flores, I. Naili, and A. Segall, unpublished results]) were expected to inhibit excision. Peptide wrwyar was used as a control because loss of the cysteine in peptide WRWYAR reduces its 50% inhibitory concentration (IC50) by about 10-fold relative to that of WRWYCR in inhibiting phage lambda excision in vitro (4; R. Saha and A. Segall, unpublished results); its potency is regained if the Cys-less peptide is dimerized by means other than a disulfide bridge (4). When the lambda lysogen G889 was treated with peptide wrwycr or wrwyrggrywrw, no significant change in bacterial survival at 42°C was seen compared to when it was treated with DMSO or wrwyar (Table 3). Significantly higher concentrations of peptide were not used because it became difficult to obtain sufficient numbers of colonies to test heat resistance due to the killing effect of the peptides (note that E. coli is somewhat more sensitive to peptide than is Salmonella [see below], with an MIC between 32 and 64 μM). After the temperature upshift to induce the prophages, we added chloroform to an aliquot of each culture in order to detect any effects on phage production; if the peptide inhibited excision and/or phage replication, we should see evidence of lower phage production in these treated cultures. Indeed, when spot titers of lysates made from the treated G889 lysogens were determined on MG1655, we observed a five- to sevenfold drop in the titers of lysates from wrwycr- and wrwyrggrywrw-treated bacteria compared to the titers of DMSO- or from wrwyar-treated bacteria (Table 3).
TABLE 3.
Effect of peptide treatment on survival and phage production by a lambda lysogen
| Treatment (concn) | % Survival ± SD (no. of independent colonies) | Phage titer (no. of colonies tested) | Fraction of phagea |
|---|---|---|---|
| DMSO (0.29%) | 59.4 ± 7.7 (9) | 8.9 × 109 (4) | 1 |
| wrwycr (24 μM) | 69.1 ± 8.5 (15) | 1.7 × 109 (4) | 0.18 |
| wrwyrggrywrw (12 μM) | 70.7 ± 6.3 (6) | 1.2 × 109 (4) | 0.14 |
| wrwyar (24 μM) | 58.5 ± 5.7 (12) | 8.3 × 109 (3) | 0.9 |
Fraction of phages whose titers were determined in lysates from bacteria treated with peptides relative to those whose titers were determined in lysates from bacteria treated with DMSO.
Induction and excision of phage P22.
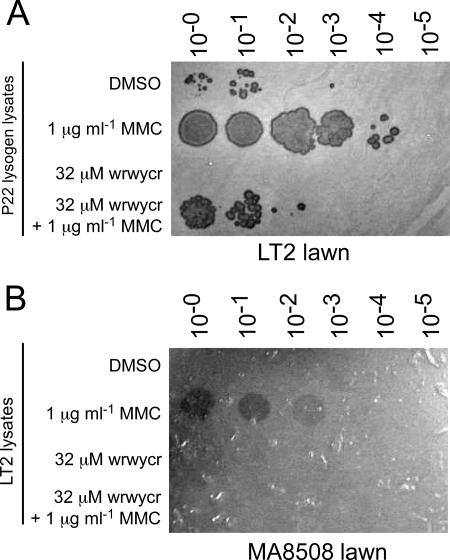
The mechanism of excision by tyrosine recombinase-mediated site-specific recombination is shared by many temperate phages (10, 27). To determine if the peptides inhibit the excision of prophages other than lambda, we first tested the ability of a Salmonella phage P22 lysogen to produce PFU after treatment with peptide wrwycr (Fig. 2A). The P22 lysogen was treated with peptide, 1 μg/ml MMC to induce phage excision, or both, and lysates were spotted onto a lawn of LT2 cells. While some phages are produced even in the DMSO (mock)-treated sample, MMC induced approximately 1,000-fold more P22 phages (Fig. 2A). A sublethal concentration of the peptide, 32 μM, inhibited the production of phages ∼100-fold relative to the DMSO-treated culture. The peptide also inhibited the number of phages produced by MMC treatment 100- to 1,000-fold. This assay does not distinguish between two possible effects of the peptide—inhibition of excision per se or inhibition of phage replication after the prophage has excised. Phage P22 uses a similar mechanism to perform site-specific recombination as phage lambda (24, 33), and thus, if excision of one is inhibited, it is likely that excision of the other is also inhibited.
FIG. 2.
Peptide wrwycr inhibits excision of Salmonella prophage. (A) Chloroform lysates of LT2 P22 lysogen cultures treated with MMC, peptide, or both (as indicated on the left). (B) Chloroform lysates of wild-type LT2 cultures treated with MMC or peptide (as indicated). Lysates were serially diluted 10-fold and plated onto a lawn of MA8508, an LT2-derived strain cured of three of its four naturally occurring prophages and deleted for a large fraction of the fourth prophage (Gifsy-1, Gifsy-2, Fels-1, and Fels-2). Two microliters of each lysate, either undiluted or subsequent 10-fold dilutions, was spotted onto the bacterial lawns. These plates are representative of four to eight independent induction experiments.
Induction and excision of Gifsy and Fels phages.
Four prophages within the chromosome of Salmonella strain LT2—Gifsy-1, Gifsy-2, Fels-1 and Fels-2—are capable of producing viable phages. Each of these phages form plaques on strains that have had the respective prophages deleted or whose repressor of lytic growth has been inactivated (e.g., MA8508). Phage plaques were produced when the culture was treated with MMC but not when treated with peptide by itself, and the peptide prevented the MMC-dependent formation of phage plaques (Fig. 2B).
Since DNA damage can trigger activation of prophages, Salmonella's prophages might be induced by the peptide treatment itself. This induction, however, may be masked by the inhibition of excision by the peptide. Phage induction can lead to two forms of phage replication: (i) the prophage may first excise and subsequently replicate extrachromasomally, or (ii) the phage may initiate “onion skin” or “escape” replication if it does not or cannot excise after induction of the lytic cycle (14, 35). Frye and colleagues have shown that the induction of Fels-1 can result in both types of replication: (i) “onion skin” replication extending beyond the ends of the integrated phage, generating linear chromosomal replication products as long as 900 kb, or (ii) the more classic replication following excision, generating replicating DNA that comprises only phage sequences (12). To detect inhibition of excision as well as inhibition of “onion skin” replication, we used qPCR to measure the relative amounts of attR or attL and attB for each of the prophages after MMC induction with or without peptide treatment (Table 4). The attR and attL sites are present only when the prophage is still integrated, while the attB site is created only once the phage excises from the bacterial chromosome. The attP site was not directly measured because it is significantly longer than the attB, attL, or attR sites and is, thus, less amenable to quantifying PCR products. The qPCR-determined copy numbers of each att site were first normalized to the averaged copy number of two non-prophage-associated loci, rpe and sulA, obtained under the same treatment conditions (for details, see Materials and Methods and Fig. 1). Subsequently, the change in copy number resulting from independently treating cultures with MMC or peptide was calculated in comparison to DMSO (mock) treatment (Table 4). The effect of the combined treatment of MMC and peptide on copy numbers was then compared to that of the MMC treatment alone and expressed as the inhibition by the peptides on MMC-induced excision (Table 4); this further factors out any MMC effect on template quality (as discussed in Materials and Methods). Peptide WRWYCA, which has an IC50 of ∼0.1 μM for in vitro HJ-trapping activity versus the IC50 of 0.01 μM for peptide WRWYCR and also has a >10-fold higher MIC than does peptide wrwycr or WRWYCR (4; R. Saha and A. Segall, unpublished results, and A. Flores and A. Segall, unpublished results), was compared with peptide wrwycr; if the activity of peptide wrwycr in vivo is indeed related to its in vitro activity, peptide WRWYCA should have correspondingly lesser effects.
TABLE 4.
Fold change in att site copy number after treatment with 1 μg ml−1 MMC and/or 32 μM peptide wrwycr or WRWYCAa
| Prophage | Site | Fold change in copy no. with MMC or peptide treatment
|
Fold inhibition of MMC-induced replication or excision
|
|||
|---|---|---|---|---|---|---|
| MMC/DMSO | wrwycr/DMSO | WRWYCA/DMSO | MMC/MMC + wrwycr | MMC/MMC + WRWYCA | ||
| Gifsy-1 | attR | 1.2 | 0.9 | 3.1 | 1.1 | 1.8 |
| attB | 102 | 0.1 | 2.7 | 316 | 29 | |
| Gifsy-2 | attR | 2.5 | 0.8 | 3.5 | 2.0 | 3.5 |
| attB | 266 | 0.2 | 4.2 | 242 | 7.7 | |
| Fels-1 | attL | 11 | 1.2 | 3.4 | 6.9 | 4.0 |
| attB | 24 | 3.6 | 1.3 | 3.5 | 0.4 | |
| Fels-2 | attR | 1.5 | 1.2 | 7.3 | 0.5 | 0.1 |
| attB | 107 | 5 | 18 | 5.6 | 0.4 | |
The data represent the averages of the results for at least two independent experiments, each experiment with two independent colonies.
As expected for an inducer of the SOS response, MMC caused a 24- to 266-fold increase in the copy number of the attB sites of all four prophages (Table 4). The concomitant 11-fold increase in the Fels-1 attL copy number agrees with the observation that Fels-1 exhibits significant “onion skin” replication prior to excising from the Salmonella genome (12). In contrast, peptide wrwycr alone had little effect in inducing the prophages of Salmonella and, in most cases, had even less of an effect than peptide WRWYCA (Table 4). This difference is consistent with the lesser potency of peptide WRWYCA in inhibiting site-specific recombination, but the data suggest that this peptide does induce the SOS response to a low extent.
In order to test inhibition of excision of the prophages, they first have to be induced to enter the lytic cycle. However, β-galactosidase measurements of a peptide-treated Salmonella recN::lacZ reporter strain indicated that wrwycr induces the SOS response moderately only, ∼4- to 6-fold inhibition (18). We thus worried that with peptide treatment alone, we would be unable to see the full inhibition of excision or phage replication because the prophages may be only weakly induced to resume lytic growth. Therefore, we compared the att site copy number after MMC treatment (an efficient inducer of lytic growth) to that after the combined treatment of the lysogens with MMC and peptides. When we treated LT2 with both MMC and wrwycr, excision of all prophages was inhibited by the peptide. Gifsy-1 and Gifsy-2 prophage excisions were most sensitive to inhibition by peptide wrwycr, 316-fold and 240-fold inhibitions, respectively (Table 4). By comparison, peptide WRWYCA inhibited the excision of MMC-induced phages much less (Table 4). Peptide wrwycr also inhibited the Fels-1 in situ replication ∼7-fold (as discussed below), while WRWYCA inhibited in situ replication 4-fold (Table 4).
Trapping HJs in bacteria.
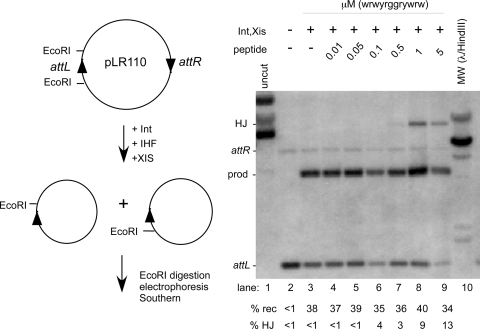
The results of the earlier experiments show that peptide wrwycr does inhibit tyrosine recombinase-mediated recombination inside bacteria but do not address whether the peptide can stabilize the HJ intermediate. This question was addressed specifically by inducing Int-dependent recombination between a plasmid-borne pair of lambda attachment sites, attL and attR (plasmid pLR110 [1]) (Fig. 3). Strain EDT1084 is a lysogen which retains only the left side of a lambda prophage, and it expresses both Int and Xis from pL when the cI857 repressor is heat inactivated (17, 32). The peptide used for this experiment is a single-chain dodecamer, wrwyrggrywrw, designed to mimic the active dimer form of the wrwycr peptide while being resistant to the reducing environment of the cell (J. L. Boldt, A. Flores, and A. Segall, unpublished results). This peptide gave more consistent results than did wrwycr in the experiment described below, although wrwycr gave similar results (data not shown). The attL site is flanked by EcoRI sites (Fig. 3, left top panel). When Int and Xis are expressed (IHF protein is provided by the host), recombination deletes the segment of the plasmid between the two att sites, putting one of the EcoRI sites on a separate DNA circle (Fig. 3, left bottom panel). Thus, substrates and recombinant products can be distinguished by their different electrophoretic mobilities on an agarose gel after digestion with EcoRI. The strain was subcultured in MHB and grown at 30°C into mid-late log phase (optical density at 600 nm, 0.5 to 0.6), then peptide wrwyrggrywrw was added at the indicated concentrations, and the culture was diluted 1:1 with MHB prewarmed to 65°C or 37°C. After 15 min at either 42°C or 37°C, incubation was continued for 30 min at 37°C (37°C permits somewhat better excisive recombination than does 42°C in E. coli). When Int and Xis expression were induced, we detected 3% accumulation of HJ intermediates in the presence of 0.5 μM dodecamer and accumulation of 13% HJ at 5 μM dodecamer (Fig. 3). Thus, the dodecamer peptide is able to trap HJ.
FIG. 3.
Trapping of phage lambda excisive recombination junction intermediates inside E. coli. The left side of the figure shows the structure of plasmid pLR110 and the orientation of the attL and attR sites. The right panel displays the results of a Southern analysis of plasmid DNA isolated from cells in which Int and Xis expression was induced or not, which were treated with the indicated amounts of peptide wrwyrggrywrw. The probe used was a 496-bp fragment encoding attL from pHN872 (32). The position of the markers for recombination substrate and product fragments and HJs is based on a parallel in vitro reaction. prod, products; rec, recombination; MW, molecular weight; +, present; −, absent.
DISCUSSION
To examine the role of the peptides in prophage excision and replication within bacterial cells, we used several approaches. First, we used an E. coli phage lambda lysogen to monitor prophage excision after exposing lysogens to the peptides; in this case, prophage induction was mediated by temperature inactivation of the λ cI857 repressor. Second, we monitored excision of the Salmonella Gifsy and Fels prophages and that of the temperate Salmonella phage P22 by determining the titers of lysates made from peptide-treated lysogens, using the DNA-damaging agent MMC as the prophage-inducing treatment. Third, we assayed the excision of the Fels and Gifsy prophages in response to peptide treatment by qPCR. Finally, we showed that peptide wrwyrggrywrw stabilizes HJ intermediates of phage λ excision in vivo.
Peptide wrwycr and the related peptides were identified in an in vitro screen and deconvolution of a combinatorial peptide library for inhibitors of lambda site-specific recombination (4, 7, 22). The peptides have been characterized extensively and inhibit recombination by binding to the HJ intermediate and by preventing further catalysis (4, 20, 21), probably by altering the three-dimensional structure of the junction, moving the path of the phosphodiester backbone away from Int's active site (16). Because it binds HJ and, with lesser affinity and stability, other branched DNA intermediates (21), the peptide wrwycr inhibits a number of HJ processing enzymes in a structure-selective manner (20, 21). This peptide has also been shown to have antimicrobial activity (18) and inhibits Salmonella growing inside macrophages (L. Su, D. Hall, and A. Segall, unpublished data).
Our goal was to test whether peptide wrwycr inhibited recombination by site-specific tyrosine recombinases and whether it traps HJ inside bacteria, as it does in vitro. Here, we have shown that peptide wrwycr inhibits excision of several prophages (phages lambda, P22, Fels-1 and Fels-2, and Gifsy-1 and Gifsy-2) that use tyrosine recombinase-mediated excision to leave the bacterial chromosome upon induction. In turn, inhibition of excision results in a reduction in phage titer and/or generation of recombinant genomes. Using phage production as a measurement, the peptide by itself is not as effective at inducing the SOS response as is MMC (Table 4). A dodecameric version of the peptide, wrwyrggrywrw, is capable both of inhibiting excision and/or replication of phage lambda and of inhibiting recombination in vivo between attL and attR sites located on a plasmid, trapping HJ intermediates of the reaction. The most active form of peptide wrwycr is a disulfide-bridged dimer (4); the dodecamer peptide may be more effective because its “dimerized” state is not diminished in the reducing environment of the bacterial cell. In the case of the lambda lysogen, peptide wrwycr inhibited phage lambda production somewhat less (∼5-fold) than did peptide wrwyrggrywrw (∼7-fold), while peptide wrwyar, which cannot dimerize via a disulfide bridge, did not inhibit phage production at all.
Whether the active peptides inhibit only excision or both excision and phage replication remains to be tested. It is highly unlikely that replication is inhibited while excision is not because of the higher affinity of the peptide for HJ substrates than for replication fork substrates in vitro and because at least some HJ intermediates of lambda excision were trapped inside bacteria (Fig. 3). Note that peptide inhibition is evidenced by lower phage titers but not by significantly greater percent survival of lysogens. This is presumably due to two related factors. First, given sufficient time at sublethal concentrations of peptide, most lysogens will eventually escape the peptide inhibition of excision, and the peptide will be diluted. Second, the reduction in phage titers but lack of increased lysogen survival may be a consequence of incomplete inhibition of excision together with the fact that phage growth amplifies the response, since every induced lysogen produces about 100 phages (thus, the decrease in phage titer will be more easily detectable when affecting the number of phages produced than the 100-fold fewer lysogens “protected”). Escape from peptide inhibition could be due to the dissociation of the peptide followed by resolution of the junctions either by Int itself or by the junction resolvase RuvC. Over time, the peptide is effluxed from cells (18; S. Orchard and A. Segall, unpublished data), and the concentrations used may thus be limiting in that the dissociated peptide would not be replenished. The amount of peptide available may also account for a discrepancy observed between the inhibition of excisive recombination observed in the lambda lysogen experiment and the lack of inhibition observed when testing recombination between plasmid-borne attL and attR sites, despite detecting trapped junction intermediates. The plasmid-based assay may both underestimate the number of junctions trapped and overestimate the amount of recombination. In the plasmid recombination assay, at most 5 μM wrwyrggrywrw peptide was used (compared to 12 μM in the lysogen induction assay), since larger amounts made it impossible to isolate sufficient plasmid DNA to assay recombination. At 5 μM peptide, it is possible that the majority of HJ intermediates were not stably trapped by peptide and that Int and its accessory factors resolved most of the HJ to products. Added to this is the difficulty in quantitatively maintaining HJs intact during the DNA isolation process. Under the conditions used, the plasmid assay is thus a more qualitative than quantitative measure of trapping HJ intermediates.
Excisive recombination reactions of different prophages were inhibited by peptide wrwycr with different efficiencies; the excision of the Fels prophages was inhibited less than the excision of the Gifsy prophages. As described above, these differences may reflect the extent of induction both of different phages and of different recombination pathways (fewer substrates recombined would generate fewer intermediates to trap), as well as potentially distinct geometries of the HJ intermediate in different pathways (discussed in references 4, 7, and 22). Such differences are even more substantial in the case of recombination reactions catalyzed by different tyrosine recombinases; for example, peptide WRWYCR inhibits Int-mediated excision about 10-fold more than Cre-mediated loxP × loxP recombination (4).
In the case of Fels-1, the peptide inhibited the in situ “onion skin” replication of the integrated prophage, which had been detected previously in a microarray study of UV- or MMC-induced Salmonella strain LT2 (12). The inhibitory effect, ∼7-fold, was moderate but significant. In contrast, the peptides have little direct effect on bulk bacterial DNA synthesis when measured by thymidine incorporation (18) or in a reconstituted in vitro replication system of an OriC-dependent plasmid (J. Kaguni, personal communication). Thus, either the phage replisome is more sensitive to interference by the peptide than is the bacterial replisome, or the inhibition is through an indirect (and so far undetermined) mechanism of peptide action. This question will be pursued for this and other prophage replisomes in the future. The difficulty in isolating plasmid DNA at high concentrations of peptide suggests that ColE1 plasmid replisomes may also be equally sensitive to peptide.
Unexpectedly, our qPCR control experiments showed that peptide wrwycr, like MMC, reduces the efficiency of DNA as a template for PCR. This is not the case for peptide WRWYCA. Since the templates used for PCR were purified genomic DNA, any modification must have occurred during the treatment of bacterial cells. We do not yet know what the peptide-induced modification may be, or whether it is a direct action of the peptide or an indirect consequence of the peptide on bacterial physiology.
The effects that the peptides have on bacterial cells, including DNA breaks and chromosome fragmentation, and the changes in sensitivity seen with different recombination mutations have provided indirect evidence that the peptides act on targets within the cell (18; Gunderson and Segall, unpublished). Here, we have shown that the peptides can inhibit the site-specific recombination events that result in the excision of phage lambda and other prophages. As a result of this work, we now know that the peptides are able to enter bacterial cells, retaining an active form that is capable of their in vitro activity. Thus, we can be more confident that since the peptides can inhibit site-specific recombination in vivo, they are also available to inhibit recombination and DNA repair, their proposed mechanism of antimicrobial activity (18; Gunderson and Segall, unpublished). This novel mode of inhibition may be exploited in the future to develop a new class of antibiotics.
Acknowledgments
We are very grateful to Robert Weisberg for his extremely thorough constructive criticisms of the manuscript and to him, Lionello Bossi, Nara Figueroa-Bossi, Jeffrey Gardner, and John Roth for providing the strains used in this study. We thank Rob Edwards for helping to bioinformatically identify the att sites of the four prophages in LT2. We appreciate Samantha Orchard's editorial help. Elysa Brown and Samantha Orchard constructed the Fels-1 knockout. The students of the 2007 Advanced Bacterial Genetics course at Cold Spring Harbor Laboratory performed the initial Salmonella prophage induction and qPCR experiments. We thank Jennifer Boylston for designing and testing the qPCR primers for the sulA and rpe genes.
This work was supported by Public Health Service grant AI-58253 from the National Institute for Allergy and Infectious Diseases to A.M.S.
Footnotes
Published ahead of print on 30 January 2009.
REFERENCES
- 1.Abremski, K., and S. Gottesman. 1982. Purification of the bacteriophage lambda xis gene product required for lambda excisive recombination. J. Biol. Chem. 2579658-9662. [PubMed] [Google Scholar]
- 2.Azaro, M. A., and A. Landy. 2002. λ Integrase and the λ Int family, p. 118-148. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 3.Becerril, C., M. Ferrero, F. Sanz, and A. Castano. 1999. Detection of mitomycin C-induced genetic damage in fish cells by use of RAPD. Mutagenesis 14449-456. [DOI] [PubMed] [Google Scholar]
- 4.Boldt, J. L., C. Pinilla, and A. M. Segall. 2004. Reversible inhibitors of lambda integrase-mediated recombination efficiently trap Holliday junction intermediates and form the basis of a novel assay for junction resolution. J. Biol. Chem. 2793472-3483. [DOI] [PubMed] [Google Scholar]
- 5.Brumby, A. M., I. Lamont, I. B. Dodd, and J. B. Egan. 1996. Defining the SOS operon of coliphage 186. Virology 219105-114. [DOI] [PubMed] [Google Scholar]
- 6.Bunny, K., J. Liu, and J. Roth. 2002. Phenotypes of lexA mutations in Salmonella enterica: evidence for a lethal lexA null phenotype due to the Fels-2 prophage. J. Bacteriol. 1846235-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassell, G., M. Klemm, C. Pinilla, and A. Segall. 2000. Dissection of bacteriophage lambda site-specific recombination using synthetic peptide combinatorial libraries. J. Mol. Biol. 2991193-1202. [DOI] [PubMed] [Google Scholar]
- 8.Craig, N. L., and J. W. Roberts. 1980. E. coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature 28326-30. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 253605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueroa-Bossi, N., E. Coissac, P. Netter, and L. Bossi. 1997. Unsuspected prophage-like elements in Salmonella typhimurium. Mol. Microbiol. 25161-173. [DOI] [PubMed] [Google Scholar]
- 12.Frye, J. G., S. Porwollik, F. Blackmer, P. Cheng, and M. McClelland. 2005. Host gene expression changes and DNA amplification during temperate phage induction. J. Bacteriol. 1871485-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimoto, D. F., C. Pinilla, and A. M. Segall. 2006. New peptide inhibitors of type IB topoisomerases: similarities and differences vis-a-vis inhibitors of tyrosine recombinases. J. Mol. Biol. 363891-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukasawa, T., K. Hirai, T. Segawa, and K. Obonai. 1978. Regional replication of the bacterial chromosome induced by derepression of prophage lambda. IV. Escape synthesis of gal operon in phage 82. Mol. Gen. Genet. 16783-93. [DOI] [PubMed] [Google Scholar]
- 15.Gemski, P., Jr., L. S. Baron, and N. Yamamoto. 1972. Formation of hybrids between coliphage lambda and Salmonella phage P22 with a Salmonella typhimurium hybrid sensitive to these phages. Proc. Natl. Acad. Sci. USA 693110-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh, K., C. K. Lau, F. Guo, A. M. Segall, and G. D. Van Duyne. 2005. Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination. J. Biol. Chem. 2808290-8299. [DOI] [PubMed] [Google Scholar]
- 17.Goodman, S. D., S. C. Nicholson, and H. A. Nash. 1992. Deformation of DNA during site-specific recombination of bacteriophage lambda: replacement of IHF protein by HU protein or sequence-directed bends. Proc. Natl. Acad. Sci. USA 8911910-11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunderson, C. W., and A. M. Segall. 2006. DNA repair, a novel antibacterial target: Holliday junction-trapping peptides induce DNA damage and chromosome segregation defects. Mol. Microbiol. 591129-1148. [DOI] [PubMed] [Google Scholar]
- 19.Hotta, K., C. B. Zhu, P. Phomsuwari, J. Ishikawa, S. Mizuno, M. Hatsu, and S. Imai. 1995. PCR inhibition assay for DNA targeted antibiotics. J. Antibiot. 481267-1272. [DOI] [PubMed] [Google Scholar]
- 20.Kepple, K. V., J. L. Boldt, and A. M. Segall. 2005. Holliday junction-binding peptides inhibit distinct junction-processing enzymes. Proc. Natl. Acad. Sci. USA 1026867-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kepple, K. V., N. Patel, P. Salamon, and A. M. Segall. 2008. Interactions between branched DNAs and peptide inhibitors of DNA repair. Nucleic Acids Res. 365319-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klemm, M., C. Cheng, G. Cassell, S. Shuman, and A. M. Segall. 2000. Peptide inhibitors of DNA cleavage by tyrosine recombinases and topoisomerases. J. Mol. Biol. 2991203-1216. [DOI] [PubMed] [Google Scholar]
- 23.Lamont, I., A. M. Brumby, and J. B. Egan. 1989. UV induction of coliphage 186: prophage induction as an SOS function. Proc. Natl. Acad. Sci. USA 865492-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leong, J. M., S. Nunes-Duby, C. F. Lesser, P. Youderian, M. M. Susskind, and A. Landy. 1985. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J. Biol. Chem. 2604468-4477. [PubMed] [Google Scholar]
- 25.Little, J. W. 1984. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. USA 811375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mardanov, A. V., and N. V. Ravin. 2007. The antirepressor needed for induction of linear plasmid-prophage N15 belongs to the SOS regulon. J. Bacteriol. 1896333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunes-Düby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phizicky, E. M., and J. W. Roberts. 1980. Kinetics of RecA protein-directed inactivation of repressors of phage lambda and phage P22. J. Mol. Biol. 139319-328. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Rajeev, L., A. Segall, and J. Gardner. 2007. The bacteroides NBU1 integrase performs a homology-independent strand exchange to form a Holliday junction intermediate. J. Biol. Chem. 28231228-31237. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Segall, A. M., and H. A. Nash. 1996. Architectural flexibility in lambda site-specific recombination: three alternate conformations channel the attL site into three distinct pathways. Genes Cells 1453-463. [DOI] [PubMed] [Google Scholar]
- 33.Smith-Mungo, L., I. T. Chan, and A. Landy. 1994. Structure of the P22 att site. Conservation and divergence in the lambda motif of recombinogenic complexes. J. Biol. Chem. 26920798-20805. [PubMed] [Google Scholar]
- 34.Weisberg, R. A. 1970. Requirements for curing of λ lysogens. Virology 41195-199. [DOI] [PubMed] [Google Scholar]
- 35.Willard, M., and H. Echols. 1968. Role of bacteriophage DNA replication in lambda-dg escape synthesis. J. Mol. Biol. 3237-46. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto, N. 1969. Genetic evolution of bacteriophage. I. Hybrids between unrelated bacteriophages P22 and Fels 2. Proc. Natl. Acad. Sci. USA 6263-69. [DOI] [PMC free article] [PubMed] [Google Scholar]