Abstract
Mutational analysis revealed that members of the Clp system, specifically the ClpL chaperone and the ClpXP proteolytic complex, modulate the expression of important virulence attributes of Streptococcus mutans. Compared to its parent, the ΔclpL strain displayed an enhanced capacity to form biofilms in the presence of sucrose, had reduced viability, and was more sensitive to acid killing. The ΔclpP and ΔclpX strains displayed several phenotypes in common: slow growth, tendency to aggregate in culture, reduced autolysis, and reduced ability to grow under stress, including acidic pH. Unexpectedly, the ΔclpP and ΔclpX mutants were more resistant to acid killing and demonstrated enhanced viability in long-term survival assays. Biofilm formation by the ΔclpP and ΔclpX strains was impaired when grown in glucose but enhanced in sucrose. In an animal study, the average number of S. mutans colonies recovered from the teeth of rats infected with the ΔclpP or ΔclpX strain was slightly lower than that of the parent strain. In Bacillus subtilis, the accumulation of the Spx global regulator, a substrate of ClpXP, has accounted for the ΔclpXP phenotypes. Searching the S. mutans genome, we identified two putative spx genes, designated spxA and spxB. The inactivation of either of these genes bypassed phenotypes of the clpP and clpX mutants. Western blotting demonstrated that Spx accumulates in the ΔclpP and ΔclpX strains. Our results reveal that the proteolysis of ClpL and ClpXP plays a role in the expression of key virulence traits of S. mutans and indicates that the underlying mechanisms by which ClpXP affect virulence traits are associated with the accumulation of two Spx orthologues.
Streptococcus mutans, a common inhabitant of dental biofilms, is considered a major etiologic agent of human dental caries. The abilities of this organism to form biofilms on the tooth surface, metabolize a wide range of carbohydrates, and tolerate rapid and frequent environmental fluctuations are key virulence attributes of this bacterium (3, 29). In addition to dental caries, S. mutans is often a cause of subacute bacterial endocarditis, a life-threatening inflammation of heart valves.
Bacteria in dental plaque are subjected to a wide range of stresses. The intermittent ingestion of food by the host results in dramatic variations in nutrient availability and pH. In addition, bacteria also have to cope with significant fluctuations in oxygen tension and osmolarity. One of the consequences of exposure to environmental stresses is the accumulation of abnormal proteins due to increased errors in transcription and translation (9, 22, 24). The capacity to maintain protein homeostasis by stabilizing proteins that perform essential functions and by refolding or degrading misfolded or aberrant proteins is central for the viability and growth of bacterial pathogens in the host (9, 17, 22, 24).
In the gram-negative bacterial paradigm Escherichia coli, there are four classes of energy-dependent cytoplasmic proteases: Lon, FtsH, ClpAP/XP, and ClpYQ (also known as HslUV) (22). In Lon and FtsH, a single protein encodes both the ATPase and proteolytic sites, whereas the ATPase and proteolytic domains are encoded by different proteins in the ClpAP/XP and ClpYQ classes. While the metalloprotease FtsH is ubiquitous in eubacteria, low-GC gram-positive bacteria (GPB) lack the Lon protease (9, 17). In addition, the available genome sequences of streptococci indicate that they also lack ClpYQ that is found in the genome of some GPB, including the pathogenic Staphylococcus aureus (9, 18). Therefore, it has been proposed that ClpP complexes are more central for stress tolerance and global regulation in streptococci than in other bacterial groups (17). In the ClpP proteolytic system, ClpP must associate with a Clp ATPase partner that possesses nucleotide binding domains characteristic of the AAA+ superfamily of ATPases to form a functional complex (9, 17). In addition to forming protease complexes, Clp ATPases can also perform essential housekeeping functions, including protein reactivation activities typical of molecular chaperones (39).
In addition to cellular protein quality control, bacterial proteolysis also plays an important role by controlling the stability of regulatory proteins (17, 22, 24). Many bacterial stress responses are triggered by inhibition of the degradation of stress-induced transcriptional regulators or sigma factors or by the degradation of repressors of stress gene expression (17, 24). Among the regulators targeted by Clp proteolysis is Spx, suppressor of clpP and clpX, a global transcriptional regulator of oxidative stress commonly found in low-GC GPB (42). Studies with Bacillus subtilis revealed that Spx is a substrate of Clp proteolysis (35) and that the accumulation of Spx was responsible for the pleiotropic phenotypes associated with clpP or clpX mutations (34). Similar observations were made for Lactococcus lactis, for which the inactivation of an Spx homologue alleviated the effects conferred by the clpP and clpX mutations (19).
The genome of S. mutans UA159 encodes orthologs of five Clp ATPases (ClpB, ClpC, ClpE, ClpL, and ClpX), and a single ClpP peptidase (2, 30). Among the five Clp ATPases, ClpC, ClpE, and ClpX have the recognition tripeptide that allows the proteins to interact with ClpP, while ClpB and ClpL are expected to function as protein chaperones independent of ClpP (17). Previously, we showed that a strain lacking ClpP exhibited impaired growth under stress conditions, formed long chains, had a strong tendency to aggregate in culture, had reduced genetic transformation efficiencies, and showed major defects in its capacity to form biofilms in medium containing glucose as the primary carbohydrate source (30). More recently, it was demonstrated that clpP mutants exhibited increased sensitivity to several environmental stress challenges and that ClpP is critical in the organism's adaptive response to oral care products such as sodium fluoride, hydrogen peroxide, and chlorhexidine (4, 13).
Despite the progress that has been made in demonstrating the relevance of Clp proteolysis in S. mutans, the molecular mechanisms underlying these phenotypes remain mostly undetermined. To better define the role of the Clp system in S. mutans, clpB, clpC, clpE, clpL, clpP, and clpX mutants were constructed by allelic replacement. Phenotypic characterization of the mutants has shown that the ΔclpL, ΔclpP, and ΔclpX strains presented the most striking phenotypes compared to those of the parent strain. In addition, we have shown that many phenotypes typical of clpP or clpX mutations were, in great part, associated with the accumulation of two putative Spx orthologues.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. mutans UA159 and its derivatives were grown in brain heart infusion (BHI) medium at 37°C (except for growth under heat stress at 42°C) in a 5% CO2 atmosphere. When appropriate, kanamycin (1 mg ml−1), spectinomycin (1.5 mg ml−1), or erythromycin (10 μg ml−1) was added to the medium. The BHI medium was adjusted to pH 5.5 with HCl where indicated. Growth curves were determined by measuring the change in the optical density at 600 nm (OD600) in BHI broth. To evaluate the capacity of the mutant strains to grow under different stress conditions, the strains were grown overnight in BHI broth and diluted 20-fold in fresh BHI broth buffered to pH 7 and incubated at 37°C (control) or 42°C (heat stress) or in BHI broth buffered to pH 5.5 (acid stress, 37°C).
TABLE 1.
S. mutans strains used in this study
| Strains | Relevant genotypea | Source or reference |
|---|---|---|
| UA159 | Wild type | Laboratory stock |
| UA159StR | Spontaneous Str | 12 |
| JL4 (ΔclpB) | clpB::Ω-Km | This study |
| JL5 (ΔclpC) | clpC::Sp | This study |
| JL3 (ΔclpE) | clpE::nonpolar Km | This study |
| JL10 (ΔclpL) | clpL::Em | This study |
| JL1 (ΔclpP) | clpP::nonpolar Km | This study |
| JL2 (ΔclpX) | clpX::nonpolar Km | This study |
| JL12 (ΔspxA) | spxA::Sp | This study |
| JL13 (ΔspxB) | spxB::Em | This study |
| JL15 (ΔclpP ΔspxA) | clpP::nonpolar Km, spxA::Sp | This study |
| JL16 (ΔclpP ΔspxB) | clpP::nonpolar Km, spxB::Em | This study |
| JL17 (ΔclpX ΔspxA) | clpX::nonpolar Km, spxA::Sp | This study |
| JL18 (ΔclpX ΔspxB) | clpX::nonpolar Km, spxB::Em | This study |
| JL20 (ΔclpP ΔspxA ΔspxB) | clpP::nonpolar Km, spxA::Sp, spxB::Em | This study |
| JL19 (ΔclpX ΔspxA ΔspxB) | clpX::nonpolar Km, spxA::Sp, spxB::Em | This study |
Str, streptomycin resistance.
Construction of mutant strains.
Standard DNA manipulation techniques were used as previously described (30, 38). The primers used to isolate the mutants are listed in Table 2. S. mutans strains lacking the various clp genes (clpB, clpC, clpE, clpP, and clpX) or putative spx genes (smu1142c or smu2084c) were constructed using a PCR ligation mutagenesis approach (27). Briefly, PCR fragments flanking each target gene were obtained and ligated to a nonpolar kanamycin (Kmr), erythromycin (Emr), or spectinomycin (Spr) resistance marker, and the ligation mix was used to transform S. mutans UA159. A strain lacking clpL was made by the insertion of an Emr marker in a naturally occurring BglII restriction site located 324 bp from the start codon. Mutant strains were isolated on BHI plates supplemented with the appropriate antibiotic(s). Double and triple mutants were also constructed by further transformations using chromosomal DNA isolated from single mutants. The deletions were confirmed as correct by PCR sequencing of the insertion site and flanking sequences.
TABLE 2.
Primers used for gene inactivation
| Primer | Sequencea | Application |
|---|---|---|
| 5′clpBArm1SphI | 5′-GTTGAGGAAGCATGCCGTTCGGGC-3′ | clpB deletion |
| 3′clpBArm1BamHI | 5′-CATTTTATCGGATCCAAAGTCACC-3′ | clpB deletion |
| 5′clpBArm2BamHI | 5′-GGTGACTTTGGATCCGATAAAATG-3′ | clpB deletion |
| 3′clpBArm2SpeI | 5′-CACCTGAATACTAGTCCAAAACGG-3′ | clpB deletion |
| 5′clpCArm1 | 5′-GGGATAAAATCATTCCTGGT-3′ | clpC deletion |
| 3′clpCArm1SphI | 5′-TAATCGGTCATGCATGCTATCCTTTTCT-3′ | clpC deletion |
| 5′clpCArm2SphI | 5′-GGACAGACTGCATGCATAGGTATTC-3′ | clpC deletion |
| 3′clpCArm2 | 5′-ACCCGTTTGGGCATCACG-3′ | clpC deletion |
| 5′clpEArm1 | 5′-GATCTGATAAGGCAGTAATAAA-3′ | clpE deletion |
| 3′clpEArm1BamHI | 5′-GACAGAGCATGGATCCAATACCCCT-3′ | clpE deletion |
| 5′clpEArm2BamHI | 5′-CTTCAGATTAGGATCCTCAGGTCTTTC-3′ | clpE deletion |
| 3′clpEArm2 | 5′-GAATGAAGCTGTCTAAG-3′ | clpE deletion |
| clpLFwdSphI | 5′-AGCTGGTGCGCATGCATCTTCATT-3′ | clpL mutation |
| clpLRevSpeI | 5′-GCCGATTGGACTAGTGCCTTCATC-3′ | clpL mutation |
| 5′clpPArm1 | 5′-GAAACGCCGATTACTAAGACTGTAC-3′ | clpP deletion |
| 3′clpPArm1BamHI | 5′-ACTGTATTTCGGATCCAATTCAATCTTGG-3′ | clpP deletion |
| 5′clpPArm2BamHI | 5′-CGCTTCTAGCGGATCCAAAGGAAAACG-3′ | clpP deletion |
| 3′clpPArm2 | 5′-CAAATTTAAGAGCACCAATC-3′ | clpP deletion |
| 5′clpXArm1 | 5′-GGCATGTACAGTGAAC-3′ | clpX deletion |
| 3′clpXArm1BamHI | 5′-CAGCCATTTGGATCCCTAACTTCTAA-3′ | clpX deletion |
| 5′clpXArm2BamHI | 5′-CTAAGAAAGCGGATCCAGGGACCGA-3′ | clpX deletion |
| 3′clpXArm2 | 5′-CTGACCTTCATTTTCAAA-3′ | clpX deletion |
| 5′spx1Arm1 | 5′-GTATTGTAAACGTACATG-3′ | spxA deletion |
| 3′spx1Arm1SphI | 5′-GGTAACCATGCATGCACTACCCCTT-3′ | spxA deletion |
| 5′spx1Arm2SphI | 5′-AGAAGATGACGCATGCGAATTACGC-3′ | spxA deletion |
| 3′spx1Arm2 | 5′-CAACTTCATTTTAGGAAT-3′ | spxA deletion |
| 5′spx2Arm1 | 5′-GTTTTATTCTTCTGTGCG-3′ | spxB deletion |
| 3′spx2Arm1EcoRI | 5′-GTTTTAATCGAATTCTCCTTACAG-3′ | spxB deletion |
| 5′spx2Arm2HindIII | 5′-GAATGTTCGAAGCTTTGAAAAGTC-3′ | spxB deletion |
| 3′spx2Arm2 | 5′-CTTTCTTTTTCTATAAGCAAAAAG-3′ | spxB deletion |
Restriction sites used to facilitate ligation are highlighted in bold.
Long-term survival.
The ability of the S. mutans strains to survive a period of several days was assessed via a long-term survival assay, in which an overnight culture of cells was diluted 1:20 in tryptone-yeast extract (TY) medium containing excess glucose (50 mM). The growth of the cultures was monitored until stationary phase was reached, at which point an aliquot was removed for serial dilution and plating on BHI agar. The cultures were incubated in TY medium containing 50 mM glucose at 37°C and 5% CO2 for several days, with serial dilutions of the cultures plated daily until growth was no longer detected. The plates were incubated for 48 h before colonies were counted.
Acid-killing experiments.
For acid killing, strains were grown in BHI medium to mid-logarithmic growth phase (OD600 ≈ 0.5), washed once with 0.1 M glycine buffer (pH 7), and resuspended in 1/5 of the growth volume in 0.1 M glycine buffer (pH 2.85) for up to 90 min. Every 30 min, aliquots were serially diluted, plated on BHI plates and incubated for 48 h before colonies were counted.
Autolysis assay.
An autolysis assay was performed as described previously (1). Briefly, S. mutans strains were grown in BHI medium to late exponential phase (OD600 ≈ 0.7), harvested, and washed twice with phosphate-buffered saline. The cells were then resuspended to an OD550 of 0.9 in 20 mM potassium phosphate buffer (pH 6.5), containing 1 M KCl, 1 mM CaCl2, 1 mM MgCl2, and 0.4% sodium azide. Autolysis was measured at 30-min intervals at 44°C using a Bioscreen C growth monitor (Oy Growth Curves AB Ltd., Helsinki, Finland) by measuring the OD540. Sterile mineral oil was added over the cell suspension to create an anaerobic environment, and the plate was shaken for 15 s prior to each reading.
Biofilm assay.
The ability of S. mutans strains to form biofilms was assessed by growing cells in wells of polystyrene microtiter plates using a semidefined biofilm medium (BM) (33). The wells of the plates were first coated for 30 min with 100 μl of sterile, clarified, pooled human saliva. Strains grown in BHI medium to an OD600 of approximately 0.5 were diluted 1:100 in BM containing either 1% glucose or 1% sucrose and added to the wells of the microtiter plate. The plates were incubated at 37°C in a 5% CO2 atmosphere for 24 h. After incubation, the plates were washed twice with water to remove planktonic and loosely bound bacteria, and adherent cells were stained with 0.1% crystal violet for 15 min. The bound dye was eluted with 33% acetic acid solution, and biofilm formation was then quantified by measuring the optical density of the solution at 575 nm.
Western blot analysis.
To detect Spx levels in the clpP and clpX mutants, whole-cell protein lysates were prepared by the homogenization of cells in the presence of glass beads with a Bead Beater (Biospec, Bartlesville, OK). The protein concentration of the samples was determined using the bicinchoninic acid assay. The protein lysates were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto Immobilon-FL membranes (Millipore, Bedford, MA), and subjected to Western blot analysis by using standard techniques. The membranes were incubated with antibody raised against Bacillus subtilis Spx (a generous gift from Peter Zuber, Oregon Health & Science University, Beaverton, OR). Immune reactivity was visualized by the incubation of the blot with ECL detection reagents (GE Healthcare, Buckinghamshire, United Kingdom) and immediate exposure to BioMax MS film (Kodak, Rochester, NY).
Animal studies.
To test the infectivity of the clpP and clpX mutants in vivo, we conducted a study to evaluate the ability of the strains to colonize the teeth of the animals using a rodent model of dental caries (7). Briefly, specific-pathogen-free Sprague-Dawley rats were infected by means of a cotton swab with the ΔclpP and ΔclpX strains and UA159StR, a spontaneous streptomycin-resistant strain (12). At weaning, pups aged 21 days were randomly placed into three groups of eight and infected for two consecutive days with actively growing ΔclpP, ΔclpX, or UA159StR strains. The rats were fed a highly cariogenic diet (Diet-2000 containing 56% sucrose) and 5% sucrose water (wt/vol) ad libitum. On experimental days 4 and 10, the animals were screened for the successful infection of each strain by oral swabbing and plating on mitis-salivarius agar containing the appropriate antibiotic. The experiment proceeded for 16 days, at the end of which the animals were euthanized by CO2 asphyxiation and the lower jaws removed for microbiological assessment (25). The number of mutans streptococci recovered from the animals was expressed as CFU ml−1 of jaw sonicate. The data were subjected to analysis of variance for significance and Grubbs' test to detect outliers. This study was reviewed and approved by the University of Rochester Committee on Animal Resources.
RESULTS
Growth characteristics of clp mutants.
To assess the function of S. mutans Clp proteins in homeostasis and the expression of virulence properties, deletions of clpB, clpC, clpE, clpL, clpP, and clpX were created by replacing the entire corresponding gene with an antibiotic resistance cassette, or, in the case of the clpL mutant, with an Emr resistance marker at the clpL 5′ end (Table 1). Because there is precedent in the literature that ClpC and ClpE may have overlapping functions and one could compensate for the loss of the other (17), a ΔclpC ΔclpE double mutant was generated. Similarly, ClpB and ClpL, which lack the ClpP recognition tripeptide, appear to have similar function in S. aureus (17). Thus, a ΔclpB ΔclpL double mutant was also obtained.
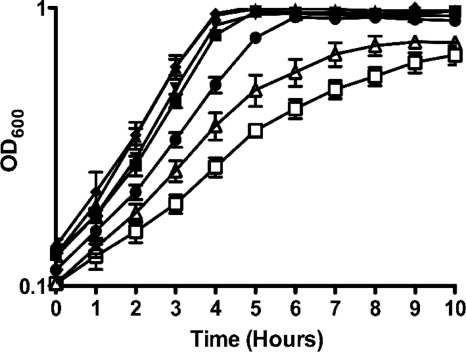
Striking phenotypes were immediately noticed in the clpP and clpX mutant strains; both of these strains formed unusually long chains and had a strong tendency to clump in broth (data not shown), characteristics that were previously observed in the clpP insertional mutant (30). Standard growth curves in BHI medium (37°C, 5% CO2 atmosphere) revealed a slow-growth phenotype for the ΔclpP and ΔclpX strains (Fig. 1). Moreover, the ΔclpP and ΔclpX strains failed to reach the same final growth yield as the other strains. The ΔclpL mutant also consistently demonstrated a longer lag phase than the parent but was able to reach a similar final yield as the parent before entering stationary phase (Fig. 1). The growth curves of the ΔclpCE strain indicated that the double mutant did not differ from its ΔclpC and ΔclpE counterparts, i.e., the ΔclpCE strain could grow as well as its parental strain (data not shown). The slow-growth phenotype of the ΔclpL strain was retained in the ΔclpBL double mutant strain (data not shown).
FIG. 1.
Growth curves of S. mutans UA159 (▪), ΔclpB (▴), ΔclpC (▾), ΔclpE (♦), ΔclpL (•), ΔclpP (□), and ΔclpX (▵) strains in BHI medium at 37°C. The curves shown are the averages with standard deviations of the results from three independent experiments.
We also tested the capacity of each clp deletion mutant to grow at 42°C or in medium acidified to pH 5.5. Under both conditions, the growth of the ΔclpP and ΔclpX strains was slower and the final growth yield was considerably lower compared to that of the wild-type strain (data not shown). Moreover, the growth of the ΔclpP and ΔclpX strains under aerobic conditions (using a rotary shaker, 150 rpm) was severely impaired (data not shown). Compared to that of the parent strain, there were no obvious differences in the growth of the ΔclpB, ΔclpC, ΔclpE, and ΔclpL strains under the stress conditions tested.
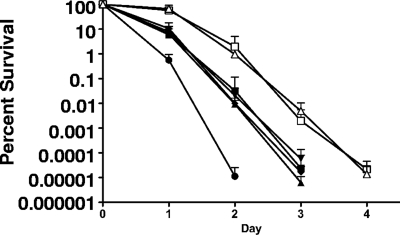
Clp mutations affect long-term survival.
In a long-term survival assay, the viability of the ΔclpL strain was significantly lower (an approximately 3-log reduction after 48 h) than that observed for UA159, whereas no significant differences were observed for the ΔclpB, ΔclpC, and ΔclpE strains. Surprisingly, there was an approximate 2-log enhancement in survival for both the ΔclpP and ΔclpX strains (Fig. 2). Additionally, the ΔclpP and ΔclpX strains survived until day 4 of the experiment, while no other strain had detectable colonies after day 3. All strains tested were capable of lowering the pH of the culture to ∼4.2 after 24 h of incubation, with the exception of the ΔclpP (24-h pH, ∼4.4) and ΔclpX (24-h pH, ∼4.3) strains. After 48 h, there was no substantial difference in the average pH of the ΔclpX strain versus that of the parent UA159 strain, but the ΔclpP strain maintained a slightly higher pH (≤0.2 pH unit) throughout the duration of the experiment. Because the ΔclpP and ΔclpX strains were unable to lower the pH as rapidly and to the same values of the other strains, it was possible that the enhanced survival of these mutants was linked to differences in the growth yield and final pH. To test this hypothesis, we repeated the experiment, but this time, upon entering stationary phase, we used 1 N HCl to artificially lower the pH of the ΔclpP and ΔclpX cultures to the same pH value attained by the parent. The culture pH remained stable for the duration of the experiment for all strains tested. The results obtained were nearly identical to those observed when the final culture pH was not adjusted (data not shown), strongly suggesting that the enhanced survival of the ΔclpP and ΔclpX mutants was not due to differences in the pH.
FIG. 2.
Long-term survival of S. mutans UA159 (▪), ΔclpB (▴), ΔclpC (▾), ΔclpE (♦), ΔclpL (•), ΔclpP (□), and ΔclpX (▵) strains in TY medium supplemented with 50 mM glucose at 37°C. Aliquots of culture were first plated on BHI agar when stationary growth phase was reached (day 0) and again each day until no surviving colonies were detected. The results presented are averages and standard deviations of results from three independent experiments.
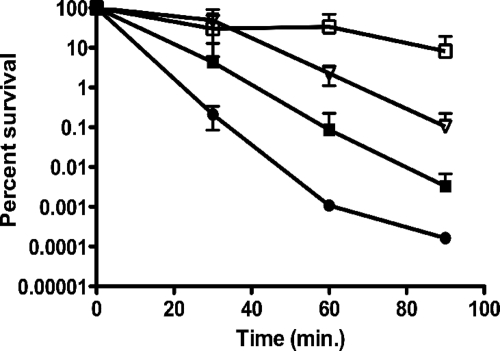
The clpL, clpP, and clpX mutations affect acid challenge survival.
The differences noted in the ΔclpL, ΔclpX, and ΔclpP strains in the long-term survival assays led us to further characterize the acid tolerance of these strains by performing acid-killing experiments. Consistent with the long-term survival results that showed that the viability of the clpL mutant was significantly reduced, the ΔclpL strain was more sensitive to acid challenge compared to the parent strain (Fig. 3). Also in agreement with the long-term survival data, we observed an enhanced ability of the ΔclpP and ΔclpX strains to survive acid challenge compared to that of UA159. At the final time point, the mutant strains displayed an approximate 2-log (ΔclpX) to 4-log (ΔclpP) enhancement of survival compared to that of UA159. Strikingly, the ΔclpP mutant suffered very little cell death over the course of the experiment.
FIG. 3.
Acid killing of S. mutans UA159 (▪), ΔclpL (•), ΔclpP (□), and ΔclpX (▿) strains. Aliquots were plated on BHI agar immediately upon suspension in 0.1 M glycine (pH 2.85) and after 30, 60, and 90 min of incubation. The curves shown are the averages with standard deviations of the results from three independent experiments.
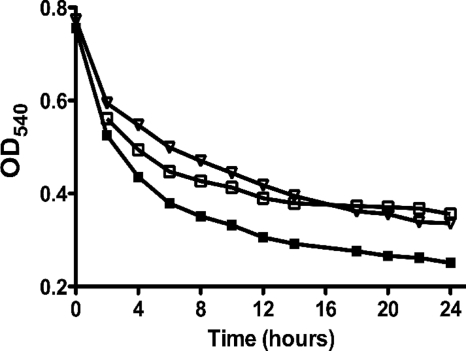
Mutations in clpP and clpX reduce S. mutans autolysis.
The results of the long-term and acid-survival assays led us to question if the enhanced survival of strains bearing mutations in clpP and clpX could be due to reductions in autolytic rates. This hypothesis was confirmed as the ΔclpP and ΔclpX strains underwent autolysis at a slower rate than the parental strain (Fig. 4).
FIG. 4.
Autolysis of S. mutans UA159 (▪), ΔclpP (□), and ΔclpX (▿) strains. A typical result representative of three independent experiments is shown.
Clp proteins have a large role in biofilm formation.
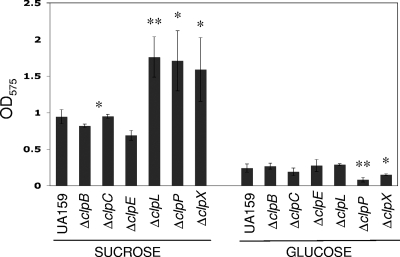
The capacity of the clp deletion strains of S. mutans to form biofilms in the presence of glucose or sucrose was evaluated. When provided with glucose, the ΔclpP and ΔclpX strains showed significantly less biofilm mass than the parent strain, whereas no significant differences were observed in the ΔclpB, ΔclpC, ΔclpE, and ΔclpL strains (Fig. 5). In sucrose, biofilm formation by the ΔclpL, ΔclpP, and ΔclpX mutants was significantly enhanced, and a small reduction in biomass was observed in the ΔclpE strain (Fig. 5). To exclude the possibility that the differences observed were due to small variations in the growth capacity of the strains, the biofilm data were normalized by the final OD600 growth.
FIG. 5.
Biofilm formation by S. mutans UA159 (wild type) and its derivatives. Cultures were grown in a microtiter plate containing BM supplemented with 1% glucose or 1% sucrose at 37°C for 24 h. Biofilm formation was normalized by total growth to exclude apparent differences due to the growth abilities of each strain. Absorbance at 575 nm of crystal violet is shown as averages with standard deviations of the results from three independent experiments. *, P ≤ 0.05; **, P ≤ 0.005 (t test).
The double inactivation of clpCE or clpBL did not alter phenotypes associated with the single mutants.
As mentioned above, studies conducted in other organisms have suggested redundant roles between the ClpC/ClpE and ClpB/ClpL ATPases (17). Biofilm and long-term survival assays with the ΔclpCE and ΔclpBL mutants indicated that the double mutant strains did not differ phenotypically from the single mutant counterparts (data not shown). Although we cannot exclude that there may be functional redundancies among the chaperones, our experimental conditions failed to identify phenotypes that were caused or enhanced by the clpCE or clpBL double mutations.
The inactivation of clpP or clpX decreases the infectivity of S. mutans.
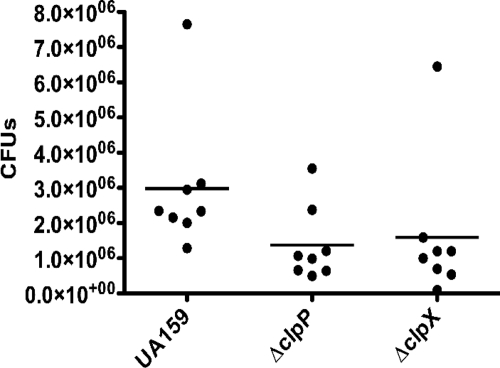
The characterization of the clp mutants revealed that the inactivation of either clpP or clpX resulted in increased resistance to acid killing and the enhanced capacity to form biofilms in sucrose. Considering that the ability to form biofilms in the presence of sucrose and the capacity to survive under low-pH conditions are critical for S. mutans to colonize the oral cavity and cause disease, one might reason that mutations in clpP or clpX render a strain more virulent. To test this possibility, we evaluated the capacity of the ΔclpP and ΔclpX strains to colonize the teeth of pathogen-free rats. However, the average recovered CFU from recipient animals infected with the wild-type strain was slightly higher than the average recovered CFU from the clpP and clpX mutants. Although both mutants showed the same trend, the differences observed were not statistically significant (P > 0.05 by analysis of variance) (Fig. 6).
FIG. 6.
Colonization of S. mutans UA159, ΔclpP, and ΔclpX strains on the teeth of rats after 16 days. The symbols shown represent the recovered bacterial colonies from each individual rat, while the line represents the mean recovery per bacterial strain. Grubbs's test did not indicate the presence of outliers among each strain.
The inactivation of putative spx genes alleviates the ΔclpP and ΔclpX phenotypes.
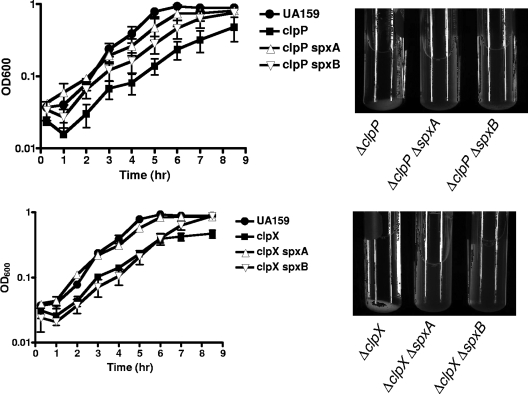
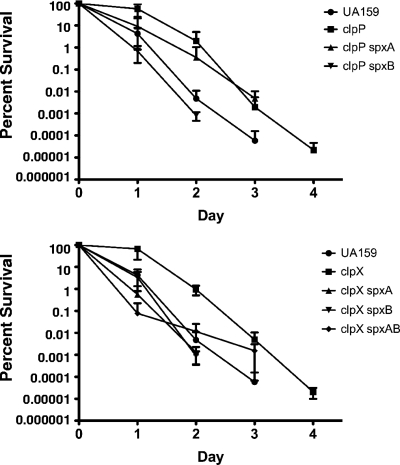
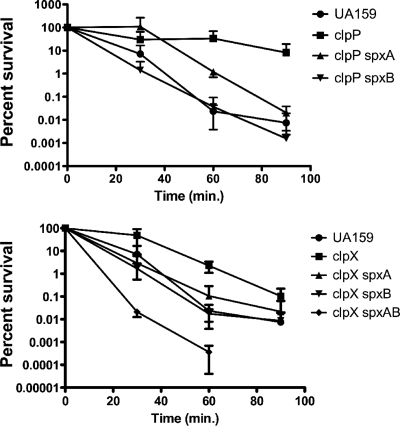
Searching the S. mutans UA159 genome (2), we identified two proteins (Smu1142c and Smu2084c, herein designated SpxA and SpxB, respectively) that shared significant homology with B. subtilis Spx (80 and 73% similarities, respectively) and contain the CxxC redox disulfide motif (see the supplemental material). To gain insight into the role of these proteins in S. mutans and to assess whether any of the phenotypes associated with the ΔclpP and ΔclpX mutations were Spx-dependent, both genes were deleted and a series of double mutants (the ΔclpP ΔspxA, ΔclpP ΔspxB, ΔclpX ΔspxA, and ΔclpX ΔspxB strains) were constructed by transforming the ΔclpP and ΔclpX single mutants with chromosomal DNA isolated from the ΔspxA and ΔspxB mutant strains. The inactivation of either spxA or spxB in the ΔclpP or ΔclpX strains reversed the tendency of these mutants to aggregate in broth (Fig. 7). The inactivation of the spxA gene in the ΔclpP or ΔclpX strains completely restored the growth defect that was initially observed in these strains, whereas the inactivation of spxB partially restored the growth defect of the ΔclpP strain but not of the ΔclpX strain (Fig. 7). Also, the inactivation of spxA or spxB altered phenotypes seen in the ΔclpP and ΔclpX mutants during long-term survival (Fig. 8) and acid-killing assays (Fig. 9). More specifically, the ΔclpP ΔspxA strain survived similarly to the ΔclpP strain until day 3 of the experiment. However, different than the ΔclpP single mutant but similar to the parent strain, the ΔclpP ΔspxA strain had no survivors on day 4 (Fig. 8). Interestingly, the inactivation of spxB in the ΔclpP strain not only reversed the original phenotype of ΔclpP, but the viability of the ΔclpP ΔspxB double mutant was significantly lower than that observed for the wild-type strain under the same conditions (Fig. 8). In the ΔclpX mutant, the inactivation of either spxA, spxB, or both eliminated the increased viability of the ΔclpX mutant, and the viabilities of the ΔclpX ΔspxA and ΔclpX ΔspxB strains were reduced by 1 day compared to that of its parent strain (Fig. 8). In the acid-killing experiments, the deletion of spxA or spxB in the ΔclpP or ΔclpX strain reversed the stress-resistance phenotype observed in the ΔclpP and ΔclpX single mutants (Fig. 9). In addition to the Δclp Δspx double mutants, triple mutant strains (ΔclpP ΔspxAB and ΔclpX ΔspxAB) were created. While there were no difficulties in isolating a ΔclpX ΔspxAB strain, the ΔclpP ΔspxAB mutant was recovered only when plates were incubated in an anaerobic atmosphere, suggesting that the strain is unable to cope with oxidative stress. Thus, because the ΔclpP ΔspxAB triple mutant was able to grow only in an anaerobic environment, this strain was not included in any further experiments. The characterization of the ΔclpX ΔspxAB triple mutant indicated that the strain displayed a behavior similar to the ΔclpX Δspx double mutant strains during long-term survival and acid-killing assays (Fig. 8 and 9). Collectively, these data indicate that the introduction of a mutation in spx, either on spxA or spxB, resulted in the complete or partial reversal of the ΔclpP or ΔclpX phenotypes.
FIG. 7.
The inactivation of spxA or spxB alleviates the slow-growth and aggregation phenotypes of the ΔclpP and ΔclpX strains.
FIG. 8.
Long-term survival of S. mutans strains bearing mutations in both the clp and spx genes in TY medium supplemented with 50 mM glucose at 37°C. Aliquots of culture were first plated on BHI agar when stationary growth phase was reached (day 0) and again each day until no surviving colonies were detected. The results presented are a representative of three independent experiments.
FIG. 9.
Acid killing of S. mutans strains bearing mutations in both the clp and spx genes. Aliquots were plated on BHI agar immediately upon suspension in 0.1 M glycine (pH 2.85) and after 30, 60, and 90 min of incubation. The curves shown are the averages with standard deviations of the results from three independent experiments.
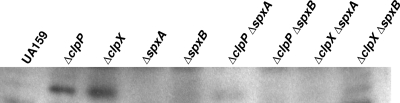
Spx accumulates in the ΔclpP and ΔclpX strains.
As the Spx protein is highly conserved in low-GC GPB, we used antibodies raised against B. subtilis Spx to assess Spx levels in the ΔclpP and ΔclpX strains. As expected, Spx clearly accumulated in stationary-phase lysates of the ΔclpP and ΔclpX mutants even though a band was not detected in the parent strain (Fig. 10). The inactivation of spxA or spxB resulted in the disappearance of the Spx protein, suggesting that the protein band detected in the ΔclpP and ΔclpX extracts was a result of the accumulation of both SpxA and SpxB.
FIG. 10.
Western blot analysis of Spx levels in S. mutans UA159 and its derivatives. The total cell protein (25 μg per lane) of the stationary-phase lysates was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western blotting using an anti-B. subtilis Spx antibody diluted 1:250.
DISCUSSION
Previous reports have demonstrated that ClpP participates in key physiologic processes of S. mutans, including stress tolerance and biofilm formation (4, 13, 30). Here, we expanded our analysis by systematically deleting each clp gene (clpB, clpC, clpE, clpL, clpP, and clpX) identified in the S. mutans genome. Our results revealed that members of the Clp system, specifically ClpL, ClpP, and ClpX, modulate the expression of important virulence attributes of S. mutans.
Although it lacks the recognition tripeptide that permits interaction with ClpP, the ΔclpL strain showed several important phenotypes. The clpL mutant displayed an enhanced capacity to form biofilms in sucrose, had reduced viability in a long-term survival assay, and was more sensitive to acid killing than the parent strain. In a previous report, the S. mutans ClpL protein was induced nearly fourfold in continuous cultures grown at pH 5 compared to that of cells grown at pH 7 (32). A role for ClpL in the acid tolerance of lactic acid bacteria is further illustrated by studies with Lactobacillus species. For Lactobacillus reuteri, clpL was upregulated during acid shock, and a clpL mutant was significantly more sensitive to low pH than the parental strain (40). Similarly, a proteomic approach identified ClpL as upregulated during the acid adaptation of Lactobacillus bulgaricus (14).
The ΔclpP and ΔclpX strains displayed several phenotypes in common, including those that support previous findings with a clpP insertional mutant such as a strong tendency to clump in broth, slow growth rates, poor growth yields, and impaired growth under low-pH or elevated-temperature conditions (30). In another study, an S. mutans clpP knockout mutant was also more sensitive to stress challenges, including growth at low pH or in the presence of compounds commonly used in products for oral care (13). With the exception of an S. aureus clpX mutant that showed improved heat tolerance (16), the inactivation of clpP or clpX of low-GC GPB has resulted in stress-sensitive phenotypes (15, 21, 37). Therefore, it was surprising that the S. mutans clpP and clpX mutants showed enhanced cell viability at a nonlethal pH and increased capacity to survive lethal acidification. There are two, not mutually exclusive, possible explanations for this result. First, the ΔclpP and ΔclpX mutants are intrinsically more resistant to acid killing due to downstream effects in gene regulation caused by the loss of proteolytic control of regulatory proteins targeted by ClpXP, such as the Spx regulator (see below for more detail). Another possibility is that strains with slower metabolisms, such as the ΔclpP and ΔclpX strains, become less susceptible to damages caused by acidification. This appears to be the case for a DnaK knockdown strain of S. mutans that was completely unable to grow at pH 5 but more resistant to acid killing (31). In support of this hypothesis, it has been demonstrated that fast-growing E. coli cells were more sensitive to stress than slow-growing cells (5). Another result that may tie in to either of these possibilities is the reduced autolysis seen for the ΔclpP and ΔclpX mutants that indicates that ClpXP proteolysis may regulate the stability or activity of autolytic enzymes in the cell.
In a previous report, we observed that a clpP insertional mutant showed major defects in its capacity to form biofilms in medium containing glucose (30). However, Deng and colleagues demonstrated increased biofilm formation of an S. mutans clpP mutant grown in sucrose (13). Here, we confirm the findings above, as biofilm formation by the ΔclpP strain was impaired in the presence of glucose but enhanced when sucrose was used as the sole carbohydrate source. The ΔclpX strain also showed a reduced capacity to form biofilms in glucose and an enhanced capacity in sucrose, suggesting that ClpXP proteolysis regulates biofilm formation in S. mutans. In S. mutans, there are two distinct mechanisms to colonize tooth surfaces, sucrose-independent and sucrose-dependent mechanisms (3). Sucrose-independent mechanisms are considered to play an important role in the initial interactions between the bacteria and components of the enamel pellicle (28). The sucrose-dependent pathway involves the production of extracellular polymers of glucose, glucans, formed through the action of three glucosyltransferase enzymes and glucan binding proteins (3). Notably, the production of glucans via glucosyltransferase enzymes is directly associated with the development of dental caries in rats (41). The increased capacity of the ΔclpL, ΔclpP, and ΔclpX strains to form biofilms when grown in sucrose suggests that glucan production is enhanced in these strains. Of note, microarray analysis indicated that the expression of gtfB, responsible for establishing the extracellular polysaccharide matrix along with gtfC, was upregulated more than fourfold in the ΔclpP and ΔclpX strains (J. K. Kajfasz and J. A. Lemos, unpublished data).
A correlation between Clp proteolysis and bacterial pathogenesis has been previously proposed (9, 17). A number of studies have demonstrated that ClpP proteolysis is required for virulence and the progression of disease by bacterial pathogens, including gram-positive organisms (16, 20, 23, 26). Immunization with ClpP was shown to elicit an immune response that protected mice against pneumococcal infections (10, 26). The relevance of ClpP complexes in pathogenesis has been further illustrated by the discovery of new drugs that target ClpP of GPB. In one study, acyldepsipeptides were shown to bind specifically to ClpP, leading to uncontrolled proteolysis by ClpP that resulted in self-digestion of the bacteria (8). More recently, cell-permeable trans-β-lactones were shown to specifically bind to ClpP, causing the complete inhibition of the peptidase activity of ClpP in S. aureus (6). The enhanced capacity to form biofilms in sucrose and increased resistance to acid killing of the S. mutans ΔclpP or ΔclpX strains suggested that the inactivation of clpP or clpX could potentially increase the virulence of this dental pathogen. The finding of a hypervirulent ΔclpP strain was unforeseen and could make targeting ClpP to control diseases potentially unsafe. To test whether the inactivation of clpP or clpX increased the infectivity of S. mutans, a study was conducted to evaluate the ability of the mutants to implant on the teeth of rats. The results obtained from this study did not indicate that the inactivation of clpP (or clpX) enhanced the infectivity of S. mutans. On the contrary, a small reduction in the levels of infection was observed for both mutants.
One of the key roles of bacterial proteolysis is to control the stability of regulatory proteins involved in a variety of cellular processes, including stress responses, sporulation, cell division, and transition between growth phases (17, 24). In B. subtilis, the inactivation of the transcriptional regulator Spx, a known target of ClpXP proteolysis, suppressed phenotypes associated with clpP or clpX mutations, including poor growth, genetic competence, and sporulation frequency (34). Similar findings have been made in L. lactis and S. aureus (19, 36), suggesting that the linkage of Spx accumulation with ClpXP phenotypes may be conserved between low-GC GPB. Here, we showed that the mutation of either of two spx orthologs, named spxA and spxB, was responsible, at least in part, for the attenuation of phenotypes associated with the loss of ClpXP proteolysis in S. mutans. Both SpxA and SpxB contain the characteristic CxxC redox disulfide motif and share significant homology to B. subtilis Spx (80% and 73%, respectively). Thus, the underlying mechanisms by which ClpXP affect virulence traits in S. mutans appear to be, in great part, associated with the accumulation of two Spx orthologues. While the high levels of homology and the comigration of SpxA and SpxB coupled with the very low concentrations of Spx in the cell lysates made it difficult to demonstrate that both Spx proteins are subject to ClpXP degradation, our physiology experiments demonstrated clearly that both SpxA and SpxB contribute to the phenotypes characteristic of the ΔclpP and ΔclpX strains.
Despite the strong association of Spx accumulation with phenotypes resulting from clpP and clpX mutations, the effects of global regulation by Spx in pathogenic bacteria have been largely overlooked. In L. monocytogenes, an spx homologue was found upregulated during the intracellular growth of the bacteria, suggesting that Spx might play a role in intracellular invasion, survival, or both (11). More recently, Spx was shown to affect growth, general stress tolerance, and biofilm formation in S. aureus (36). Notably, unsuccessful attempts to isolate a clpP spx double mutant in S. aureus were reported, suggesting that the strain was not viable (36). The finding of the dual Spx regulators is particularly novel and may have broader implications since two or more copies of putative spx genes are also found in the genome of related streptococci, enterococci, and lactococci. Studies to understand how the Clp and Spx regulons interact and exert an effect on phenotypes associated with virulence are ongoing.
Supplementary Material
Acknowledgments
We thank Kathleen Scott-Anne for assistance with rat experiments, and Peter Zuber for kindly providing the anti-Spx antibody.
This study was partially supported by the NIDCR training program in oral science grant T32DE007202 and by grant DE017425 to R.G.Q.
Footnotes
Published ahead of print on 30 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahn, S. J., and R. A. Burne. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 1896293-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banas, J. A., and M. M. Vickerman. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 1489-99. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, A., and I. Biswas. 2008. Markerless multiple-gene-deletion system for Streptococcus mutans. Appl. Environ. Microbiol. 742037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berney, M., H. U. Weilenmann, J. Ihssen, C. Bassin, and T. Egli. 2006. Specific growth rate determines the sensitivity of Escherichia coli to thermal, UVA, and solar disinfection. Appl. Environ. Microbiol. 722586-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottcher, T., and S. A. Sieber. 2008. Beta-lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J. Am. Chem. Soc. 13014400-14401. [DOI] [PubMed] [Google Scholar]
- 7.Bowen, W. H., S. K. Pearson, and D. A. Young. 1988. The effect of desalivation on coronal and root surface caries in rats. J. Dent. Res. 6721-23. [DOI] [PubMed] [Google Scholar]
- 8.Brotz-Oesterhelt, H., D. Beyer, H. P. Kroll, R. Endermann, C. Ladel, W. Schroeder, B. Hinzen, S. Raddatz, H. Paulsen, K. Henninger, J. E. Bandow, H. G. Sahl, and H. Labischinski. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 111082-1087. [DOI] [PubMed] [Google Scholar]
- 9.Butler, S. M., R. A. Festa, M. J. Pearce, and K. H. Darwin. 2006. Self-compartmentalized bacterial proteases and pathogenesis. Mol. Microbiol. 60553-562. [DOI] [PubMed] [Google Scholar]
- 10.Cao, J., D. Chen, W. Xu, T. Chen, S. Xu, J. Luo, Q. Zhao, B. Liu, D. Wang, X. Zhang, Y. Shan, and Y. Yin. 2007. Enhanced protection against pneumococcal infection elicited by immunization with the combination of PspA, PspC, and ClpP. Vaccine 254996-5005. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 741323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clancy, K. A., S. Pearson, W. H. Bowen, and R. A. Burne. 2000. Characterization of recombinant, ureolytic Streptococcus mutans demonstrates an inverse relationship between dental plaque ureolytic capacity and cariogenicity. Infect. Immun. 682621-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, D. M., J. M. ten Cate, and W. Crielaard. 2007. The adaptive response of Streptococcus mutans towards oral care products: involvement of the ClpP serine protease. Eur. J. Oral Sci. 115363-370. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez, A., J. Ogawa, S. Penaud, S. Boudebbouze, D. Ehrlich, M. van de Guchte, and E. Maguin. 2008. Rerouting of pyruvate metabolism during acid adaptation in Lactobacillus bulgaricus. Proteomics 83154-3163. [DOI] [PubMed] [Google Scholar]
- 15.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 541445-1462. [DOI] [PubMed] [Google Scholar]
- 16.Frees, D., S. N. Qazi, P. J. Hill, and H. Ingmer. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 481565-1578. [DOI] [PubMed] [Google Scholar]
- 17.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 631285-1295. [DOI] [PubMed] [Google Scholar]
- 18.Frees, D., L. E. Thomsen, and H. Ingmer. 2005. Staphylococcus aureus ClpYQ plays a minor role in stress survival. Arch. Microbiol. 183286-291. [DOI] [PubMed] [Google Scholar]
- 19.Frees, D., P. Varmanen, and H. Ingmer. 2001. Inactivation of a gene that is highly conserved in Gram-positive bacteria stimulates degradation of non-native proteins and concomitantly increases stress tolerance in Lactococcus lactis. Mol. Microbiol. 4193-103. [DOI] [PubMed] [Google Scholar]
- 20.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 351286-1294. [DOI] [PubMed] [Google Scholar]
- 21.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28787-802. [DOI] [PubMed] [Google Scholar]
- 22.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19565-587. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim, Y. M., A. R. Kerr, N. A. Silva, and T. J. Mitchell. 2005. Contribution of the ATP-dependent protease ClpCP to the autolysis and virulence of Streptococcus pneumoniae. Infect. Immun. 73730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenal, U., and R. Hengge-Aronis. 2003. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6163-172. [DOI] [PubMed] [Google Scholar]
- 25.Koo, H., S. K. Pearson, K. Scott-Anne, J. Abranches, J. A. Cury, P. L. Rosalen, Y. K. Park, R. E. Marquis, and W. H. Bowen. 2002. Effects of apigenin and tt-farnesol on glucosyltransferase activity, biofilm viability and caries development in rats. Oral Microbiol. Immunol. 17337-343. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, H. Y., A. D. Ogunniyi, M. H. Choi, S. N. Pyo, D. K. Rhee, and J. C. Paton. 2004. The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect. Immun. 725646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49193-205. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. F., A. Progulske-Fox, G. W. Erdos, D. A. Piacentini, G. Y. Ayakawa, P. J. Crowley, and A. S. Bleiweis. 1989. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II). Infect. Immun. 573306-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemos, J. A., and R. A. Burne. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 1543247-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 1846357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemos, J. A., Y. Luzardo, and R. A. Burne. 2007. Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J. Bacteriol. 1891582-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 1501339-1351. [DOI] [PubMed] [Google Scholar]
- 33.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 1821374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano, M. M., F. Hajarizadeh, Y. Zhu, and P. Zuber. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42383-394. [DOI] [PubMed] [Google Scholar]
- 35.Nakano, S., G. Zheng, M. M. Nakano, and P. Zuber. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 1843664-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pamp, S. J., D. Frees, S. Engelmann, M. Hecker, and H. Ingmer. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 1884861-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson, G. T., W. L. Ng, R. Gilmour, and M. E. Winkler. 2003. Essentiality of clpX, but not clpP, clpL, clpC, or clpE, in Streptococcus pneumoniae R6. J. Bacteriol. 1852961-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Schirmer, E. C., J. R. Glover, M. A. Singer, and S. Lindquist. 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21289-296. [PubMed] [Google Scholar]
- 40.Wall, T., K. Bath, R. A. Britton, H. Jonsson, J. Versalovic, and S. Roos. 2007. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 733924-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 613811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuber, P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 1861911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.