Abstract
Copper is an essential cofactor for many enzymes, and at over a threshold level, it is toxic for all organisms. To understand the mechanisms underlying copper homeostasis of the gram-positive bacterium Bacillus subtilis, we have performed microarray studies under copper-limiting conditions. These studies revealed that the ycnJ gene encodes a protein that plays an important role in copper metabolism, as it shows a significant, eightfold upregulation under copper-limiting conditions and its disruption causes a growth-defective phenotype under copper deprivation as well as a reduced intracellular content of copper. Native gel shift experiments with the periplasmic N-terminal domain of the YcnJ membrane protein (135 residues) disclosed its strong affinity to Cu(II) ions in vitro. Inspection of the upstream sequence of ycnJ revealed that the ycnK gene encodes a putative transcriptional regulator, whose deletion caused an elevated expression of ycnJ, especially under conditions of copper excess. Further studies demonstrated that the recently identified copper efflux regulator CsoR also is involved in the regulation of ycnJ expression, leading to a new model for copper homeostasis in B. subtilis.
Transition metals such as copper, iron, and zinc play important roles in bacterial metabolism as essential cofactors for numerous enzymes. However, when the concentrations of these metals increase in the cell, they undergo undesirable redox reactions or bind inappropriately to the metal binding sites of several enzymes, thereby altering their specificity and finally leading to toxic effects (4, 24). Thus, the acquisition of these metals has to be strictly balanced in the cells, and hence bacteria tend to evolve in response to the bioavailability of metals to sustain the metal homeostasis.
Copper homeostasis is well studied in gram negative bacteria such as Enterococcus hirae and Escherichia coli. In E. hirae, the process occurs at the plasma membrane and includes four genes, i.e., copY, copZ, copA, and copB. copA and copB encode two integral membrane P-type ATPases that are necessary for the transport of copper into the cells under copper-limiting conditions (20). CopA, which serves to import copper, interacts with CopZ, which acts as a copper chaperone. CopZ then chaperones the metal atom to the transcriptional repressor CopY, thereby releasing the repression of copper homeostasis genes (28, 29). In E. coli, there are several sets of genes which are responsible for copper homeostatic functions. The cusRS genes form a sensor-regulatory pair which senses copper and activates the cusCFBA genes (19). CusF is a periplasmic copper binding protein, while cusCBA gene products are homologous to a family of proton/cation antiporter complexes (7). In a second copper efflux system, regulated by CueR, a MerR-like transcriptional activation controls two copper efflux genes, copA and cueO (19), whereas cueO encodes a multicopper oxidase. In addition, the cutABCDEF genes are also believed to be involved in copper uptake, storage, delivery, and efflux (21, 23). Copper efflux is carried out mainly by heavy metal exporters which belong primarily to the integral membrane protein family of P-type ATPases (8, 9, 22, 27, 33), whose expression is controlled mainly at the level of transcription. These P-type ATPases are functional in translocating Cu(I) across the cytoplasmic membrane. The recently discovered copper-specific repressor CsoR in Mycobacterium tuberculosis belongs to an entirely new set of copper-responsive repressors, whose homologs are widely spread in all major classes of eubacteria (16). CsoR from Bacillus subtilis, which is encoded upstream of the copZA operon, is 37% homologous to M. tuberculosis CsoR, and elevated copper levels in B. subtilis are sensed by CsoR, which leads to derepression of the copZA copper efflux operon (16, 26).
In contrast, in Saccharomyces cerevisiae, high-affinity copper uptake is mediated by two transmembrane transport proteins, Ctr1p and Ctr3p. Prior to uptake, Cu(II) is reduced to Cu(I) by Cu(II)/Fe(III)-specific reductases Fre1p and Fre2p (11). The CTR1, CTR3, and FRE1 genes are activated under copper starvation and repressed under copper repletion by the copper-sensing transcription factor Mac1p (6, 36). Studies undertaken to understand the copper resistance in Pseudomonas syringae strains that infect tomato revealed that the copper resistance operon copABCD is plasmid encoded and is regulated by the two-component system copRS (3, 4, 15). Similar copper resistance (pcoABCD) and regulatory (pcoRS) genes are also plasmid encoded in E. coli (14, 31, 35). In spite of high homology between these two systems, the resistance mechanisms dealing with an excess of copper inside the cell are completely different. Copper resistance in E. coli is achieved mainly by a copper efflux mechanism, whereas P. syringae performs sequestration of excess cytosolic copper (15).
Here, we explore the role of B. subtilis YcnJ, which is a homolog of P. syringae CopCD, in copper homeostasis. The ycnJ gene from B. subtilis is highly induced under copper-limiting conditions, and a ΔycnJ mutant shows reduced growth under copper-limiting conditions. Uptake components for copper in B. subtilis have not been reported so far, and we demonstrate that YcnJ is a candidate for such a function. The ycnK gene located upstream from ycnJ was investigated and shown to encode a transcriptional regulator which acts, in addition to the investigated regulator CsoR, as a copper-specific repressor for ycnJ.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacillus subtilis ATCC 21332 (wild type [WT]) was grown in Belitsky minimal medium (BMM) supplemented with 0.5% (wt/vol) glucose as a carbon source and with all essential nutrients required (30). Freshly prepared CuCl (0.5 mM) was used to maintain the copper excess conditions, and 0.25 mM bathocuprione disulfonate (BCS) (Sigma-Aldrich) was used as a Cu(I)-specific chelator for maintaining copper-limiting conditions. Unless otherwise indicated, liquid medium was inoculated from an overnight preculture and incubated at 37°C with constant shaking at 225 rpm. All glassware were washed with 0.1 M HCl and double-distilled water before autoclaving. The antibiotics erythromycin (1 μg ml−1) and lincomycin (25 μg ml−1), both for testing macrolide-lincosamide-streptogramin B resistance, and spectinomycin (100 μg ml−1) were used for the selection of various B. subtilis mutants after construction. For selection of E. coli Top10 strains with transformed plasmids, the antibiotic kanamycin (50 μg ml−1) was used. E. coli strain BL-21 was used for protein overexpression. For RNA preparations, bacteria were harvested at mid-log phase.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Function | Source or reference |
|---|---|---|---|
| B. subtilis strains | |||
| ATCC 21332 | WT | 5a | |
| CSP100 | ΔcsoR::erm | Copper efflux transcriptional regulator | This study |
| CSP101 | ΔycnJ::spc | Copper homeostasis | This study |
| CSP 102 | ΔycnK::erm | Transcriptional regulation | This study |
| CSP 103 | ΔcsoR::erm ΔycnK::spc | Transcriptional regulation | This study |
| E. coli strains | |||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC)φ80lacZΔM15 ΔlacX74 deoR nupG recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 λ− | Transformation | Invitrogen |
| BL21(DE3) | F−ompT gal dcm lon hsdSB (rB− mB−) λ(DE3) | Overexpression | Novagen |
| Plasmids | |||
| pUS 19 | Spcr | Antibiotic resistance cassette Spcr | 3a |
| pMUTIN | Ermr | Gene disruption vector; antibiotic resistance | 31a |
| pET28a+ | Kanr | Expression vector | Novagen |
| pCSP01 | pET28a+ containing N-terminal 135 codons of ycnJ as C-terminal His6 tag fusion | Possible copper import | This study |
| pCSP02 | pET28a+ containing yvgZ as a C-terminal His6 tag fusion | Transcriptional regulation | This study |
| pCSP03 | pET28a+ containing ycnK as a C-terminal His6 tag fusion | Possible copper import transcriptional regulator | This study |
DNA manipulations and genetic techniques.
DNA preparations and transformations were carried out as described previously (12, 25). Electroporation was used for the transformation of plasmids into E. coli Top10 cells. Homologous recombination was used for transforming the B. subtilis ATCC 21332 strain for mutant construction. Restriction enzymes, T4 DNA ligase, and calf intestinal phosphatase were used according to the manufacturer's instructions (New England Biolabs).
Mutant construction.
Deletion mutants were generated by the long flanking homology PCR method (34). In the first-round PCR, long flanking homologous PCR fragments were amplified from upstream and downstream regions of the gene to be deleted. The 3′ ends of the resulting homologous PCR products were designed to be complementary to the resistance cassette and were used in the second-round PCR, generating a fusion construct which replaces the gene of interest with a resistance marker to facilitate the selection of the mutant on antibiotic plates. The primers used in generating the mutants are listed in Table 2. All PCRs were performed using Platinum Pfx DNA polymerase (Invitrogen). Chromosomal DNA of B. subtilis ATCC 21332 was used as a template to amplify the corresponding upstream and downstream flanking regions. The Expand long-template PCR system was used to fuse the homologous flanks with the corresponding resistance markers. PCR fusion products were used directly for the transformation of B. subtilis strain ATCC 21332 to generate the ΔcsoR, ΔycnK, ΔycnJ, and ΔcsoR ΔycnK mutants. Transformants were selected on the respective antibiotic-containing LB plates. Chromosomal DNA was isolated from all the mutants, and the recombinations were confirmed by PCR. The erythromycin resistance cassettes used for the construction of the ΔcsoR and ΔycnK single mutants were amplified from the pMUTIN vector. To generate the ΔycnJ mutant, the spectinomycin resistance cassette was amplified from the pUS19 vector, while the resistance cassettes for the ΔcsoR ΔycnK double mutant were amplified from pMUTIN and pUS19, respectively.
TABLE 2.
Primers
| Primer (5′ → 3′) | Sequencea |
|---|---|
| Csp100-Us-FP | CCA CAT GAC GAA GCA ACT TCG TAC AG |
| Csp100-Us-RP | GCA AGT CAG CAC GAA CAC GAA CC GCT TTT ATG GTT TAA TGT TTT ATG TTC GTT ATG CTT TTC CAT |
| Csp100-Ds-FP | GTC TAT TTT TAA TAG TTA TCT ATT ATT TAA CGG GAG GAA A TAA GGG GAA CAG GCC ATT TCT GAG C |
| Csp100-Ds-RP | GGC TTC CCG TTT GTC ACG GTT CC |
| Csp101-Us-FP | GGC AAG GAG CAG GCA AAA GTG G |
| Csp101-Us-RP | CTC TTG CCA GTC ACG TTA CGT TAT TAG GGC CTG ACC GGC GAC TTT AAC G |
| Csp101-Ds-FP | CTA TAA ACT ATT TAA ATA ACA GAT TAA AAA AAT TAT AA GCA GAA GGA TGA TCC ACC ATC TGT TTC G |
| Csp101-Ds-RP | CGC TCA AAT AAC TCC CAA AGC GTT GC |
| Csp102-Us-FP | CCG ATC CTA CAA TCA CCC CAA TTG C |
| Csp102-Us-RP | GCA AGT CAG CAC GAA CAC GAA CC CAG CCA CTT CAG TAT GTG TTG CTG TC |
| Csp102-Ds-FP | CTA TAA ACT ATT TAA ATA ACA GAT TAA AAA AAT TAT AA GCA GAA GGA TGA TCC ACC ATC TGT TTC G |
| Csp102-Ds-RP | GAG AGC AGC AAA ACG AGT GCC G |
| Csp103 ΔycnK-Us-FP | CCG ATC CTA CAA TCA CCC CAA TTG C |
| Csp103 ΔycnK-Us-RP | CTC TTG CCA GTC ACG TTA CGT TAT TAG CAG CCA CTT CAG TAT GTG TTG CTG TC |
| Csp103 ΔycnK-Ds-FP | CTA TAA ACT ATT TAA ATA ACA GAT TAA AAA AAT TAT AA CGA AAG AAA CGC TGC ACC TGC ACC |
| Csp103 ΔycnK-Ds-RP | GAG AGC AGC AAA ACG AGT GCC G |
| ycnJ pET28a+FP | ATA TAT CCA TGG ACA TGA AGC GAA ACA GAT GGT GG |
| ycnJ pET28a+RP | TAT ATA CTC GAG TGA ATC GGC TGC TTT TTG GC |
| csoR pET28a+FP | ATA TAC CAT GGA AAA GCA TAA CGA ACA TAA AAC ATT AAA CCA TAA AAG C |
| csoR pET28a+RP | ATA TAC TCG AGT GAT TTT GTG AAC TTT TTA AAT ACG TCC AAA AGC TCA G |
| ycnK pET28a+FP | ATA TAC CAT GGA CAT GCT TCC G AT TAA TAG ACA GCA ACA C |
| ycnK pET28a+RP | TAT ATC TCG AGT TTC TTG GTG CAG GTG CAG CG |
Underlining indicates the flanking regions of resistance cassette and restriction sites.
Purification of CsoR, YcnK, and the N-terminal 135 amino acids (aa) of YcnJ.
The open reading frames of genes csoR, ycnK, and ycnJ were amplified by PCR using chromosomal DNA isolated from B. subtilis strain ATCC 21332 as the template and cloned with NcoI and XhoI sites into the pET28a+ vector for overexpression. The ligated plasmids were then transformed into E. coli DH5α cells. The resulting plasmids were confirmed by restriction analysis for the integration of the fragment and transformed into BL21 cells using electroporation. Overnight cultures were grown by inoculating an isolated colony from a plate into 5 ml LB medium containing kanamycin (50 μg ml−1). The overnight cultures were used to inoculate 2 liters of LB medium containing kanamycin (50 μg ml−1) with a starting optical density at 600 nm (OD600) of 0.05. Inoculated cultures were grown under continuous shaking at 225 rpm at 35°C, and the temperature was decreased to 30°C after 1 hour. When the OD600 of the cells reached 0.5 to 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 0.1 mM was added to the culture flasks and growth was continued for 4 hours. Cells were harvested by centrifugation (Sorvall RC 5B plus), and the resulting pellets were resuspended in HEPES-A buffer (50 mM HEPES, 300 mM NaCl [pH 7.5], 1 mM dithiothreitol [DTT]) and lysed with EmulsiFlex-C5 Avestin. The lysate obtained was clarified by centrifugation (Sorvall RC 26 plus) at 17,000 rpm for 30 min, and an Ni-nitrilotriacetic acid (NTA) column was used to purify the protein using a linear gradient with HEPES-B buffer (50 mM HEPES, 300 mM NaCl, 250 mM imidazole [pH 7.5], 1 mM DTT). The purified protein was dialyzed against HEPES-A buffer containing 100 mM NaCl. Protein purity of more than 90% was observed by Coomassie blue staining of a sodium dodecyl sulfate-polyacrylamide gel.
Native gel electrophoresis.
Purified protein samples (∼100 μM) were incubated with 200 μM Cu(I) or Cu(II) for about 15 to 30 min at room temperature and loaded onto 6% continuous native polyacrylamide gels. Samples were run at 100 V and 10 mA for about 4 to 5 h until the bromophenol blue ran out of the gel. Gels were stained using Coomassie blue and destained using 10% acetic acid.
Microarray analysis.
For microarray analysis, both the WT (ATCC 21332) and the ΔcsoR mutant were grown in BMM under copper-replete conditions (growth in the presence of 0.5 mM CuCl) and copper-depleted conditions (growth in the presence of the copper-specific chelator BCS at 0.25 mM). Cultures were harvested in the mid-log phase for RNA extraction. The Macaloid/Roche method was used for the RNA extraction (13). Concentrations of RNA were measured using the nanodrop method. For reverse transcription, a solution containing 18 μl annealing mix (10 to 20 μg total RNA and 2 μl random nonamers adjusted to a final volume of 18 μl with nuclease-free water) was incubated for 5 min at 70°C and 10 min at 4°C for annealing. Reverse transcription was performed by addition of this annealing mix to a solution containing 6 μl Superscript III buffer (supplied with the reverse transcriptase), 10 mM DTT, nucleotide master mix containing aminoallyl-dUTP, and 300 U Superscript III reverse transcriptase (Invitrogen). cDNA was synthesized overnight at 42°C in a total volume of 30 μl. The aminoallyl-modified cDNA was incubated with the CyDye NHS ester (Cy3 or Cy5 monoreactive dye) at room temperature in the dark for 60 to 90 min. Labeled cDNAs were purified using Nucleo Spin Extraxt II columns, and the dye incorporation was measured using the nanodrop method. Equal quantities of Cy3/Cy5-incorporated cDNAs were mixed, dried in a Speedvac, and then dissolved in 5 μl H2O, followed by heating at 94°C for 2 min. The heated sample was subsequently mixed with 30 μl preheated hybridizing buffer (68°C), hybridized onto microarray glass slides, and incubated overnight at 68°C in a hybridization oven.
Microarray data analysis.
Hybridized DNA arrays were read using a GenePix 4200AL autoloader (Axon Instruments), and the data obtained were processed with ArrayPro 4.5 (Media Cybernetics Inc., Silver Spring, MD) ArrayVision software. Expression levels were processed and normalized (Lowess method) with Micro-Prep (10, 32). The ln-transformed ratios of the expression levels of mutant versus WT were subject to a t test using the Cyber-T tool (2). Three independent measurements for each condition along with a dye swap were analyzed. The results obtained were averaged, the raw data were processed to Cyber-T web interface software for the calculation of the expression ratios, and the data were exported to Microsoft Excel.
Estimation of copper concentrations inside cells.
Total cytoplasmic copper concentrations were measured using inductively coupled plasma mass spectrometry. B. subtilis strain ATCC 21332 and the ΔcsoR mutant were grown in BMM overnight. Fresh BMM (100 ml) either with 0.5 mM copper in excess or under copper-limiting conditions achieved by addition of 0.25 mM BCS was inoculated with overnight cultures with a starting OD600 of 0.05, and the cells were harvested in mid-log phase. Cells were centrifuged at 13,000 rpm (18,000 × g) for 5 min, and the pellets were washed three times with buffer containing 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA and finally with MilliQ water. Cells were dried overnight at 85°C, and the total copper concentration was determined by breaking the cells using nitric acid.
Dot blot analysis.
B. subtilis strains were grown under normal (BMM), copper limited (BMM plus 0.25 mM BCS), and copper-replete (BMM plus 0.5 mM copper in excess) conditions. Overnight cultures were inoculated into fresh medium to an initial OD600 of 0.05, and cells were harvested at an OD600 of 0.25. Total RNA was isolated from these cells using the RNAZol method (http://microarrays.nki.nl/download/protocols.html). RNA concentrations were measured using the nanodrop method at 260 nm/280 nm. The ratios of RNA concentration to protein concentration were above 1.65 in all samples. Denaturing gel electrophoresis was run to test the quality of RNA for 16S and 23S RNAs. Two micrograms of RNA from each sample was subsequently dotted onto a nylon membrane using a dot blot apparatus and hybridized after UV cross-linking with a UTP-11-digoxigenin-labeled antisense RNA probe specific for ycnJ mRNA. The riboprobe was synthesized by in vitro transcription using T7 RNA polymerase. The T7 promoter sequence was introduced into the PCR product of the ycnJ gene by using primers 5′-AGCTCGTCAAACGGACGAGACG-3′ and 5′-TAATACGACTCACTATAGGGGAAATCCATCAAAATGCCGACCG-3′ (the T7 promoter extension is underlined). After hybridization and washing, the filters were treated with a digoxigenin-specific antibody fragment conjugated with alkaline phosphatase (Roche) and AttoPhos (Amersham Biosciences) as an enhanced chemifluorescence substrate. The hybridization signals were detected with a Storm860 fluorescence imager, and relative signal quantification was performed with ImageQuant software.
RESULTS AND DISCUSSION
Microarray analyses reveal ycnJ as a putative copper uptake determinant.
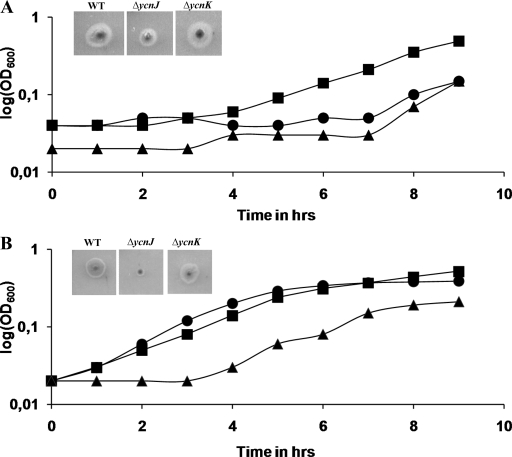
In order to gain insights into the transcriptional response under various copper conditions, microarray experiments were performed with the following combinations: (i) WT (BMM without Cu) compared with WT (BMM), (ii) ΔcsoR mutant (BMM without Cu) compared with WT (BMM without Cu), (iii) WT (BMM plus Cu) compared with WT (BMM), and (iv) ΔcsoR mutant (BMM plus Cu) compared with WT (BMM plus Cu). The essential results from these experiments are summarized in Table 3. B. subtilis WT cultures grown under copper-limiting conditions (without Cu) show a significant increase in the upregulation of the copper-responsive genes ycnI, ycnJ, ycnK, and ycnL. In contrast, in the WT the same genes are significantly downregulated in copper-replete medium (plus Cu). Thus, we speculated that these genes play a possible role in copper acquisition. To investigate this hypothesis, deletion mutants were constructed and checked for their ability to grow under different copper availability conditions. The ΔycnK mutant showed no or little difference in growth under copper-limiting conditions (Fig. 1B). In contrast, it exhibited enhanced growth under copper excess conditions (Fig. 1A). On the other hand, the ΔycnJ mutant exhibited a growth-defective phenotype under copper-limiting conditions (Fig. 1B), suggesting an important role for YcnJ in copper import. In case of the ΔycnL mutant, no effect on growth under both conditions was observed (data not shown).
TABLE 3.
Microarray results
| Genetic background | Ratio of expression levelsa
|
|||
|---|---|---|---|---|
| WT (BMM−Cu)/WT (BMM) | ΔcsoR (BMM−Cu)/WT (BMM−Cu) | WT (BMM+Cu)/WT (BMM) | ΔcsoR (BMM+Cu)/WT (BMM+Cu) | |
| copA | 0.90 | 7.80 | 0.93 | 4.20 |
| copZ | 0.98 | 16.64 | 2.61 | 6.53 |
| ycnJ | 8.17 | 0.93 | 0.40 | 2.31 |
| ycnI | 6.36 | 0.29 | ||
| ycnK | 10.9 | 0.34 | ||
| ycnL | 1.77 | 0.68 | ||
Less than 1, downregulation; greater than 2, upregulation.
FIG. 1.
(A) Analysis of copper-dependent growth in BMM with 1 mM copper in excess. Growth curves of the WT (•), the ΔycnJ mutant (▴), and the ΔycnK mutant (▪) are shown. The insets show the phenotypes of the WT and the ΔycnJ and ΔycnK mutants on BMM with 1 mM copper in excess. (B) Analysis of copper-dependent growth in BMM with 0.5 mM BCS. Growth curves of the WT (•), the ycnJ mutant (▴), and the ΔycnK mutant (▪) are shown. The insets show the phenotypes of the WT and the ΔycnJ and ΔycnK mutants on BMM agar with 0.5 mM BCS.
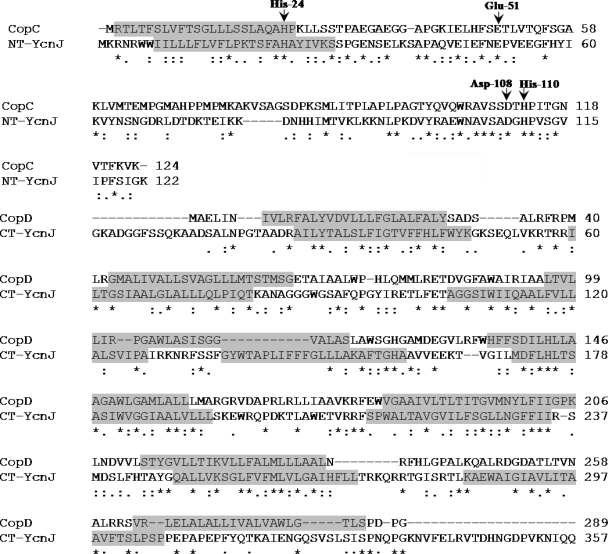
The CopC and CopD gene products of P. syringae exhibit a high sequence identity (28%) with YcnJ. The N-terminal region of YcnJ (∼135 aa) is homologous to the periplasmic protein CopC, a copper binding protein which is involved in copper uptake in P. syringae (1, 5). The putative copper binding amino acid residues (His-24, Glu-51, Asp-108, and His-110) identified within the N-terminal region of YcnJ are highly homologous to the Cu(II) binding sites in CopC (1). The C-terminal region of YcnJ (401 aa) represents a transmembrane domain that is homologous to the inner membrane copper transport protein CopD of P. syringae (1, 5). Alignments of the N- and C-terminal regions of YcnJ with CopC and CopD are shown (Fig. 2). For the genes ycnK and ycnL, encoding a putative transcriptional regulator and a putative signal permease, respectively, several metal binding sites (Cys-x-x-Cys and Cys-x-Cys) were identified. YcnK was identified as a putative transcriptional regulator with a conserved helix-turn-helix motif in its N-terminal region and a C-terminal region with a sensor function. The sensor function is defined by a NosL superfamily motif which indicates specific binding of Cu(I) (17). Apart from these genes, the copper-inducible copZA operon, encoding a copper chaperone and an efflux ATPase, was found to be strongly upregulated in the ΔcsoR mutant, suggesting a considerable role of CsoR in copper efflux as recently described (26).
FIG. 2.
Alignment of amino acid sequences of P. syringae CopC and CopD and B. subtilis YcnJ (ClustalW). NT, N terminal; CT, C terminal. Arrows indicate conserved copper binding residues. Highlighted amino acids represent the predicted transmembrane domains.
Regulation of ycnJ by YcnK and CsoR.
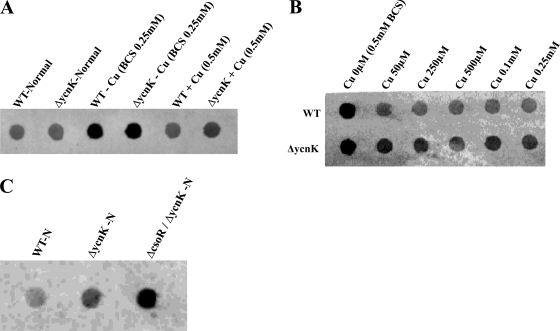
The initial transcriptome studies were followed by a closer examination of ycnJ regulation with respect to different copper concentrations and possibly involved transcription factors. As a first candidate, we addressed the putative transcriptional regulator-encoding gene ycnK, which is located directly upstream from ycnJ. A ycnK deletion mutant was constructed and grown under different copper conditions along with the WT. Total RNA was isolated at mid-log phase, and ycnJ gene expression was estimated semiquantitatively by use of dot blots (Fig. 3). As expected, transcription of ycnJ in the WT was elevated under copper-limiting conditions. In comparison to the WT, the ycnK mutant showed an upregulation of ycnJ expression, especially under copper excess conditions (Fig. 3A). Further in this context, the effect of increasing copper concentrations on ycnJ gene expression was tested, and the ΔycnK mutant was found to upregulate ycnJ expression during copper excess approximately twofold compared to the WT (Fig. 3B). This points to a function of YcnK in which it acts as a negative regulator of ycnJ. Since this function is present mainly under conditions of high copper concentrations, YcnK is predicted to use copper as a corepressor. In addition, enhanced growth of the ycnK mutant under copper excess conditions was observed (Fig. 1A). Thus, as long as toxic copper concentrations are compensated for by copper efflux detoxification systems such as CopZA, induction of the predicted copper uptake system YcnJ is not detrimental for cell growth without carbon and energy source limitation.
FIG. 3.
Transcriptional analysis of ycnK- and csoR-dependent ycnJ expression. Two micrograms of total RNA isolated from the WT and mutants under normal, 0.25 mM BCS, and 0.5 mM copper excess conditions was blotted onto a nylon membrane by dot blotting and hybridized with a digoxigenin-UTP-labeled antisense riboprobe specific for the ycnJ transcript. Hybridization signals were detected by using a digoxigenin-specific antibody fragment conjugated with alkaline phosphatase and AttoPhos as chemifluorescence substrate. (A) ycnK-dependent expression of ycnJ with different copper concentrations. (B) ycnK-dependent expression of ycnJ under increasing copper concentrations. (C) Expression of ycnJ in the WT and the ΔycnK and ΔcsoR ΔycnK mutants under normal conditions.
As a second putative candidate for ycnJ regulation, we examined the recently described copper efflux regulator CsoR (16, 26), according to the finding of ycnJ upregulation in the ΔcsoR background during the microarray studies. Since absence of the CsoR repressor leads to strong derepression of the copZA copper efflux system (26), effects of CsoR on ycnJ expression might be regarded as rather indirect via modulating the intracellular copper concentration and, in the course of that, also YcnK activity. However, when the ΔcsoR and ΔycnK backgrounds were combined in a ΔcsoR ΔycnK double mutant which was tested for ycnJ expression, an additional elevation of ycnJ expression compared to that in the ΔycnK mutant was observed (Fig. 3C), suggesting also a direct participation of CsoR in ycnJ regulation.
Estimation of intracellular copper content.
To further verify the predicted roles of ycnJ and ycnK as a copper uptake mediator and corresponding regulator, respectively, total intracellular copper contents were measured by using inductively coupled plasma mass spectrometry analysis. These studies revealed that the ΔycnK mutant contains approximately double the amount of cytoplasmic copper (511.1 ppb) in the WT (278,87 ppb) and the ΔycnJ mutant (205.0 ppb). These high values are a result of copper accumulation under copper excess conditions. In agreement with the dot blot results, these findings suggest that YcnK may act as a negative transcriptional regulator of copper uptake that is active in the presence of copper. In addition, the entire copper contents within the WT and ΔycnJ cells under normal and copper-limiting conditions were determined. The results revealed that the amount of copper in WT cells grown in the presence of the copper-specific chelator BCS is approximately twofold higher (3.58 ppb) than that in cells those grown without the addition of BCS (1.84 ppb). This might be a possible consequence of ycnJ upregulation under copper-limiting conditions, where the ΔycnJ mutant under normal conditions holds only 1.2 ppb (Fig. 3A). The total copper content measured in the ΔycnJ mutant when grown in the presence of BCS was found to be 1.57 ppb and thus more than twofold less than that in the WT under these conditions. Together with the growth experiments, this supports the supposed role for YcnJ in copper acquisition under copper-limiting conditions.
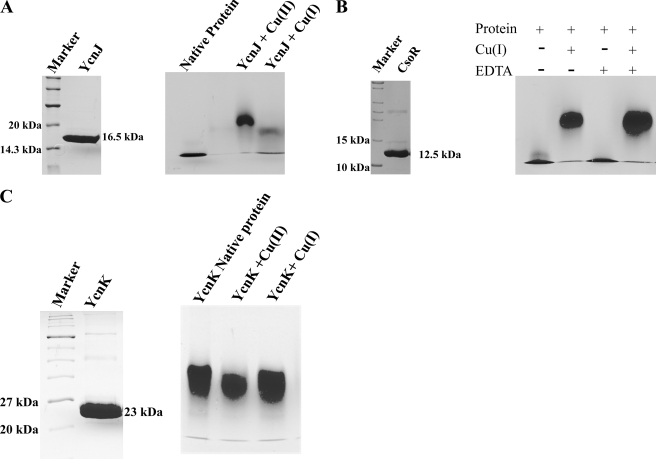
Copper-induced oligomerization of CsoR and YcnJ.
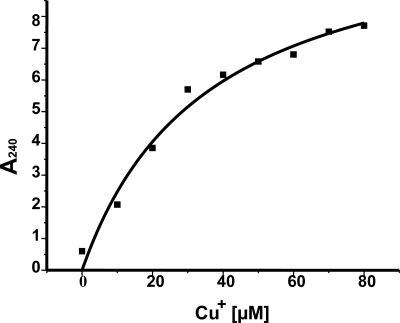
We cloned and overexpressed CsoR and the cytosolic domain (N-terminal 135 aa) of YcnJ to examine possible formation of oligomeric complexes in the presence of copper by using native gel electrophoresis. Polyacrylamide gels (6%) were loaded with assay mixtures containing protein samples that were incubated with and without 0.1 mM CuCl and 0.1 mM DTT. DTT was added to minimize the conversion of Cu(I) to Cu(II). A gel shift of the CsoR protein sample was observed in the presence of 0.1 mM CuCl and 0.1 mM DTT (Fig. 4B). Binding was also tested in the presence of the strong metal chelator EDTA. While incubation of a protein sample with 0.5 mM EDTA alone resulted in no shift, the shift was not abolished in the presence of EDTA and copper (Fig. 4B), indicating that CsoR has higher affinity to Cu(I) than EDTA. A similar observation has been made previously in binding competition experiments with CsoR and BCS (16). To define the half-maximal copper concentration (Cu0.5), it was necessary to saturate the CsoR protein; different copper concentrations of between 10 and 80 μM were added to the assay mixture, keeping the protein concentration constant at 80 μM. The formation of cysteine thiolate-copper complex was measured at 240 nm (Fig. 5), and the calculated Cu0.5 was found to be at 35.5 μM, suggesting a Cu(I)-CsoR complex stoichiometry of 1:1.
FIG. 4.
Copper-induced oligomerization. (A) The N-terminal part (135 aa) of recombinant YcnJ protein, purified using Ni-NTA chromatography, was loaded on denaturing 17% sodium dodecyl sulfate gel along with markers. The molecular mass of the protein was observed to be 16.5 kDa, including the His6 tag and two additional amino acids before the start codon (left panel). A native 6% polyacrylamide gel was loaded with 15 μg of purified protein either without metal preincubation as a control (first lane) or after incubation with either 0.2 mM CuCl or 0.2 mM CuSO4 (right panel). (B) CsoR recombinant protein, purified using Ni-NTA chromatography, was loaded on a denaturing 17% gel along with markers, and the molecular mass of the protein was observed to be 12.5 kDa, including the His6 tag (left panel). Copper binding studies were performed in a 6% native gel, in which lanes were loaded with 80 μg purified protein incubated without or with 200 μM Cu(I) and/or 100 μM EDTA as indicated (right panel). (C) YcnK recombinant protein, purified using Ni-NTA chromatography, was loaded on a denaturing 17% gel along with markers, and the molecular mass of the protein was observed to be 23 kDa, including the His6 tag (left panel). Metal binding studies with purified proteins were performed in 6% native gels. Lane 1 is loaded with 100 μg purified protein. Protein samples incubated with 200 μM Cu(I) and 200 μM Cu(II) are loaded in lanes 2 and 3.
FIG. 5.
Estimation of cysteine thiolate bond formation. Purified CsoR protein (80 μM) was incubated with different copper concentrations ranging from 10 μM to 80 μM, and the corresponding cysteine thiolate-copper complex formation was measured at 240 nm. The values obtained were plotted on a graph, and the Cu0.5 was determined.
Experiments performed with the recombinant periplasmic domain of YcnJ (N-terminal 135 aa) exhibited oligomerization specifically in the presence of Cu(II), which resulted in a clear shift of the preincubated protein under these conditions (Fig. 4A). The shift observed upon incubation with Cu(I) was rather weak and might also result from partial oxidation of Cu(I) to Cu(II). However, these findings suggest that YcnJ recruits into an oligomeric state if copper sensing and/or transport into the cell is mediated. The specific response of the predicted periplasmic N-terminal domain of YcnJ to Cu(II) further supports its role in uptake of extracellular oxidized copper, whereas copper-responsive regulators such as CsoR are specific for the intracellular copper in its reduced state.
In the case of YcnK, which was also recombinantly expressed and tested for copper-induced oligomerization, no such oligomerization in the presence of copper was observed (Fig. 4C). However, since oligomerization may also take place in the absence of copper and might additionally become further stabilized but not altered by addition of the metal, as already suggested for M. tuberculosis CsoR (16), there is yet no indication that YcnK does not oligomerize or does not bind copper at all.
Conclusions.
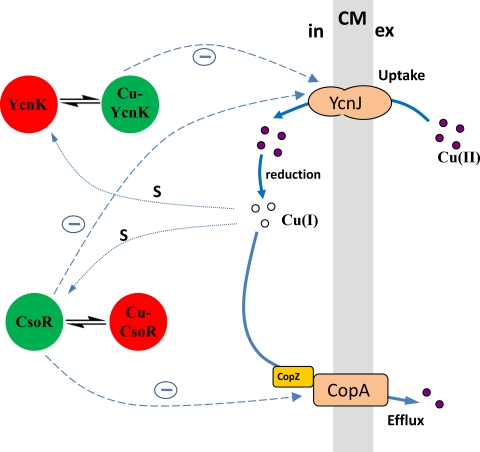
Transcriptome studies were performed to identify genes involved in copper homeostasis in B. subtilis. So far, copper transport in B. subtilis is known only as a function of efflux, which is mediated by the CsoR-dependent CopZA system and the nonspecific cation diffusion facilitator CzcD, which is repressed by the ArsR homolog CzrA (18, 26). Components and mechanisms of copper uptake have not yet been identified. In our approach, we have identified genes that were upregulated by copper deprivation and downregulated under copper excess. The ycnJ gene was especially highly upregulated under copper-limiting conditions. The sequence homology studies reveal that YcnJ is highly homologous at its N terminus to CopC and at its C terminus to CopD from P. syringae. Unlike P. syringae, in which copper uptake is assisted by two distinct proteins (CopC and CopD), B. subtilis YcnJ is organized in a single polypeptide chain that facilitates the direct transfer of copper ions across the membrane. Studies undertaken with P. syringae demonstrate either that the CopC protein could interact with CopA and perhaps the outer membrane protein CopB to perform copper sequestration or that CopC along with CopD may function in copper uptake. Disruption of the ycnJ gene in B. subtilis resulted in a growth-defective phenotype under copper-limiting conditions and a reduced intracellular copper content. Thus, these findings indicate that the primary role of YcnJ in B. subtilis is associated with copper uptake. A putative resistance function as observed for CopC in P. syringae cannot be excluded. However, since this resistance was described for the periplasmic compartment, this might be a rather secondary function in B. subtilis. In an attempt to find the possible transcriptional factors that regulate the expression of ycnJ, mutants with a deletion of the unknown transcriptional regulator gene ycnK in combination with a deletion of the recently identified copper-sensing repressor gene csoR were constructed and the ycnJ expression was quantified using dot blots. The ΔycnK mutant showed elevated expression of ycnJ compared to the WT, especially under copper excess conditions. Expression was further elevated in the background of the ΔcsoR ΔycnK double mutant, suggesting that both regulators participate in ycnJ expression control. The current model for copper homeostasis for B. subtilis (Fig. 6) shows the novel components presented here and indicates the regulatory connection between copper uptake and efflux systems. As far as this interplay has been investigated, it points out the demands on a system that is developed to maintain the essential accurate levels of copper inside the cell and, at the same time, to avoid passage over the critical threshold of copper toxification in order to allow proper physiological function.
FIG. 6.
Current model of copper homeostasis in B. subtilis, including functional components for copper uptake and efflux as well as their cognate repressors. Depending on their association with copper, the active state of transcriptional repressors YcnK and CsoR is shown in green, and the inactive state of transcriptional repressors is shown in red. The negative regulation of components (−) is indicated with dashed arrows. Copper sensing (s) is indicated with dotted arrows. Cu, copper; CM, cytoplasmic membrane; in, intracellular; ex, extracellular.
Acknowledgments
We gratefully acknowledge the Deutsche Forschungsgemeinshaft and Fonds der Chemischen industry for financial support.
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Arnesano, F., L. Banci, I. Bertini, S. Mangani, and A. R. Thompsett. 2003. A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc. Natl. Acad. Sci. USA 1003814-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17509-519. [DOI] [PubMed] [Google Scholar]
- 3.Bender, C. L., and D. A. Cooksey. 1986. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J. Bacteriol. 165534-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of sigma B levels and activity in Bacillus subtilis. J. Bacteriol. 1752347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cha, J., and D. A. Cooksey. 1991. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 888915-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooksey, D. A. 1994. Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol. Rev. 14381-386. [DOI] [PubMed] [Google Scholar]
- 5a.Cooper, D. G., C. R. Macdonald, S. J. B. Duff, and N. Kosaric. 1981. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation addition. Appl. Environ. Microbiol. 42408-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dancis, A., D. Haile, D. S. Yuan, and R. D. Klausner. 1994. The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 26925660-25667. [PubMed] [Google Scholar]
- 7.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 1853804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaballa, A., M. Cao, and J. D. Helmann. 2003. Two MerR homologues that affect copper induction of the Bacillus subtilis copZA operon. Microbiology 1493413-3421. [DOI] [PubMed] [Google Scholar]
- 9.Gaballa, A., and J. D. Helmann. 2003. Bacillus subtilis CPx-type ATPases: characterization of Cd, Zn, Co and Cu efflux systems. Biometals 16497-505. [DOI] [PubMed] [Google Scholar]
- 10.Garcia de la Nava, J., S. Van Hijum, and O. Trelles. 2003. PreP: gene expression data pre-processing. Bioinformatics 192328-2329. [DOI] [PubMed] [Google Scholar]
- 11.Georgatsou, E., L. A. Mavrogiannis, G. S. Fragiadakis, and D. Alexandraki. 1997. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 27213786-13792. [DOI] [PubMed] [Google Scholar]
- 12.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204305-320. [DOI] [PubMed] [Google Scholar]
- 13.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis, requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216281-291. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. M., G. Grass, C. Rensing, S. R. Barrett, C. J. D. Yates, J. V. Stoyanov, and N. L. Brown. 2002. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295616-620. [DOI] [PubMed] [Google Scholar]
- 15.Lim, C. K., and D. A. Cooksey. 1993. Characterization of chromosomal homologs of the plasmid-borne copper resistance operon of Pseudomonas syringae. J. Bacteriol. 1754492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, T., A. Ramesh, Z. Ma, S. K. Ward, L. Zhang, G. N. George, A. M. Talaat, J. C. Sacchettini, and D. P. Giedroc. 2007. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 360-68. [DOI] [PubMed] [Google Scholar]
- 17.McGuirl, M. A., J. A. Bollinger, N. Cosper, R. A. Scott, and D. M. Dooley. 2001. Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. J. Biol. Inorg. Chem. 6189-195. [DOI] [PubMed] [Google Scholar]
- 18.Moore, C. M., A. Gaballa, M. Hui, R. W. Ye, and J. D. Helmann. 2005. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol. Microbiol. 5727-40. [DOI] [PubMed] [Google Scholar]
- 19.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 1825864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odermatt, A., H. Suter, R. Krapf, and M. Solioz. 1992. An ATPase operon involved in copper resistance by Enterococcus hirae. Ann. N. Y. Acad. Sci. 671484-486. [DOI] [PubMed] [Google Scholar]
- 21.Outten, F. W., C. E. Outten, J. Hale, and T. V. O'Halloran. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J. Biol. Chem. 27531024-31029. [DOI] [PubMed] [Google Scholar]
- 22.Palmgren, M. G., and K. B. Axelsen. 1998. Evolution of P-type ATPases. Biochim. Biophys. Acta 136537-45. [DOI] [PubMed] [Google Scholar]
- 23.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27197-213. [DOI] [PubMed] [Google Scholar]
- 24.Rutherford, J. C., and A. J. Bird. 2004. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell 31-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Smaldone, G. T., and J. D. Helmann. 2007. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 1534123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solioz, M., and C. Vulpe. 1996. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 27237-241. [PubMed] [Google Scholar]
- 28.Solioz, M., and J. V. Stoyanov. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27183-195. [DOI] [PubMed] [Google Scholar]
- 29.Strausak, D., and M. Solioz. 1997. CopY is a copper-inducible repressor of Enterococcus hirae copper ATPases. J. Biochem. 2728932-8936. [DOI] [PubMed] [Google Scholar]
- 30.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 1392041-2045. [DOI] [PubMed] [Google Scholar]
- 31.Tetaz, T. J., and R. K. Luke. 1983. Plasmid-controlled resistance to copper in Escherichia coli. J. Bacteriol. 1541263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 1443097-3104. [DOI] [PubMed] [Google Scholar]
- 32.Van Hijum, S., N. J. Garcia, O. Trelles, J. Kok, and O. P. Kuipers. 2003. MicroPreP: a DNA microarray data preprocessing framework. Appl. Bioinformatics 2241-244. [PubMed] [Google Scholar]
- 33.Vulpe, C., B. Levinson, S. Whitney, S. Packman, and J. Gitschier. 1993. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper-transporting ATPase. Nat. Genet. 37-13. [DOI] [PubMed] [Google Scholar]
- 34.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology 15 regions for gene disruptions in S. cerevisiae. Yeast 12259-265. [DOI] [PubMed] [Google Scholar]
- 35.Williams, J. R., A. G. Morgan, D. A. Rouch, N. L. Brown, and B. T. Lee. 1993. Copper-resistant enteric bacteria from United Kingdom and Australian piggeries. Appl. Environ. Microbiol. 592531-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, Z., S. Labbe, M. M. O. Pena, and D. J. Thiele. 1998. Copper differentially regulates the activity and degradation of yeast MacI transcription factor. J. Biol. Chem. 2731277-1280. [DOI] [PubMed] [Google Scholar]