Abstract
Previous transcript mapping of the bacteriophage Aeh1 nrd operon revealed a predicted RNA hairpin upstream of the homing endonuclease mobE gene. We enzymatically mapped the hairpin, showing that the mobE ribosome binding site is sequestered. Cloning of the hairpin upstream of lacZ resulted in reduced β-galactosidase activity, consistent with translational regulation.
Homing endonucleases are a unique class of site-specific, sequence-tolerant DNA endonucleases that promote the mobility of their coding region and flanking sequences by a recombination-dependent process termed homing (2). Homing endonucleases are associated with self-splicing group I or II introns (2, 16) or inteins (10, 11), but they are also found as freestanding versions in the intergenic regions separating evolutionarily conserved genes (6, 15, 21, 28). Whereas the mobility pathways of freestanding endonucleases are likely similar to those used by intron-encoded versions (3, 17, 22, 25), relatively little is understood about the mechanisms regulating the expression of free-standing endonucleases. Control of endonuclease function is crucial to the successful integration of a free-standing endonuclease into host transcriptional units upon invasion of a genome, because unregulated expression of the endonuclease may perturb expression patterns of essential neighboring genes required for viability.
As a model system for studying the regulation of homing endonuclease function, we have focused on the aerobic ribonucleotide reductase (nrd) genomic region of T-even-like phage, which is a hot spot for endonuclease insertion (20, 21, 23, 26, 27). The genome of the Aeromonas hydrophila bacteriophage Aeh1 contains a putative freestanding endonuclease gene mobE, that has inserted into the large subunit gene (nrdA) of aerobic ribonucleotide reductase (Fig. 1A) (8). In other T-even phage, mobE is inserted between the nrdA and nrdB (small subunit of aerobic ribonucleotide reductase) genes (23, 26). The insertion of mobE in Aeh1 has split nrdA into two smaller genes, nrdA-a and nrdA-b, but remarkably, the ribonucleotide reductase of Aeh1 is functional (8).
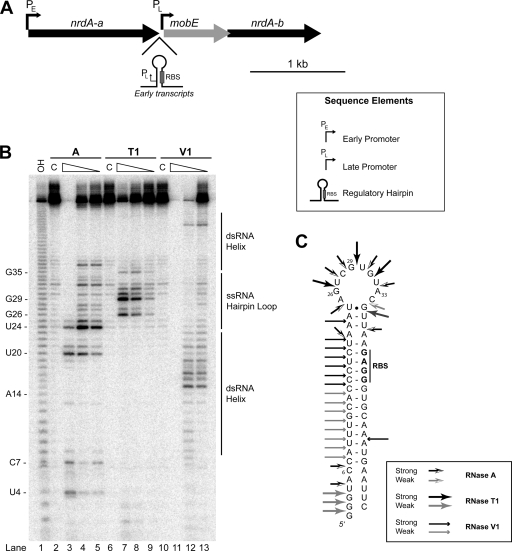
FIG. 1.
Enzymatic probing of the Aeh1 mobE regulatory hairpin. (A) Genomic organization and regulatory elements of the Aeh1 nrd genomic region (9). Genes are indicated by arrows indicating the direction of transcription, with the gene names designated above the arrows. (B) Enzymatic cleavage sites generated by single-stranded nucleases RNase T1 (G specific) and RNase A (U and C specific) and double-stranded nuclease RNase V1. Lane 1, sequencing ladder generated by alkaline hydrolysis; lanes 2, 6, and 10, undigested RNA (labeled C); lanes 3 to 5, RNase A; lanes 7 to 9, RNase T1; lanes 11 to 13, RNase V1. Each series of digestions are shown by decreasing concentrations of the RNase. (C) Model of secondary structure of mobE regulatory hairpin based on enzymatic mapping and Mfold (29) predictions.
A previous transcriptional profiling study of the Aeh1 nrd operon revealed a complex, multilayered mechanism to regulate the expression of the nrd genes and the embedded homing endonuclease gene, mobE (9). Early transcripts that initiate upstream of the nrdA-a gene are polycistronic and include a predicted RNA hairpin that sequesters the mobE ribosome binding site (RBS), presumably preventing MobE translation at early stages in Aeh1 infection. However, transcripts that initiate at the late promoter immediately upstream of mobE do not include sufficient sequence to form this secondary structure, likely facilitating translation of MobE (Fig. 1A). Here, we address the structure and function of the RNA hairpin that sequesters the mobE RBS.
The proposed model for translational repression of Aeh1 mobE predicts key canonical base pairs between the mobE RBS and the upstream RNA strand in a single internal loop hairpin. To obtain experimental evidence supporting the proposed hairpin structure, we enzymatically mapped in vitro-transcribed hairpin RNA. Enzymatic structural mapping is commonly used to confirm computer-predicted secondary structure models of RNA folds. For our study, three ribonucleases were chosen for secondary structure probing, as follows: RNase A, which cleaves 3′ to unpaired cytosine and uracil residues; RNase T1, which cleaves 3′ to unpaired guanine residues; and RNase V1, which cleaves base-paired or stacked regions of RNA. To generate a hairpin RNA substrate for enzymatic mapping, we ligated annealed oligonucleotides DE-286 (5′-CATTTGCACCCTCTAAATAGTCGTGTACGTTAAGAGGGTGCAATG) and DE-287 (5′-AATTCATTTGCACCCTCTTAACGTACACGACTATTTAGAGGGTGCAAATGGTAC) into the EcoRI and KpnI sites of pBluescript SK(−) to create pBS-Pin. Hairpin RNA of uniform length was transcribed from PstI-digested plasmid DNA by using T7 RNA polymerase (NEB), end labeled with γ-32P, gel purified, and resuspended in 1× structural buffer (Ambion). The end-labeled RNA was renatured at 30°C and digested with RNase A, RNase T1, or RNase V1. This temperature was chosen for structural studies because it is the optimal growth temperature for the Aeh1 host, A. hydrophila (5). All digestions were performed according to the manufacturer's instructions (Ambion), and the products were resolved on a 12% (wt/vol) denaturing polyacrylamide gel (19:1 acrylamide-to-bisacrylamide ratio) alongside a size marker generated by alkaline hydrolysis.
The results of partial digestion with RNase A, RNase T1, and RNase V1 are summarized in Fig. 1B. Partial digestions with RNase V1 revealed numerous cleavages from residues 7 to 25, suggesting that nucleotides in this region were base paired (Fig. 1B, lanes 11 to 13). Most importantly, strong RNase V1 cleavage signals were observed from residues 15 to 19, which are predicted to be involved in base-pairing the mobE RBS, while these same residues were found to be insensitive to cleavage by RNase A and RNase T1 (Fig. 1B, lanes 3 to 10). Residue 20 was found to be very sensitive to RNase A in addition to RNase V1, which suggests that this residue is transiently paired. In contrast, the single-stranded loop region predicted at the top of the hairpin was highly susceptible to cleavage by RNase A and RNase T1 (Fig. 1B, lanes 3 to 10). Specifically, the U, C, and G residues from positions 24 to 35 were cleaved by RNase A and RNase T1 (Fig. 1B, lanes 3 to 10). When the enzymatic digestion data were mapped to the predicted hairpin, we found that our data supported the model in which the mobE RBS is sequestered by an RNA structure (Fig. 1C). To our knowledge, this is the first report of the structural mapping of an RNA hairpin that functions in translational repression of a homing endonuclease.
To test the capacity of the mobE hairpin to repress translation, we constructed a series of plasmids encoding lacZ under the control of a constitutive Escherichia coli promoter. We amplified the lacZ gene from E. coli strain HB101 by using primers DE-333 (5′-CCGGTACCTTGACAATTAATCATCGGCTCGTATAATGCTAGCAGGGTACATGACTATGATTACGGATCC) and DE-334 (5′-CCTCTAGATTATTTTTGACACCAGACCAACTGG) and ligated the PCR product into pSP72 (Promega) by using KpnI and XbaI to create pLacZ. DE-333 introduces a BamHI site just downstream of the start codon of lacZ and a NheI site immediately upstream of it. The NheI and BamHI sites were used to create a series of hairpin lacZ constructs by ligating annealed oligonucleotides into pLacZ. Each pair of oligonucleotides represented a structural variant of the mobE hairpin. For each construct, we maintained the sequence of the mobE RBS (5′-GAGG-3′), because the translation initiation region of other A. hydrophila genes functions in E. coli (13). The wild-type hairpin construct, pHP, was created using DE-335 (5′-CTAGCAGGGTACATTTGCACCCTCTAAATAGTCGTGTACGTTAAGAGGGTGCAAATATGACTATGATTACG) and DE-336 (5′-GATCCGTAATCATAGTCATATTTGCACCCTCTTAACGTACACGACTATTTAGAGGGTGCAAATGTACCCTG); a mutant hairpin construct, pMutHP, in which the RBS is intact but the hairpin is unable to form, was generated using DE-337 (5′-CTAGCAGGGTACTAAACGTGGGAGATTTATCAGCTGTACGTTAAGGGGTGCAAATATGACTATGATTACG) and DE-338 (5′-GATCCGTAATCATAGTCATATTTGCACCCTCTTAACGTCAGCTGATAAATCTCCCACGTTTAGTACCCTG); a restored mutant, pRmutHP, in which the mutant hairpin has compensatory mutations to restore the hairpin structure, was generated with DE-339 (5′-CTAGCAGGGTACTAAACGTGCCTCATTTATCAGCTGTAGCAATTGAGGCACGTTTAATGACTATGATTACG) and DE-340 (5′-GATCCGTAATCATAGTCATTAAACGTGCCTCAATTGCTACAGCTGATAAATGAGGCACGTTTAGTACCCTG); and a control construct, pRBS(+), closely resembling the late initiated mobE transcripts that would not contain sufficient sequence to sequester the RBS, was generated using DE-341 (5′-CTAGCAGGGTACGTACGTTAAGAGGGTGCAAATATGACTATGATTACG) and DE-342 (5′-GATCCGTAATCATAGTCATATTTGCACCCTCTTAACGTACGTACCCTG) (Fig. 2B). All plasmids were transformed into DH5α and grown in LB supplemented with 100 μg/ml ampicillin and verified by digests and sequencing.
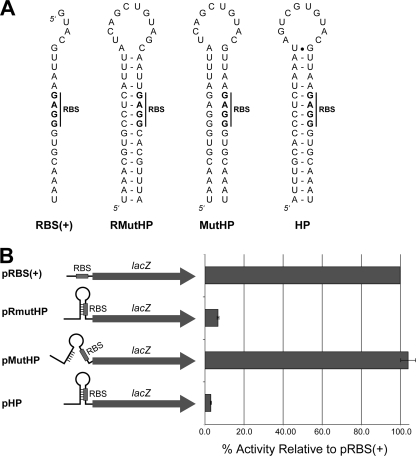
FIG. 2.
Effect of mobE hairpin on β-galactosidase activity. (A) Sequences compared to the native Aeh1 structure used in this study. RBS(+), ribosome binding site; RMutHP, restored mutant hairpin; MutHP, mutant hairpin; HP, wild-type hairpin found in Aeh1. (B) Summary of β-galactosidase activity for the HP-LacZ constructs expressed as percentage normalized to pRBS(+). Assays were performed in triplicate for each plasmid construct.
The constructs were assayed for β-galactosidase activity using Miller assays. In the HP-LacZ constructs, β-galactosidase activity was decreased by ∼97% for pHP with a wild-type hairpin relative to that in the pRBS(+) control (Fig. 2B). Disrupting base pairing in the stem region to free the RBS (pMutHP) resulted in increased activity compared to that for pHP, as expected. However, the pMutHP activity was 4% higher than that of the pRBS(+), possibly due to an additional translational initiation from an RBS-like sequence (5′-GGAG-3′) found upstream of the native RBS (Fig. 2A). Restoring base pairing in the stem to sequester the RBS (pRmutHP) reduced β-galactosidase activity to 7% of that of pRBS(+). The pLacZ constructs lacking an RBS had 2% β-galactosidase activity relative to that of pRBS(+) (data not shown). Collectively, our enzymatic mapping and lacZ fusion data suggest a model where the translation of MobE is limited by an RNA hairpin in which the RBS is sequestered by the secondary structure.
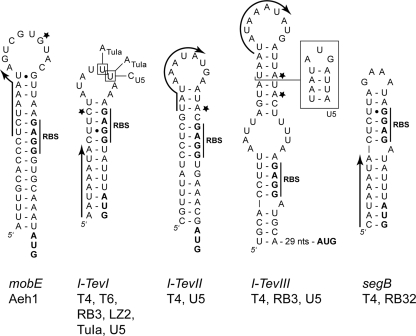
Many T4 late genes are cotranscribed with early genes on polycistronic messages, but translation of these late genes is repressed due to RNA secondary structures that are predicted to sequester the RBS. Examples of this arrangement include soc (capsid protein) (18), e (T4 lysozyme) (19), and gene 49 (endonuclease VII) (1) all of which are translationally repressed by an RNA hairpin on early transcripts. Similarly, translational repression regulates the expression of intron-encoded homing endonucleases I-TevI, I-TevII, and I-TevIII (7, 12) and has been suggested for the freestanding homing endonuclease gene segB (Fig. 3) (4). In the case of the homing endonucleases, however, the direct involvement of an RNA hairpin in regulating expression was not shown. Importantly, the hairpin structures found upstream of the intron-encoded endonucleases all possess middle or late promoters that generate structure-free mRNAs, therefore allowing the ribosomes access to the RBS (Fig. 3). In the case of segB, no middle or late promoters were predicted upstream of segB that would allow transcription in such a way as to prevent sequestration of the RBS; hence, translation of segB would be continually repressed throughout a T4 infection (4). However, we identified a potential late promoter forming the initial stem of the predicted hairpin with the sequence 5′-CATAAATA-3′ (Fig. 3), which is very similar to the consensus T4 late promoter sequence 5′-TATAAATA-3′ (21). If this promoter were functional, segB would be regulated by a mechanism analogous to that of mobE in Aeh1 and the intron-encoded endonucleases in T4.
FIG. 3.
Examples of hairpins upstream of homing endonuclease genes in T-even-like bacteriophage. Arrows indicate the position of late promoters in corresponding DNA sequence and the direction of transcription for each endonuclease gene. A star indicates the initiating nucleotide(s) in cases where it has been mapped (9, 12). The RBS and start codons are shown in boldface type. Nucleotide variants in the I-TevI hairpin are indicated for phages TuIa and U5 (27), and the alternative structure of the phage U5 I-TevIII hairpin is indicated by a box.
It is intriguing that similar translational regulatory mechanisms are employed by diverse T-even phage to regulate the expression of unrelated homing endonucleases that are embedded within different transcriptional units. Strong negative regulation of endonuclease function may limit spurious cleavage of the phage genome during critical stages of replication and gene expression. For instance, overexpression of cloned T4 mobE in E. coli resulted in nonspecific DNA degradation within 15 min of induction (24). Similarly, attempts to clone the Aeh1 mobE gene in a wild-type form have not yet been successful, likely due to extreme toxicity of the enzyme (E. A. Gibb and D. R. Edgell, unpublished data). It is also possible that regulating endonuclease expression to late stages in the infective cycle may enhance endonuclease-mediated mobility because sufficient genome equivalents would be available to initiate and repair a homing event. Moreover, in phage, T4 endonuclease-mediated homing is dependent on key phage-encoded recombination and replication proteins (14, 22, 25), and limiting expression to late stages to coincide with the recombination-dependent mode of DNA replication may further enhance mobility.
Acknowledgments
This work was supported by an operating grant (MOP77779) from the Canadian Institute of Health Research, a discovery grant (311610-2005) to D.R.E. from the Natural Sciences and Engineering Council of Canada, and an Early Researcher Award to D.R.E. from the Government of Ontario.
Footnotes
Published ahead of print on 30 January 2009.
REFERENCES
- 1.Barth, K. A., D. Powell, M. Trupin, and G. Mosig. 1988. Regulation of two nested proteins from gene 49 (recombination endonuclease VII) and of a lambda RexA-like protein of bacteriophage T4. Genetics 120329-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belfort, M., V. Derbyshire, B. Cousineau, and A. Lambowitz. 2002. Mobile introns: pathways and proteins, p. 761-783. In N. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 3.Belle, A., M. Landthaler, and D. A. Shub. 2002. Intronless homing: site-specific endonuclease SegF of bacteriophage T4 mediates localized marker exclusion analogous to homing endonucleases of group I introns. Genes Dev. 16351-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brok-Volchanskaya, V. S., F. A. Kadyrov, D. E. Sivogrivov, P. M. Kolosov, A. S. Sokolov, M. G. Shlyapnikov, V. M. Kryukov, and I. E. Granovsky. 2008. Phage T4 SegB protein is a homing endonuclease required for the preferred inheritance of T4 tRNA gene region occurring in co-infection with a related phage. Nucleic Acids Res. 362094-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow, M. S., and M. A. Rouf. 1983. Isolation and partial characterization of two Aeromonas hydrophila bacteriophages. Appl. Environ. Microbiol. 451670-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgell, D. R. 2005. Free-standing endonucleases of T-even phages: free-loaders or functionaries?, p. 147-160. In M. Belfort, B. L. Stoddard, D. W. Wood, and V. Derbyshire (ed.), Homing endonucleases and inteins. Springer-Verlag, Heidelberg, Germany.
- 7.Edgell, D. R., V. Derbyshire, P. Van Roey, S. LaBonne, M. J. Stanger, Z. Li, T. M. Boyd, D. A. Shub, and M. Belfort. 2004. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nat. Struct. Mol. Biol. 11936-944. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich, N. C., E. Torrents, E. A. Gibb, M. Sahlin, B. M. Sjoberg, and D. R. Edgell. 2007. Insertion of a homing endonuclease creates a genes-in-pieces ribonucleotide reductase that retains function. Proc. Natl. Acad. Sci. USA 1046176-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibb, E. A., and D. R. Edgell. 2007. Multiple controls regulate the expression of mobE, an HNH homing endonuclease gene embedded within a ribonucleotide reductase gene of phage Aeh1. J. Bacteriol. 1894648-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gimble, F. S. 2000. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett. 18599-107. [DOI] [PubMed] [Google Scholar]
- 11.Gogarten, J. P., A. G. Senejani, O. Zhaxybayeva, L. Olendzenski, and E. Hilario. 2002. Inteins: structure, function, and evolution. Annu. Rev. Microbiol. 56263-287. [DOI] [PubMed] [Google Scholar]
- 12.Gott, J. M., A. Zeeh, D. Bell-Pedersen, K. Ehrenman, M. Belfort, and D. A. Shub. 1988. Genes within genes: independent expression of phage T4 intron open reading frames and the genes in which they reside. Genes Dev. 21791-1799. [DOI] [PubMed] [Google Scholar]
- 13.Howard, S. P., and J. T. Buckley. 1986. Molecular cloning and expression in Escherichia coli of the structural gene for the hemolytic toxin aerolysin from Aeromonas hydrophila. Mol. Gen. Genet. 204289-295. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y. J., M. M. Parker, and M. Belfort. 1999. Role of exonucleolytic degradation in group I intron homing in phage T4. Genetics 1531501-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kutter, E., K. Gachechiladze, A. Poglazov, E. Marusich, M. Shneider, P. Aronsson, A. Napuli, D. Porter, and V. Mesyanzhinov. 1995. Evolution of T4-related phages. Virus Genes 11285-297. [DOI] [PubMed] [Google Scholar]
- 16.Lambowitz, A. M., and S. Zimmerly. 2004. Mobile group II introns. Annu. Rev. Genet. 381-35. [DOI] [PubMed] [Google Scholar]
- 17.Liu, Q., A. Belle, D. A. Shub, M. Belfort, and D. R. Edgell. 2003. SegG endonuclease promotes marker exclusion and mediates co-conversion from a distant cleavage site. J. Mol. Biol. 33413-23. [DOI] [PubMed] [Google Scholar]
- 18.Macdonald, P. M., E. Kutter, and G. Mosig. 1984. Regulation of a bacteriophage T4 late gene, soc, which maps in an early region. Genetics 10617-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McPheeters, D. S., A. Christensen, E. T. Young, G. Stormo, and L. Gold. 1986. Translational regulation of expression of the bacteriophage T4 lysozyme gene. Nucleic Acids Res. 145813-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 1855220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 6786-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller, J. E., J. Clyman, Y. J. Huang, M. M. Parker, and M. Belfort. 1996. Intron mobility in phage T4 occurs in the context of recombination-dependent DNA replication by way of multiple pathways. Genes Dev. 10351-364. [DOI] [PubMed] [Google Scholar]
- 23.Nolan, J. M., V. Petrov, C. Bertrand, H. M. Krisch, and J. D. Karam. 2006. Genetic diversity among five T4-like bacteriophages. Virol. J. 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nord, D. 2007. Homing endonucleases and horizontal gene transfer in bacteria and bacteriophages. Stockholm University, Stockholm, Sweden.
- 25.Parker, M. M., M. Belisle, and M. Belfort. 1999. Intron homing with limited exon homology. Illegitimate double-strand-break repair in intron acquisition by phage T4. Genetics 1531513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandegren, L., D. Nord, and B. M. Sjoberg. 2005. SegH and Hef: two novel homing endonucleases whose genes replace the mobC and mobE genes in several T4-related phages. Nucleic Acids Res. 336203-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandegren, L., and B. M. Sjoberg. 2004. Distribution, sequence homology, and homing of group I introns among T-even-like bacteriophages: evidence for recent transfer of old introns. J. Biol. Chem. 27922218-22227. [DOI] [PubMed] [Google Scholar]
- 28.Sharma, M., R. L. Ellis, and D. M. Hinton. 1992. Identification of a family of bacteriophage T4 genes encoding proteins similar to those present in group I introns of fungi and phage. Proc. Natl. Acad. Sci. USA 896658-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]