Abstract
Pea plants incubated in 15N2 rapidly accumulated labeled γ-aminobutyrate (GABA) in the plant cytosol and in bacteroids of Rhizobium leguminosarum bv. viciae 3841. Two pathways of GABA metabolism were identified in R. leguminosarum 3841. In the first, glutamate is formed by GABA aminotransferase (GabT), transferring the amino group from GABA to 2-oxoglutarate. In the second, alanine is formed by two ω-aminotransferases (OpaA and OpaB), transferring the amino group from GABA to pyruvate. While the gabT mutant and the gabT opaA double mutant grew on GABA as a nitrogen source, the final triple mutant did not. The semialdehyde released from GABA by transamination is oxidized by succinate semialdehyde dehydrogenase (GabD). Five of six potential GabD proteins in R. leguminosarum bv. viciae 3841 (GabD1, -D2, -D3, -D4, and -D5) were shown by expression analysis to have this activity. However, only mutations of GabD1, GabD2, and GabD4 were required to prevent utilization of GABA as the sole nitrogen source in culture. The specific enzyme activities of GabT, Opa, and GabD were highly elevated in bacteroids relative to cultured bacteria. This was due to elevated expression of gabT, opaA, gabD1, and gabD2 in nodules. Strains mutated in aminotransferase and succinate semialdehyde dehydrogenases (gabT, opaA, or opaB and gabD1, gabD2, or gabD4, respectively) that cannot use GABA in culture still fixed nitrogen on plants. While GABA catabolism alone is not essential for N2 fixation in bacteroids, it may have a role in energy generation and in bypassing the decarboxylating arm of the tricarboxylic acid cycle.
Rhizobia are alphaproteobacteria that form beneficial symbioses with higher plants, mainly of the legume family. The symbiosis occurs in root structures, called nodules, which provide appropriate conditions, such as a low oxygen concentration, for nitrogen fixation and proliferation of bacteria. To establish a root nodule, a highly specific exchange of signaling compounds is required; rhizobia attach to root hairs and grow down plant-derived infection threads into the root cortex (29). Bacteria are engulfed by plant cells and surrounded by a plant-derived membrane (symbiosome membrane) with the whole structure called a symbiosome (13). Within symbiosomes, bacteria differentiate into bacteroids that reduce N2 to ammonia, which is supplied to the plant in exchange for a dicarboxylate (e.g., l-malate, succinate, and fumarate) for use as a carbon source (1, 47). Consistent with this, dicarboxylate transport via DctA is essential for bacteroid function (9, 34), and labeling studies support a high turnover of malate (35). Recently, it was shown that nutrient exchange is more complex, with amino acid movement between Rhizobium leguminosarum bv. viciae 3841 and the plant cytosol essential in peas (Pisum sativum) for an effective symbiosis (26). This was apparent because mutation of the two main broad-range amino acid ABC transporters (AapJQMP and BraDEFGC) led to a severely impaired symbiotic phenotype. The nitrogen fixation rate of this double transport mutant was substantially reduced, as was the plant weight (dry weight). However, bacteroids of the mutant still fixed nitrogen at around 30% of the wild-type rate, while building up the carbon storage compound polyhydroxybutyrate. Bacteroids in indeterminate pea nodules do not normally store polyhydroxybutyrate, which suggests that blocking amino acid movement also affects carbon metabolism. To explain this, it was proposed that glutamate or a derivative of it, such as γ-aminobutyrate (GABA), is imported into the bacteroid and another amino acid, such as aspartate or alanine, is secreted (25, 32, 47). Major questions arising from this model include the question which amino acids are essential for exchange between the two symbiotic partners and in which direction do they move? Indeed, it can also be asked whether a two-way exchange is essential or whether amino acids might only need to move in one direction. Another question is how are nitrogen and carbon metabolism balanced? To address these questions, we used 15N2 labeling to identify the amino acids into which fixed nitrogen is incorporated in bacteroids. This indicated that GABA is rapidly labeled in bacteroids, and therefore, its metabolism was investigated in detail.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains, plasmids, and primers used in this study are detailed in Table 1. Rhizobium strains were grown at 28°C in either tryptone yeast extract (TY) (3) or acid minimal salts medium (AMS) (30) with 10 mM d-glucose or 10 mM disodium succinate as the carbon source and 10 mM sodium γ-aminobutyrate (GABA) or 10 mM NH4Cl as the nitrogen source. The following antibiotics were used at the following concentrations (in micrograms per milliliter): streptomycin (Str), 500; kanamycin (Km), 20; neomycin (Nm), 80; tetracycline (Tet), 5; gentamicin (Gm), 20; and spectinomycin (Spec), 100. Doubling times were determined in triplicate 10-ml cultures with shaking at 200 rpm on a horizontal shaker at 30°C over a maximum time of 30 h.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequencea | Reference |
|---|---|---|
| Strains | ||
| R. leguminosarum bv. viciae 3841 | 20 | |
| A34 | Rhizobium leguminosarum bv. viciae strain | 7 |
| RU1816 | R. leguminosarum bv. viciae 3841 mutant, overexpresses gabD4 | 47 |
| RU2516 | R. leguminosarum bv. viciae 3841 gabT::ΩTet | This work |
| RU2517 | R. leguminosarum bv. viciae 3841 gabD2::ΩTet | This work |
| RU2518 | R. leguminosarum bv. viciae 3841 ΔgabT | This work |
| RU2519 | R. leguminosarum bv. viciae 3841 ΔgabD2 | This work |
| RU3845 | R. leguminosarum bv. viciae 3841(pRU1818) | This work |
| RU3846 | R. leguminosarum bv. viciae 3841(pRU1819) | This work |
| RU3847 | R. leguminosarum bv. viciae 3841(pRU1820) | This work |
| RU3849 | R. leguminosarum bv. viciae 3841(pRU1822) | This work |
| RU3850 | R. leguminosarum bv. viciae 3841(pRU1823) | This work |
| RU3922 | R. leguminosarum bv. viciae 3841 ΔgabT opaA::ΩSpec | This work |
| RU3924 | R. leguminosarum bv. viciae 3841 ΔgabD2 gabD4::ΩTet | This work |
| RU3930 | R. leguminosarum bv. viciae 3841 ΔgabD2 pK19mobgabD1 gabD4::ΩTet | This work |
| RU4011 | R. leguminosarum bv. viciae 3841 ΔgabT ΔopaA | This work |
| RU4012 | R. leguminosarum bv. viciae 3841 ΔgabT ΔopaA opaB::ΩSpec | This work |
| RU4016 | R. leguminosarum bv. viciae 3841 ΔgabD2 pK19mobgabD1 | This work |
| RU4104 | R. leguminosarum bv. viciae 3841 pK19mobgabD1 | This work |
| Plasmids | ||
| pJQ200SK | pACYC derivative, P15A origin of replication; Gmr | 33 |
| pRK415 | Broad-host-range P-group cloning vector; Tetr | 23 |
| pK19mob | Used to generate mutants in R. leguminosarum bv. viciae 3841; Nmr | 40 |
| pRU1818 | gabD1 cloned in pRK415 | This work |
| pRU1819 | gabD2 cloned in pRK415 | This work |
| pRU1820 | gabD3 cloned in pRK415 | This work |
| pRU1822 | gabD5 cloned in pRK415 | This work |
| pRU1823 | gabD6 cloned in pRK415 | This work |
| pRU1882 | gabT::ΩTet cloned in pJQ200SK | This work |
| pRU1883 | ΔgabT cloned in pJQ200SK | This work |
| pRU1884 | gabD2::ΩTet cloned in pJQ200SK | This work |
| pRU1885 | ΔgabD2 cloned in pJQ200SK | This work |
| pRU1890 | gabD4::ΩTet cloned in pJQ200SK | This work |
| pRU1891 | opaA::ΩSpec cloned in pJQ200SK | This work |
| pRU1892 | gabD1cloned in pK19mob | This work |
| pRU1933 | ΔopaA cloned in pJQ200SK | This work |
| pRU1934 | opaB::ΩSpec cloned in pJQ200SK | This work |
| Primers | ||
| p676 | CGCAAGAGCGGCATTGAAGG | |
| p741 | TCTAGAGGCGGCGAGTTCGTTGCTTC | |
| p742 | AAGCTTTGCCACCGATGAAGAACAGAGCG | |
| p743 | TCTAGAGCCGCCAGAATTGGTGACAAG | |
| p744 | GGATCCAACTCCTCCCTCATTGTCAGCCG | |
| p745 | GGATCCCGCCGAGCAATGGAAGACGG | |
| p746 | TCTAGACCTCCATGCTGCAAGCAAGC | |
| p747 | AAGCTTGCGACCATGCTGCCCTATGTG | |
| p748 | TCTAGACGGTGATGGTCTCAGCGACGC | |
| p751 | TCTAGACTATGCTATGACGATGATCGAGCTG | |
| p752 | AAGCTTCTCCGCCAATATCAATCCCGA | |
| p759 | AAAAGTCCGCCAGATCGCCCGTCC | |
| p760 | ACCATCCAGGACGAGATCTTCGG | |
| p761 | CACGTCGAAAGTCTTTGTGGCATCG | |
| p762 | AGAACTGATCGCCGTCGAGCACC | |
| p767 | TTGCAGCGAAGGAAAGAGCGGCC | |
| p768 | TCGCTTGTGTCGCTCCTGTCAGG | |
| p825 | AGGCGAGCCGCTGGGGGTGGTGC | |
| p826 | TTCGATCATCAGCCGGTGCC | |
| p859 | TGACGGCTGAACAGGGCAAG | |
| p860 | CGCCTTGCGTGTGATCATTG | |
| p861 | CGGGGCGATCTTGCGAGTGA | |
| p862 | CGAAATGGGCAAGCCGTTCC | |
| p890 | CCAGCAGACCGAAGTGACGA | |
| p891 | GGCACCATGTAGAAGCCGTG | |
| p899 | GGCGAAGATCATTAGCCACC | |
| p900 | ATAGTAGGCGTCACCCGCTC | |
| p992 | TTCCTGCTGGCAAAGACCGC | |
| p993 | GGTTCTGCCTGCGAAGGGCT | |
| p994 | CCTCTGGTGCGTCAATGCCG | |
| p995 | AGCGCGGGTTCGACGGATTC | |
| p1000 | AGGAATTTGCAGCCCGACCG | |
| p1240 | CTCGTCCTGGATGGTGATTGG | |
| p1241 | GGAATCAATGCGCGAGACG |
Restriction sites are shown in bold type.
Genetic modification of bacterial strains.
All DNA cloning and analysis was performed as previously described (38). PCRs were conducted in 30 μl, using RedTaq ReadyMix (Sigma), 10 to 30 ng genomic DNA, and 0.3 μM each primer. The cycling conditions were as follows: 1 step of 3 min at 95°C; 30 cycles, with 1 cycle consisting of 45 s at 95°C, 45 s at 58°C, and 2 min at 72°C; and a final extension step of 5 min at 72°C.
15N labeling and amino acid analysis.
Six plants inoculated with R. leguminosarum bv. viciae 3841 were harvested after 3 weeks and put into sealed 100-ml Schott bottles. 15N2 gas (29 ml into a total volume of 145 ml) was injected into the bottles and incubated for 30 min. Nodules (∼120 mg from each plant) were picked and macerated in 500 μl of 50 mM phosphate-300 mM sucrose buffer (pH 7.4). Plant debris was separated by centrifugation (1,000 rpm for 5 min). Supernatant (250 μl) was centrifuged (13,000 rpm for 5 min) to pellet the bacteroids. The pellet was resuspended in 500 μl of boiling ethanol and incubated for 20 min at 75°C. Cell debris was removed by centrifugation (13,000 rpm for 10 min) and reextracted with 80% ethanol at 75°C for 20 min. Supernatants were combined, vacuum dried, resuspended in 0.05 M HCl, and loaded on SP Sephadex C25 columns. The columns were washed with 3 ml of water, and amino acids were eluted with 0.2 M NH4OH. The eluates were vacuum dried and analyzed as described previously (26).
Bacteroid isolation for enzyme assays.
Bacteroids were separated on Percoll gradients. Percoll (35 ml of 57%) was prepared in 40 mM HEPES buffer (pH 7.2) and spun for 45 min at 50,000 relative centrifugal force (RCF) at 4°C to form gradients. Fresh nodules (2 to 5 g [wet weight]) were macerated in 2 ml of 40 mM HEPES buffer (pH 7.2) with a pestle and mortar. The nodule debris was pressed through four layers of muslin, and 1.5-ml portions of bacteroid solution were loaded on the Percoll gradients and spun for 15 min at 50,000 RCF at 4°C. The bacteroid layer (5 to 10 ml) was taken from the gradient, diluted with 40 mM HEPES buffer (pH 7.2), and recollected by centrifugation. The resulting bacteroid pellets were frozen in liquid nitrogen and kept at −80°C.
Preparation of cell-free protein extracts.
Cell extracts were prepared from frozen bacteroid pellets or from cell pellets harvested from overnight cultures (50 ml). Pellets were resuspended in 1 ml of 40 mM HEPES buffer (pH 7.5) with 1 mM dithiothreitol. The cell suspension was added to matrix B tubes (Bio 101) and disrupted in a FastPrep FP120 ribolyzer (Bio 101). The ribolyzer was run twice at a speed of 6.5 for 30 s with intermediate cooling on ice for 2 min. The debris was spun down at 17,500 RCF at 4°C for 45 min. Protein concentrations were determined using Bradford assays and a bovine serum albumin standard (0 to 5 μg) (4).
Enzyme assays.
GABA:2-oxoglutarate aminotransferase (50), GABA:pyruvate aminotransferase, and succinate semialdehyde dehydrogenase (SSDH) were measured as described elsewhere (31). Glutamate:pyruvate aminotransferase (GPT) was assayed in 100 mM phosphate (pH 7.5) with 0.2 mM NADH, 0.05 mM pyridoxal phosphate, 1.8 U glutamate dehydrogenase (catalog no. G-2626; Sigma), 100 mM NH4Cl, and 75 mM pyruvate; finally, 50 mM glutamate was added to start the reaction. Nodule plant cytosol samples were prepared by spinning nodule debris at 2,000 RCF for 10 min.
Microarray and qRT-PCR analysis.
Bacteroids were harvested from nodules of 28-day-old pea plants inoculated with R. leguminosarum bv. viciae 3841. Cultures of strain R. leguminosarum bv. viciae 3841 were grown in AMS plus succinate and NH4Cl (AMS-succinate-NH4Cl) and harvested in log phase (optical density at 600 nm of ∼0.3). To isolate RNA, cells were resuspended in RNAprotect (RNA stabilization reagent) as described by the manufacturer (Qiagen), and contaminating DNA was removed by on-column treatment with RNase-free DNase (Qiagen). The RNA was quantified using an Experion automated electrophoresis station (Bio-Rad). RNA (250 ng) was amplified with the SenseAmp RNA amplification kit (Genisphere). The resulting RNA was again quantified and diluted to a concentration of 80 ng/μl. Primers were designed using VectorNTI 9 (Invitrogen) to yield amplicon sizes of ∼200 bp. The primers p1240/p1241, p992/p993, p994/p995, p859/p860, p861/p862, and p825/p826 (Table 1) were used for quantitative reverse transcription-PCR (qRT-PCR) analysis of the gabT, opaA, opaB, gabD1, gabD2, and gabD4 genes, respectively. The qRT-PCRs were performed in triplicate using the QuantiTect SYBR green RT-PCR kit (Qiagen, Germany) in reaction mixtures (20 μl) which contained primers at 0.5 μM and 80 ng of RNA. Controls lacking reverse transcriptase were included for each RNA sample. The PCR program was as follows: 30 min at 50°C, 15 min at 95°C, and 35 cycles, with 1 cycle consisting of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. The induction of each gene was calculated as 2 to the power of the ΔCT (difference of threshold cycle [CT] values between the two conditions analyzed). Samples were calibrated against the absolute level of RNA.
Duplicate independent cultures of R. leguminosarum bv. viciae 3841 were grown on AMS-succinate-NH4Cl or AMS plus succinate and GABA (AMS-succinate-GABA) and harvested at mid-logarithmic growth (optical density at 600 nm of 0.4), and RNA was isolated as described above. Microarray analysis was then performed as previously described (12).
GabD expression.
Five gabD genes (gabD1 to gabD3 and gabD5 and gabD6) were amplified with primers p743/p744, p745/p746, p747/p748, p751/p752, and p741/742, respectively, using the high-fidelity polymerase BIO-X-ACT (Bioline) and cloned into pCR2.1 (Invitrogen). The primers included restriction sites (Table 1) that were used in cloning the PCR products into pRK415 to generate pRU1818-1820 and pRU1822-23. They were conjugated into R. leguminosarum bv. viciae 3841 to produce strains RU3845-3847 and RU3849-3950, which were assayed for enzyme expression.
Mutant isolation.
In R. leguminosarum bv. viciae 3841, a gabT deletion mutant was isolated; this mutant has no antibiotic marker linked to the deletion. To do this, a 2,425-bp fragment was amplified, using primers p761/p762, and cloned into pCR8-GW-TOPO (Invitrogen). A deletion was made by replacing a 387-bp XmnI/SmaI fragment of the gabT open reading frame with a SmaI-digested ΩTet cassette (8). A PsiI fragment was then transferred into the SmaI-digested pJQ200SK vector (33) (forming pRU1882) and used to generate strain RU2516 (gabT::ΩTet) by selecting for recombination using the sac mutagenesis strategy as previously described (24). In parallel, the fragment was cloned into pJQ200SK without the ΩTet cassette, generating pRU1884. This plasmid was used to delete the ΩTet marker from strain RU2516, resulting in the antibiotic marker-less strain RU2518 (ΔgabT).
An opaA deletion mutant was constructed in the RU2518 (ΔgabT) background. A 2,610-bp fragment of opaA was amplified using primers p899/p900 and cloned into pGEM-T (Promega), and a deletion was made by replacing the 174-bp BglII/BclI fragment of opaA with a BamHI-digested ΩSpec cassette (8). A SphI/SpeI fragment was cloned into pJQ200SK, forming pRU1891, and used to generate strain RU3922 (ΔgabT opaA::ΩSpec). Plasmid pRU1891 was digested with SanDI and religated, deleting the Spec marker but leaving the Ω elements (pRU1933). This plasmid was used via the sac sucrose selection strategy to remove the ΩSpec cassette in strain RU3922, resulting in an antibiotic marker-less strain, RU4011 (ΔgabT ΔopaA).
An opaB deletion mutant was constructed in the RU4011 (ΔgabT ΔopaA) background. To do this, a 2,481-bp fragment of opaB was amplified using primers p676/p1000 and cloned into pGEM-T Easy (Promega). A deletion was made by replacing a 423-bp BclI fragment with a BamHI-digested ΩSpec cassette. A NotI fragment was transferred into pJQ200SK, producing pRU1934, and was used to generate strain RU4012 (ΔgabT ΔopaA opaB::ΩSpec).
A gabD2 antibiotic resistance marker-less deletion mutant was constructed in R. leguminosarum bv. viciae 3841. A 2,482-bp fragment using primers p759/p760 was amplified and cloned into pCR8-GW-TOPO (Invitrogen), and a deletion was made by replacing a 420-bp MslI fragment with a SmaI-digested ΩTet cassette (8). A PsiI fragment was then transferred into SmaI-digested pJQ200SK, producing pRU1883, and used to generate strain RU2517 (gabD2::ΩTet). In parallel, the fragment was cloned without the ΩTet cassette into pJQ200SK, generating pRU1885. The ΩTet marker was deleted by homologous recombination of RU2517 with pRU1885, forming strain RU2519 (ΔgabD2).
A 939-bp internal gabD1 fragment was amplified using primers p767/p768 and cloned into pCR2.1 (Invitrogen). A BamHI/XbaI fragment was transferred into pK19mob (40) to produce pRU1892, and this was conjugated into the RU2519 background. A single-crossover insertion mutant RU4016 (ΔgabD2 pK19mobgabD1) was isolated on TY agar with Str and Nm. The insertion was also introduced into R. leguminosarum bv. viciae 3841 to generate strain RU4104 (pK19mobgabD1).
A gabD4 mutation was introduced into the RU2519 (ΔgabD2) background. A 2,510-bp fragment of gabD4 was amplified using primers p890/p891 and cloned into pCR2.1 (Invitrogen). An EcoRI-digested ΩTet cassette was then cloned into an MfeI site within gabD4. A SacI/XbaI fragment was transferred into pJQ200SK, producing pRU1890, and this was used to generate strain RU3924 (ΔgabD2 gabD4::ΩTet) by sac selection. Conjugation of pK19mobgabD1 (pRU1892) into strain RU3924 and selection for single-crossover mutants was performed to generate strain RU3930 (ΔgabD2 gabD4::ΩTet pK19mobgabD1).
All mutations were checked by PCR using a primer specific to a region of DNA either up- or downstream of the crossover event leading to the mutation and a primer that is specific to the inserted fragment. Antibiotic resistance marker-less deletion mutants were checked with two primers specific for DNA on either side of the deletion, yielding a smaller fragment than obtained with R. leguminosarum bv. viciae 3841 template DNA.
Acetylene reduction assay.
Acetylene reduction was performed with six plants for each strain as described previously (1).
Microarray data accession number.
The microarray data were deposited in Array Express with an accession number of E-MEXP-1894.
RESULTS
15N2 labeling.
Amino acids that are actively metabolized in bacteroids should be highly labeled with 15N2. To assess the turnover of amino acids in bacteroids, whole pea plants were therefore incubated with 15N2. Whole plants were used because it is important for the system to be as undisturbed as possible. Isolated bacteroids are subject to membrane damage (11, 36), and even detached nodules undergo rapid physiological changes, such as the shutdown of the oxygen diffusion barrier (6, 48). Plants were incubated in 15N2 for 30 min to allow steady-state labeling. The amino acid concentrations obtained in this study were well within the range of those found in a previous study which used detached pea nodules and 14CO2 labeling (35) or alfalfa nodules formed by Sinorhizobium meliloti (10).
In the plant cytosol, glutamate, alanine, GABA, and glutamine (in this order) were the most highly labeled amino acids as determined by the atom percent excess (APE) of 15N over natural abundance (Table 2). The high degree of labeling was expected for glutamate and glutamine, because they are the direct ammonia assimilation products formed by the combined action of glutamine synthetase and glutamate synthase. Alanine is easily labeled with glutamate pyruvate aminotransferase (Fig. 1), which is highly active in the plant cytosol (5.71 μmol min−1 mg protein−1; standard error of the mean [SEM], 0.18 μmol min−1 mg protein−1; n = 3). The most abundant amino acids were (in order) asparagine, glutamate, alanine, and GABA (Table 2), which is consistent with previous work (10, 35). The labeling (APE) of asparagine was much lower than that of glutamate or glutamine, but this probably reflects its very large pool size.
TABLE 2.
15N enrichment in amino acids in bacteroids and the plant cytosol
| Amino acid | Plant cytosol
|
Bacteroid
|
||||
|---|---|---|---|---|---|---|
| Mean APEa | Mean total amino acidb | Total amino acid (%) | Mean APE | Mean total amino acid | Total amino acid (%) | |
| Glutamate | 6.48 (±0.29) | 2,661.1 (±220.6) | 12.4 | 4.37 (±0.28) | 51.0 (±7.7) | 10.4 |
| Alanine | 5.89 (±0.30) | 603.5 (±60.2) | 2.8 | 1.78 (±0.06) | 93.4 (±20.6) | 19.0 |
| GABA | 4.31 (±0.43) | 497.4 (±32.8) | 2.3 | 4.52 (±0.34) | 7.9 (±2.0) | 1.6 |
| Glutamine | 3.55 (±0.40) | 72.0 (±8.4) | 0.3 | 1.1 (±0.1) | 0.2 | |
| Proline | 1.07 (±0.54) | 71.4 (±5.0) | 0.3 | 0.50 (±0.67) | 3.4 (±0.5) | 0.7 |
| Isoleucine | 1.06 (±0.23) | 216.2 (±35.6) | 1.0 | 0.84 (±0.21) | 20.8 (±6.1) | 4.2 |
| Leucine | 1.02 (±0.23) | 254.5 (±41.8) | 1.2 | 0.79 (±0.21) | 24.5 (±7.2) | 5.0 |
| Asparagine | 1.01 (±0.29) | 15,711.9 (±1,179.0) | 73.2 | 0.79 (±0.08) | 223.9 (±19.2) | 45.6 |
| Phenylalanine | 0.94 (±0.89) | 232.5 (±24.1) | 1.1 | 2.68 (±0.85) | 8.2 (±2.5) | 1.7 |
| Serine | 0.91 (±0.15) | 302.4 (±25.3) | 1.4 | 0.54 (±0.24) | 7.5 (±0.7) | 1.5 |
| Glycine | 0.47 (±0.13) | 212.8 (±19.7) | 1.0 | 0.43 (±0.12) | 15.1 (±1.7) | 3.1 |
| Valine | 0.31 (±0.16) | 197.1 (±22.7) | 0.9 | 0.33 (±0.38) | 10.5 (±2.7) | 2.1 |
| Threonine | 0.26 (±0.21) | 156.0 (±8.8) | 0.7 | −0.16 (±0.20) | 5.8 (±0.9) | 1.2 |
| β-Alanine | 0.24 (±0.33) | 101.4 (±7.7) | 0.5 | 0.65 (±0.23) | 5.8 (±1.3) | 1.2 |
| Aspartate | 0.18 (±0.26) | 26.3 (±2.3) | 0.1 | 1.34 (±0.63) | 3.1 (±0.8) | 0.6 |
| Tyrosine | −0.53 (±0.35) | 162.8 (±17.4) | 0.8 | 0.41 (±0.83) | 9.2 (±2.5) | 1.9 |
| Total | 21,479.3 | 100 | 491.2 | 100 | ||
The mean atom percent excess and standard error of the mean (shown in parentheses) from six experiments are shown.
Total amino acids are given in nanomoles/gram of nodule (wet weight). The mean and standard error of the mean (shown in parentheses) from six experiments are shown.
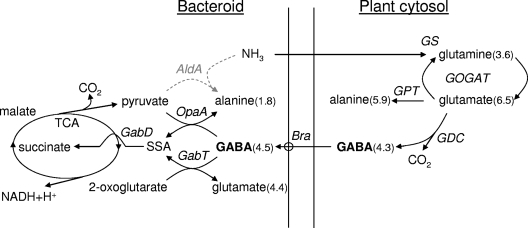
FIG. 1.
Pathway of GABA metabolism in R. leguminosarum bv. viciae 3841. Values in parentheses are the APE values taken from Table 2. Abbreviations: GS, glutamine synthetase; GPT, glutamate pyruvate aminotransferase; GOGAT, glutamate oxoglutarate amidotransferase; GDC, glutamate decarboxylase; Bra, branched-chain amino acid transporter; GabT, GABA oxoglutarate aminotransferase; OpaA, omega amino acid pyruvate aminotransferase; GabD, succinate semialdehyde dehydrogenase; AldA, alanine dehydrogenase; SSA, succinate semialdehyde.
GABA and glutamate were the most highly labeled amino acids in the bacteroid fraction, followed by alanine and aspartate (Table 2). The most abundant amino acids were found to be asparagine, alanine, and glutamate, as reported in previous work (10, 35). It is unclear whether the asparagine is derived from transport, de novo synthesis, or unavoidable contamination from the plant cytosol during the purification process. Alanine could be derived either from transamination or directly from bacteroid assimilation via alanine dehydrogenase (AldA) (Fig. 1), which is known to be active in bacteroids (1). Very similar labeling patterns were produced when peas were infected with R. leguminosarum bv. viciae strain A34, although GABA was more abundant at 8.9% of the bacteroid amino acid pool compared to 1.6% in R. leguminosarum bv. viciae 3841 (data not shown). The single discrepancy in labeling between the strains is that phenylalanine had a significant APE in R. leguminosarum bv. viciae 3841 (2.7%) but was barely labeled in A34 (0.2%).
Although the absolute amount of GABA in R. leguminosarum bv. viciae 3841 bacteroids was low, the high APE suggests that it turns over rapidly and therefore may play an important role in amino acid metabolism or amino acid cycling (26). It was also reported to be the most highly 15N-labeled amino acid in pea nodules in a previous study (41), but in that study bacteroids and plant cytosol were not separated. GABA is most likely derived from the glutamate pool of the plant cytosol via α-decarboxylation by glutamate decarboxylase (Fig. 1) (43). It is unlikely that GABA can be derived from glutamate decarboxylation inside the bacteroid, because despite the reports of some, albeit extremely low, activity (16, 19, 28, 37), there are no candidate genes available in any of the genome sequences of rhizobia that encode enzymes to carry out the reaction (13, 14, 21, 22, 49). GABA transport into the bacteroid can be performed by the BraDEFGC ABC transport system in R. leguminosarum bv. viciae 3841, which is expressed during symbiosis (18). GABA metabolism proceeds via transamination, releasing succinate semialdehyde. This is oxidized to succinate, which feeds into the tricarboxylic acid cycle (TCA) to supply carbon and energy (Fig. 1). The GABA-metabolizing enzyme GABA:2-oxoglutarate aminotransferase is highly upregulated in pea bacteroids (31, 36). In addition, GABA:pyruvate aminotransferase and succinate semialdehyde dehydrogenase (SSDH), are present in R. leguminosarum VF39 laboratory cultures grown with GABA as the sole carbon and nitrogen source (31).
Symbiotic enzyme activities.
Since labeling showed that GABA is actively metabolized by pea bacteroids, the pathways of GABA metabolism were investigated by enzyme analysis. The only known pathways of GABA utilization are via transamination of its amino group to the keto acid acceptors 2-oxoglutarate and pyruvate, producing glutamate and alanine, respectively, with release of succinate semialdehyde. Activities of potential GABA-metabolizing enzymes were determined in various pea bacteroid preparations (Table 3). GABA:2-oxoglutarate aminotransferase was very active in bacteroids (304 nmol min−1 mg protein−1), as was GABA:pyruvate aminotransferase (144 nmol min−1 mg protein−1). SSDH activity was also very high with a preference for NAD+ as the electron acceptor (NAD+-dependent 371 nmol min−1 mg protein−1; NADP+-dependent 73 nmol min−1 mg protein−1).
TABLE 3.
Enzyme activities and growth rates of R. leguminosarum bv. viciae 3841 and mutant strainsa
| Strain and preparation or genotype | Enzyme activityb
|
Doubling time (h)c | |||
|---|---|---|---|---|---|
| GABA:2-oxoglutarate transaminase | GABA:pyruvate transaminase | SSDH
|
|||
| NAD+-dependent | NADP+-dependent | ||||
| R. leguminosarum bv. viciae 3841 | |||||
| Bacteroids | 304 (±27) | 144 (±33) | 371 (±46) | 73 (±12) | N/A |
| Grown on glucose and NH4Cl | 8d | 23 (±4) | 7 (±3) | 9 (±2) | 3.3 (±0.0) |
| Grown on glucose and GABA | 20 (±4) | 183 (±19) | 59 (±3) | 78 (±3) | 4.6 (±0.1) |
| Mutants | |||||
| RU2518 (ΔgabT) | 2 (±2) | ND | ND | ND | 6.1 (±0.2) |
| RU3922 (ΔgabT opaA::ΩSpec) | ND | 73 (±11) | ND | ND | 11.3 (±0.7) |
| RU4012 (ΔgabT ΔopaA opaB::ΩSpec) | N/A | N/A | N/A | N/A | >30 |
| RU2519 (ΔgabD2) | ND | ND | 87 (±5) | 90 (±9) | 8.2 (±0.5) |
| RU4016 (ΔgabD2 pK19mobgabD1) | ND | ND | 34 (±1) | 19 (±4) | 13.7 (±1.6) |
| RU3930 (ΔgabD2 pK19mobgabD1 gabD4::ΩTet) | N/A | N/A | N/A | N/A | >30 |
Mutant cultures were grown on AMS-glucose-GABA.
Enzyme activity is shown as nmol min−1 mg protein−1. The standard errors of the means are shown in parentheses (n ≥ 3). ND, not determined; N/A, not applicable.
The standard errors of the means are shown in parentheses. N/A, not applicable.
n = 2.
Identification of the GABA aminotransferases.
Since the GABA-metabolizing enzymes were very active in pea bacteroid preparations, the proteins likely to be responsible were identified from the R. leguminosarum bv. viciae 3841 genome sequence. There are several candidates for GABA aminotransferase genes in the genome sequence (49). The GABA:2-oxoglutarate aminotransferase gene was annotated as gabT (RL0102) because of its identity to the previously identified gabT gene in R. leguminosarum bv. viciae VF39 (31). In R. leguminosarum bv. viciae 3841, gabT is part of an operon with an SSDH (gabD2; RL0101) that is conserved in S. meliloti 1021 and Rhizobium etli CFN42 (13, 14). A gabT homologue is present in Mesorhizobium loti MAFF303099 (21) but is not located near either of the two annotated gabD genes. In contrast, Bradyrhizobium japonicum USDA110 lacks an obvious gabT (22).
A gabT mutant of R. leguminosarum VF39 had no growth deficiency in medium containing GABA as a carbon and nitrogen source (31). However, this might be explained by the presence of an active GABA:pyruvate aminotransferase, although the gene(s) responsible was not identified. To identify such genes, microarray analysis was conducted on R. leguminosarum bv. viciae 3841 grown in AMS plus glucose and GABA (AMS-glucose-GABA) versus AMS plus glucose and NH4Cl (AMS-glucose-NH4Cl). The gabT gene (RL0102) was induced by GABA 7.6-fold (P value of 0.03), and the gabD2 gene was induced by GABA 4.7-fold (P value of 0.07). Additionally, pRL120419, which is annotated as coding for an ω-amino acid:pyruvate aminotransferase, was induced 8.1-fold (P value of 0.03). We tentatively named this gene opaA (omega amino acid:pyruvate aminotransferase). OpaA has 31% identity to GabT but has the highest identity to RL3538 (60%). Although RL3538 showed no induction on the array, we tentatively named it opaB. The increases in gabT and opaA expression when grown on glucose and GABA versus glucose and NH4Cl were confirmed by qRT-PCR (11.2- and 14.3-fold upregulation; P values of 0.01 and 0.08, respectively).
Growth phenotypes of GABA aminotransferase mutants.
Although R. leguminosarum bv. viciae 3841 grows poorly on GABA as both a carbon and nitrogen source, it grows well when used as a nitrogen source with either glucose or succinate as a carbon source. Therefore, AMS-glucose-GABA was used as the culture medium to determine whether mutation of the GABA aminotransferases (and subsequently, the SSDHs) disrupts GABA metabolism.
Since gabT (RL0102) was identified as the gene coding for GABA:2-oxoglutarate aminotransferase, a ΔgabT mutant (RU2518) was made. Although this mutant expressed only trace amounts of GABA:2-oxoglutarate aminotransferase activity in cultures grown in AMS-glucose-GABA (Table 3), it showed only slightly elevated doubling times during growth in the same medium. This agrees with previous results from R. leguminosarum VF39 suggesting that GABA:2-oxoglutarate transamination is not the only pathway for GABA breakdown (31).
Subsequently, a double mutant, RU3922 (ΔgabT opaA::ΩSpec), was isolated. While strain RU3922 grew in AMS-glucose-GABA, it had a significantly elevated doubling time (Table 3). GABA:pyruvate aminotransferase activity was not absent in RU3922 but dropped significantly (Table 3), making OpaA a strong candidate for a GABA:pyruvate aminotransferase. Since the remaining activity might be due to OpaB, a triple mutant (RU4012) (ΔgabT ΔopaA opaB::ΩSpec) was isolated. Growth of RU4012 was abolished in AMS-glucose-GABA. The lack of growth prevented a simple assay for complete loss of Opa activity in this strain. However, it suggests that opaB codes for a third GABA:pyruvate aminotransferase not previously described. It also demonstrates that GABA:pyruvate transamination is a major pathway for GABA utilization in cultures grown with glucose and GABA.
Identification of succinate semialdehyde dehydrogenases.
Succinate semialdehyde is released from GABA in a transamination reaction, and it is further oxidized to succinate by SSDH. There are six possible genes (gabD) coding for putative SSDHs annotated in R. leguminosarum bv. viciae 3841 (49): pRL100134 (tentatively named gabD1, following the order used in S. meliloti 1021 [13]), RL0101 (gabD2), pRL120603 (gabD3), pRL100252 (gabD4), pRL120628 (gabD5), and pRL120044 (tentatively named gabD6 and similar to SMb20424 but not annotated in S. meliloti 1021). gabD2 is downstream of the above characterized GABA:2-oxoglutarate aminotransferase (gabT [RL0102]) located on the chromosome. gabD1 and gabD4 are located on the sym plasmid, pRL10, and are both likely to form part of an operon. GabD1 of R. leguminosarum bv. viciae 3841 is very similar to AttK of Agrobacterium tumefaciens, and both genes are part of an attKLM operon, which is responsible for homoserine lactone degradation and is induced by GABA (5). GabD1, GabD2, and GabD4 are very similar to each other in both R. leguminosarum bv. viciae 3841 and S. meliloti 1021, as are GabD2 and GabD4 in R. etli CFN42 (GabD1 is missing) (14). The R. leguminosarum bv. viciae 3841 GabD3, GabD5, and GabD6 proteins have homologues in S. meliloti 1021 and R. etli CFN42, and GabD3 is similar to Mll5719 of M. loti MAFF303099.
Each gabD gene (except gabD4) was cloned into pRK415 and constitutively expressed in R. leguminosarum bv. viciae 3841 under the control of the lacZ promoter. SSDH activities were determined from cultures grown in AMS-succinate-NH4Cl, with wild-type R. leguminosarum bv. viciae 3841 used as a control (Table 4). Strain RU1816, selected from R. leguminosarum bv. viciae 3841 for fast growth on GABA as the sole carbon and nitrogen source, has constitutive expression of gabD4 (47) and was used to determine gabD4-related SSDH activity.
TABLE 4.
Succinate semialdehyde dehydrogenase activities of gabD genes when overexpressed in R. leguminosarum bv. viciae 3841
| Strain | Overexpressed genea | SSDH activityb
|
NAD+/ NADP+ SSDH activity ratio | |
|---|---|---|---|---|
| NAD+-dependent | NADP+-dependent | |||
| RU3845 | gabD1 | 137 (±4) | 174 (±5) | 0.8 |
| RU3846 | gabD2 | 337 (±11) | 65 (±4) | 5.2 |
| RU3847 | gabD3 | 55 (±13) | 38 (±8) | 1.4 |
| RU1816c | gabD4 | 189 | 61 | 3.1 |
| RU3849 | gabD5 | 139 (±15) | 21 (±1) | 6.7 |
| RU3850c | gabD6 | 6 | 7 | 0.9 |
| R. leguminosarum bv. viciae 3841 | None | 7 (±3) | 9 (±2) | 0.8 |
Overexpressed genes were cloned in pRK415 and expressed from the lacZ promoter, except for gabD4, which was expressed from its native chromosomal promoter.
Enzyme activity is given in nmol min−1 mg protein−1. The mean and standard error of the mean (shown in parentheses) of ≥3 experiments are shown.
n = 2.
All the annotated gabD genes expressed SSDH activity, with the exception of gabD6. The absence of detectable SSDH activity from gabD6 may either be real or caused by a failure to express the gene properly; however, this was not investigated further. The activities of the enzymes differed, as did the ratio of activity with NAD+ or NADP+ (Table 4). GabD2 had the highest NAD+-dependent SSDH activity (337 nmol min−1 mg protein−1), with a NAD+/NADP+ ratio similar to that measured in bacteroid preparations (5.1). GabD1 had the highest NADP+-dependent SSDH activity (174 nmol min−1 mg protein−1) together with a similar NAD+-dependent activity (137 nmol min−1 mg protein−1). GabD4 also had high NAD+-dependent activity (189 nmol min−1 mg protein−1), although this absolute value is not directly comparable, because the native gabD4 gene was expressed from its chromosomal promoter. GabD5 had high NAD+-dependent activity (139 nmol min−1 mg protein−1) with the highest NAD+/NADP+ ratio (6.7). GabD3 had relatively low values for both activities (55 and 38 nmol min−1 mg protein−1, respectively).
Growth phenotypes of SSDH mutants.
Because of the strong symbiotic expression of GabD2 reported in earlier work (36), a ΔgabD2 mutant (RU2519) was isolated initially. Strain RU2519 (ΔgabD2) had a moderately increased doubling time in cultures grown in AMS-glucose-GABA compared to that of strain R. leguminosarum bv. viciae 3841 (Table 3). This is very similar to the growth phenotype of RU2518 (ΔgabT), where the mutation might be polar on the contiguous gabD2. SSDH activities were slightly increased in RU2519 compared to R. leguminosarum bv. viciae 3841, which might be a result of overcompensation by other GabD proteins (Table 4).
There is high NADP+-dependent SSDH activity in cultures of both wild-type R. leguminosarum bv. viciae 3841 and RU2519 grown in AMS-glucose-GABA (Table 3). This suggests a major contribution by gabD1, which has a high NADP+-dependent activity. The double mutant RU4016 (ΔgabD2 pK19mobgabD1) grew in AMS-glucose-GABA (with Nm at 40 μg/ml) with a significantly elevated doubling time, and its SSDH activity was reduced (Table 3). GabD4, which is closely related to GabD1 and GabD2, was mutated, and a triple mutant was isolated (RU3930 [ΔgabD2 pK19mobgabD1 gabD4::ΩTet]). Strain RU3930 showed no growth in AMS-glucose-GABA cultures (with Nm at 40 μg/ml). This indicates that GabD4 contributes to SSDH activity in cultures grown in AMS-glucose-GABA. It also indicates that GabD3 and GabD5 are not relevant for free-living growth of R. leguminosarum bv. viciae 3841 with GABA as the sole nitrogen source.
Expression in bacteroids.
Since GabT, OpaA, OpaB, GabD2, GabD1, and GabD4 all contribute to growth in liquid cultures with GABA as the sole nitrogen source, the expression of these genes was measured in bacteroids by means of qRT-PCR. Gene expression was compared for bacteroids and free-living cultures grown on succinate and NH4Cl because dicarboxylates are the carbon sources for bacteroid respiration.
Expression of gabT and gabD2 was highly upregulated in bacteroids, compared to bacteria grown on AMS-succinate-NH4Cl (Table 5). This is consistent with both upregulation of a gabT-gusA fusion and the identification of GabT and GabD2 in the alfalfa bacteroid proteome (31, 36). qRT-PCR also showed that opaA and gabD1 were significantly upregulated, which is consistent with both GABA:pyruvate aminotransferase activity (Table 3) and SSDH activity in bacteroids of the gabD mutants (see below).
TABLE 5.
Induction of genes involved in GABA metabolism in bacteroids
| Gene | Inductiona | P valueb |
|---|---|---|
| gabT | 158 (±33) | 0.0016 |
| opaA | 4.1 (±0.7) | 0.0133 |
| opaB | 1.4 (±0.6) | 0.6754 |
| gabD1 | 2.2 (±0.1) | 0.0022 |
| gabD2 | 240 (±125) | 0.0092 |
| gabD4 | 4.0 (±2.7) | 0.3010 |
Induction of gene expression was measured by qRT-PCR in triplicate experiments and is given as the ratio for bacteroids versus free-living cultures grown in succinate and NH4Cl. The induction was calculated from the CT values of each paired experiment and given as an average (standard error of the mean shown in parentheses) from three experiments.
P values are the results of a paired t test comparing the CT values of each paired experiment.
Symbiotic phenotypes and enzyme activities in mutant bacteroids.
All the mutants generated in this work (Table 3) were tested on pea plants for their symbiotic performance. After 6 weeks of growth, plants infected with these mutants were visually indistinguishable from plants infected with R. leguminosarum bv. viciae 3841. Three-week-old pea plants infected with R. leguminosarum bv. viciae 3841, RU4012 (ΔgabT ΔopaA opaB::ΩSpec), and RU3930 (ΔgabD2 pK19mobgabD1 gabD4::ΩTet) reduced acetylene to ethylene at 4.48 μmol h−1 plant−1 (SEM, 0.76 μmol h−1 plant−1; n = 6), 5.04 μmol h−1 plant−1 (SEM, 0.65 μmol h−1 plant−1; n = 6), and 4.13 μmol h−1 plant−1 (SEM, 0.43 μmol h−1 plant−1; n = 6), respectively. This shows there was no significant change in the apparent capacity for N2 fixation.
All mutant strains were recovered from nodules and retained their antibiotic resistance markers. Bacteroids of RU4012 (ΔgabT ΔopaA opaB::ΩSpec) had undetectable GABA:2-oxoglutarate aminotransferase activity, and GABA:pyruvate aminotransferase activity dropped from 144 to 29.6 nmol min−1 mg protein−1 (n = 2). This residual GABA:pyruvate aminotransferase activity is most likely a result of background in the assay used. Uninduced cultures of R. leguminosarum bv. viciae 3841 grown on AMS-glucose-NH4Cl have a background of approximately 23 nmol min−1 mg protein−1 (Table 3).
Bacteroids of RU3930 (ΔgabD2 pK19mobgabD1 gabD4::ΩTet) did not contain any detectable NAD+- or NADP+-dependent SSDH activity. Likewise, these activities were undetectable in bacteroids of RU4016 (ΔgabD2 pK19mobgabD1). Thus, bacteroids do not have SSDH activity attributable to GabD3, GabD4, or GabD5. Bacteroids of RU2519 (ΔgabD2) had only ∼14% of NAD+- and NADP+-dependent SSDH activity compared to R. leguminosarum bv. viciae 3841. This activity is due to GabD1 and is consistent with the qRT-PCR results (Table 5). The gabD1 mutant, RU4101 (pK19mobgabD1), had ∼87% of both SSDH activities present due to GabD2.
DISCUSSION
GABA is an abundant amino acid in nodules (2, 10, 28). Furthermore, 15N2 labeling showed that GABA is rapidly labeled in whole pea nodules (41). Here we demonstrate that it is also rapidly labeled in pea bacteroids (Table 2) and the enzymes for its utilization are highly expressed (Table 3). The GabT activities reported in this study are much higher than in a previous report (31), probably because a more sensitive assay was used. It is easy to explain why GABA is labeled in plants during 15N2 incubation because it is derived from glutamate decarboxylation and glutamate itself is directly labeled by the plant's ammonia-assimilating glutamine synthetase-glutamate oxoglutarate amidotransferase system (Fig. 1). However, its origin is less clear in bacteroids because they lack glutamate decarboxylase and cannot synthesize GABA directly from glutamate. GABA can be taken up by the BraDEFGC ABC transport system located in the bacteroid membrane, which has a high affinity for GABA transport (18). This would enable GABA to be transported from the plant cytosol across the symbiosome membrane into the bacteroid. The almost identical labeling (APE) of GABA in the plant and bacteroid is consistent with this interpretation. An alternative explanation is that GabT uses glutamate as an amino group donor for transamination of succinate semialdehyde to produce GABA and the APE values of glutamate and GABA are similar in the bacteroid. This would require that succinate semialdehyde is available in the bacteroid. While this scenario is possible, succinate semialdehyde is toxic, and cells normally catabolize it very rapidly via succinate semialdehyde dehydrogenase. It would therefore be novel for R. leguminosarum bv. viciae 3841 to produce succinate semialdehyde as a transamination acceptor for synthesis of GABA. Overall, provision of GABA by the plant is easier to rationalize, and we believe more likely, but these alternatives cannot be distinguished with the available data. It might be resolved with labeling studies using bacteroid gabT and gabT opaA opaB triple mutants, but since even triple mutants exhibited unaltered N2 fixation, these experiments were not attempted.
GabT catalyzes the transfer of the amino group from GABA to 2-oxoglutarate, forming glutamate and releasing succinate semialdehyde, which is further oxidized to succinate (Fig. 1). Depending on the rate of flux, this could add significant reductant to bacteroid energy metabolism. A second pathway in which OpaA catalyzes amino group transfer from GABA to pyruvate, forming alanine, exists. Consistent with both pathways of GABA metabolism being active in bacteroid metabolism, GabT, OpaA, GabD1, and GabD2 were upregulated during symbiosis (Table 5). GabD4 (pRL100251), mutation of which was necessary to abolish growth on GABA in liquid medium, is located downstream of a GABA transport system (Gts [GABA transport system]; pRL100248-251), and its expression is transcriptionally linked. While this operon is not inducible on medium containing GABA as a nitrogen source (47), its constitutive expression can be readily selected. Mutants rapidly accumulate in any cultures grown on GABA, probably explaining why GabD4 contributes to the observed growth of cultures. However, there was no gabD4 expression (Table 3 and reference 47) or SSDH activity due to GabD4 in bacteroids of RU4016 (ΔgabD2 pK19mobgabD1).
As part of an amino acid cycle, GABA would be an ideal amino acid to be provided by the plant to the bacteroid as it is derived from glutamate, which is the first ammonium assimilation product in the plant cytosol. Bacteroids could use GABA to form glutamate and alanine by transamination. Alanine is secreted by bacteroids so it would be a good candidate to be returned to the plant cytosol (26, 32). The succinate semialdehyde released from GABA would be rapidly metabolized by SSDH and the TCA cycle. This cycle would be perfectly carbon and nitrogen balanced (C4 in; C3 and CO2 out), and the reductive part of the TCA cycle would generate additional energy. However, both triple mutants, RU4012 (ΔgabT ΔopaA opaB::ΩSpec) and RU3930 (ΔgabD2 pK19mobgabD1 gabD4::ΩTet), which cannot use GABA even as a nitrogen source, did not show any reduction in symbiotic performance on pea plants under growth room conditions. Furthermore, the high labeling of alanine observed in plant cytosol (Fig. 1) (APE, 5.89) can be explained only by glutamate:pyruvate aminotransferase transamination in the plant from the even higher labeled glutamate pool (APE, 6.48) and not from alanine secretion by the bacteroid (APE, 1.78) (1). This is a very important point because it means that alanine cannot be a transit species for exported nitrogen in pea nodules, as has been suggested for soybean nodules (46). If alanine were the transit species for nitrogen, rather than ammonia, it would have an equal or higher APE than compounds derived from it in the plant cytosol, such as glutamate. Additionally, a mutant in both the GABA-transporting Bra system and the recently described GABA-specific transport system (Gts) (47) is not impaired in nitrogen fixation (data not shown).
The high APE of GABA in 15N2 labeling experiments indicates rapid metabolism in bacteroids. While this suggests that it might be important in amino acid cycling or as a nitrogen source for the bacteroid (26, 32, 39), the mutational studies show that GABA alone is not essential for N2 fixation in bacteroids. Crucially, the roles of Aap and Bra in nitrogen fixation by R. leguminosarum bv. viciae 3841 bacteroids in pea nodules cannot be solely due to a requirement for GABA transport or metabolism. However, this does not mean that GABA metabolism is unimportant, since bacteroid metabolism may be redundant, so that mutating any one pathway, may not prevent N2 fixation. For example, it has been observed that mutating isocitrate dehydrogenase, aconitase, or 2-oxoglutarate dehydrogenase in B. japonicum still enables N2 fixation, even though this severely disrupts the decarboxylating arm (citrate synthase, aconitase, isocitrate dehydrogenase, and 2-oxoglutarate dehydrogenase) of the TCA cycle (15, 42, 44). This may be specific to B. japonicum, since 2-oxoglutarate dehydrogenase mutants of R. leguminosarum bv. viciae 3841 are Fix− on peas (45). However, this could also be because bacteroid formation is prevented (i.e., it is a developmental, rather than metabolic, mutant). Similarly, while citrate synthase activity is required for N2 fixation by Sinorhizobium meliloti and Sinorhizobium fredii bacteroids, only 7% of its maximum activity is required in S. meliloti for effective N2 fixation (17). While these studies suggest that the decarboxylating arm of the TCA cycle may not be essential, at least in soybean bacteroids, they do not necessarily mean that it is not the main pathway for carbon flux in wild-type bacteroids. The activities of enzymes in the decarboxylating arm of the TCA cycle are highly elevated in pea bacteroids (27), and their genes are also transcriptionally upregulated (R. Karunakaran, personal communication). This suggests that the enzymes are very important in bacteroids, if not always indispensable. GABA catabolism bypasses the decarboxylating arm of the TCA cycle with the semialdehyde of GABA (succinate semialdehyde) entering the cycle after oxidation to succinate. As GABA is also very effective at providing NADH, it represents an alternative to the decarboxylating arm of the TCA cycle, at least for NADH generation in bacteroids, and might even substitute for it in TCA cycle mutants. Overall, the main result of this study is the demonstration that GABA metabolism in bacteroids is not needed for N2 fixation in pea nodules. However, the caveat is that bacteroid metabolism may be redundant, so that as indicated by 15N labeling and enzyme analysis, it may make an important contribution.
Acknowledgments
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council (grant number BBS/B/02916).
We thank Vinoy Ramachandran for help with the qRT-PCR analysis.
Footnotes
Published ahead of print on 30 January 2009.
REFERENCES
- 1.Allaway, D., E. Lodwig, L. A. Crompton, M. Wood, T. R. Parsons, T. Wheeler, and P. S. Poole. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36508-515. [DOI] [PubMed] [Google Scholar]
- 2.Barsch, A., V. Tellstrom, T. Patschkowski, H. Kuster, and K. Niehaus. 2006. Metabolite profiles of nodulated alfalfa plants indicate that distinct stages of nodule organogenesis are accompanied by global physiological adaptations. Mol. Plant-Microbe Interact. 19998-1013. [DOI] [PubMed] [Google Scholar]
- 3.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84188-198. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.Chai, Y., C. S. Tsai, H. Cho, and S. C. Winans. 2007. Reconstitution of the biochemical activities of the AttJ repressor and the AttK, AttL, and AttM catabolic enzymes of Agrobacterium tumefaciens. J. Bacteriol. 1893674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denison, R. F. 1992. Mathematical modeling of oxygen diffusion and respiration in legume root nodules. Plant Physiol. 98901-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downie, J. A., G. Hombrecher, Q. S. Ma, C. D. Knight, B. Wells, and A. W. B. Johnston. 1983. Cloned nodulation genes of Rhizobium leguminosarum determine host range specificity. Mol. Genet. Genomics 190359-365. [Google Scholar]
- 8.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52147-154. [DOI] [PubMed] [Google Scholar]
- 9.Finan, T. M., J. M. Wood, and D. C. Jordan. 1983. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J. Bacteriol. 1541403-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fougere, F., D. Le Rudulier, and J. G. Streeter. 1991. Effects of salt stress on amino acid, organic acid and carbohydrate composition of roots, bacteroids, and cytosol of alfalfa (Medicago sativa L.). Plant Physiol. 961228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, A. A., J. P. White, A. H. F. Hosie, E. M. Lodwig, and P. S. Poole. 2006. Osmotic upshift transiently inhibits uptake via ABC transporters in gram-negative bacteria. J. Bacteriol. 1885304-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, M. A., R. Karunakaran, M. E. Leonard, B. Mouhsine, A. Williams, A. K. East, J. A. Downie, and P. S. Poole. 2008. Characterization of the quaternary amine transporters of Rhizobium leguminosarum bv. viciae 3841. FEMS Microbiol. Lett. 287212-220. [DOI] [PubMed] [Google Scholar]
- 13.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lalaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293668-672. [DOI] [PubMed] [Google Scholar]
- 14.González, V., R. I. Santamaría, P. Bustos, I. Hernández-González, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramírez, V. Jiménez-Jacinto, J. Collado-Vides, and G. Dávila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 1033834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, L. S., and D. W. Emerich. 1997. The formation of nitrogen-fixing bacteroids is delayed but not abolished in soybean infected by an alpha-ketoglutarate dehydrogenase-deficient mutant of Bradyrhizobium japonicum. Plant Physiol. 1141359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, L. S., Y. Z. Li, D. W. Emerich, F. J. Bergersen, and D. A. Day. 2000. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 1822838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grzemski, W., J. P. Akowski, and M. L. Kahn. 2005. Probing the Sinorhizobium meliloti-alfalfa symbiosis using temperature-sensitive and impaired-function citrate synthase mutants. Mol. Plant-Microbe Interact. 18134-141. [DOI] [PubMed] [Google Scholar]
- 18.Hosie, A. H. F., D. Allaway, H. A. Dunsby, C. S. Galloway, and P. S. Poole. 2002. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol. 1844071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, H. N., M. J. Dilworth, and A. R. Glenn. 1990. 4-Aminobutyrate is not available to bacteroids of cowpea Rhizobium MNF2030 in snake bean nodules. Arch. Microbiol. 153455-462. [Google Scholar]
- 20.Johnston, A. W. B., and J. E. Beringer. 1975. Identification of the Rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 87343-350. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7331-338. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9189-197. [DOI] [PubMed] [Google Scholar]
- 23.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., A. Bourdes, and P. S. Poole. 2005. De novo alanine synthesis by bacteroids of Mesorhizobium loti is not required for nitrogen transfer in the determinate nodules of Lotus corniculatus. J. Bacteriol. 1875493-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodwig, E., and P. Poole. 2003. Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 2237-78. [Google Scholar]
- 26.Lodwig, E. M., A. H. F. Hosie, A. Bourdes, K. Findlay, D. Allaway, R. Karunakaran, J. A. Downie, and P. S. Poole. 2003. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422722-726. [DOI] [PubMed] [Google Scholar]
- 27.McKay, I. A., M. J. Dilworth, and A. R. Glenn. 1989. Carbon catabolism in continuous cultures and bacteroids of Rhizobium leguminosarum MNF3841. Arch. Microbiol. 152606-610. [Google Scholar]
- 28.Miller, R. W., D. G. McRae, and K. Joy. 1991. Glutamate and gamma-aminobutyrate metabolism in isolated Rhizobium meliloti bacteroids. Mol. Plant-Microbe Interact. 437-45. [Google Scholar]
- 29.Oldroyd, G. E. D., and J. A. Downie. 2004. Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5566-576. [DOI] [PubMed] [Google Scholar]
- 30.Poole, P. S., N. A. Schofield, C. J. Reid, E. M. Drew, and D. L. Walshaw. 1994. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology 1402797-2809. [DOI] [PubMed] [Google Scholar]
- 31.Prell, J., B. Boesten, P. Poole, and U. B. Priefer. 2002. The Rhizobium leguminosarum bv. viciae VF39 gamma aminobutyrate (GABA) aminotransferase gene (gabT) is induced by GABA and highly expressed in bacteroids. Microbiology 148615-623. [DOI] [PubMed] [Google Scholar]
- 32.Prell, J., and P. Poole. 2006. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 14161-168. [DOI] [PubMed] [Google Scholar]
- 33.Quandt, H. J., A. Puhler, and I. Broer. 1993. Transgenic root nodules of Vicia hirsuta--a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant-Microbe Interact. 6699-706. [Google Scholar]
- 34.Ronson, C. W., P. Lyttleton, and J. G. Robertson. 1981. C4-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc. Natl. Acad. Sci. USA 784284-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosendahl, L., C. P. Vance, and W. B. Pedersen. 1990. Products of dark CO2 fixation in pea root nodules support bacteroid metabolism. Plant Physiol. 9312-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saalbach, G., P. Erik, and S. Wienkoop. 2002. Characterisation by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics 2325-337. [DOI] [PubMed] [Google Scholar]
- 37.Salminen, S. O., and J. G. Streeter. 1990. Factors contributing to the accumulation of glutamate in Bradyrhizobium japonicum bacteroids under microaerobic conditions. J. Gen. Microbiol. 1362119-2126. [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Sarma, A. D., and D. W. Emerich. 2005. Global protein expression pattern of Bradyrhizobium japonicum bacteroids: a prelude to functional proteomics. Proteomics 54170-4184. [DOI] [PubMed] [Google Scholar]
- 40.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19—selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 41.Scharff, A. M., H. Egsgaard, P. E. Hansen, and L. Rosendahl. 2003. Exploring symbiotic nitrogen fixation and assimilation in pea root nodules by in vivo 15N nuclear magnetic resonance spectroscopy and liquid chromatography-mass spectrometry. Plant Physiol. 131367-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah, R., and D. W. Emerich. 2006. Isocitrate dehydrogenase of Bradyrhizobium japonicum is not required for symbiotic nitrogen fixation with soybean. J. Bacteriol. 1887600-7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shelp, B. J., A. W. Bown, and M. D. McLean. 1999. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 4446. [DOI] [PubMed] [Google Scholar]
- 44.Thöny-Meyer, L., and P. Künzler. 1996. The Bradyrhizobium japonicum aconitase gene (acnA) is important for free-living growth but not for an effective root-nodule symbiosis. J. Bacteriol. 1786166-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walshaw, D. L., A. Wilkinson, M. Mundy, M. Smith, and P. S. Poole. 1997. Regulation of the TCA cycle and the general amino acid permease by overflow metabolism in Rhizobium leguminosarum. Microbiology 1432209-2221. [DOI] [PubMed] [Google Scholar]
- 46.Waters, J. K., B. L. Hughes, L. C. Purcell, K. O. Gerhardt, T. P. Mawhinney, and D. W. Emerich. 1998. Alanine, not ammonia, is excreted from N2-fixing soybean nodule bacteroids. Proc. Natl. Acad. Sci. USA 9512038-12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White, J. P., J. Prell, V. Ramachandran, and P. Poole. 2009. Characterization of a γ-aminobutyric acid transport system of Rhizobium leguminosarum viciae 3841. J. Bacteriol. 1911547-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witty, J. F., L. Skøt, and N. P. Revsbech. 1987. Direct evidence for changes in the resistance of legume root nodules to O2 diffusion. J. Exp. Bot. 381129-1140. [Google Scholar]
- 49.Young, J. P., L. Crossman, A. Johnston, N. Thomson, Z. Ghazoui, K. Hull, M. Wexler, A. Curson, J. Todd, P. Poole, T. Mauchline, A. East, M. Quail, C. Churcher, C. Arrowsmith, I. Cherevach, T. Chillingworth, K. Clarke, A. Cronin, P. Davis, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, S. Whitehead, and J. Parkhill. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaboura, M., and Y. S. Halpern. 1978. Regulation of γ-aminobutyric acid degradation in Escherichia coli by nitrogen metabolism enzymes. J. Bacteriol. 133447-451. [DOI] [PMC free article] [PubMed] [Google Scholar]