Abstract
The role of chromosomally encoded toxin-antitoxin (TA) loci in bacterial physiology has been under debate, with the toxin proposed as either an inducer of bacteriostasis or a mediator of programmed cell death (PCD). We report here that ectopic expression of MazFSa, a toxin of the TA module from Staphylococcus aureus, led to a rapid decrease in CFU counts but most cells remained viable as determined by differential Syto 9 and propidium iodide staining after MazFSa induction. This finding suggested that the toxin MazFSa induced cell stasis rather than cell death. We also showed that MazFSa selectively cleaves cellular mRNAs in vivo, avoiding “important” transcripts such as recA, gyrB, and sarA mRNAs in MazFSa-induced cells, while these three mRNAs can be cleaved in vitro. The results of Northwestern blotting showed that both sarA and recA mRNAs bind strongly to a putative RNA-binding protein. These data suggest that S. aureus likely undergoes stasis by protecting selective mRNA with RNA-binding proteins upon the expression of MazFSa in vivo.
Many bacteria have chromosomally encoded toxin-antitoxin (TA) loci in which the toxin and antitoxin genes exist in an operon and are coexpressed to form a TA complex. The toxin is stable, while the antitoxin is labile and can be degraded in vivo by host proteases (e.g., ClpP or Lon in Escherichia coli). Under conditions of stress whereby transcription of the TA operon is repressed and which hence preclude the continuous synthesis of the labile antitoxin, the more-stable toxin can unleash its toxic effect to inhibit cell growth. However, metabolic stresses, such as amino acid and carbon source starvation, have been shown to induce transcription of E. coli mazEF and other TA loci in E. coli (7, 9, 14). Studies with several toxin systems indicate that many toxins are probably sequence-specific endoribonucleases. For instance, MazF of E. coli cleaves mRNA at ACA sites both in vitro and in vivo (30), while the RelE toxin, also from E. coli, cleaves mRNA positioned at the ribosomal A site both in vitro and in vivo, with cleavage occurring between the second and third bases of the A site codon (UAR, where R is usually G or A) (22). The PemK toxin from plasmid R100 in E. coli also cleaves mRNA at the UAH site (where H is A, C, or U) (29). ChpBK from E. coli cleaves at ACD (where D is A, G, or U) in single-stranded mRNA (31), whereas EndoA, a Bacillus subtilis MazF homolog, cleaves mRNA at the UAC site (23). Recently, two MazF homologs from Mycobacterium tuberculosis were also found to be endoribonucleases, with one cleaving mRNA at UAC triplets and the other at U-rich regions (32).
Ectopic overexpression of MazF in E. coli has been shown to reduce cell viability as measured by CFUs (1, 2, 16). However, cell viability could be fully restored by the expression of the antitoxin MazE after induction of the MazF toxin (21). This cell viability phenomenon has been known to be reversible for up to ∼6 h after toxin induction, beyond which there is a point of no return and a gradual loss of colony formation (2). Overexpression of MazF in E. coli blocks protein synthesis by degrading many cellular mRNAs, eventually leading to growth arrest (30). More recently, Suzuki et al. (28) developed a single-protein production system based on MazF-induced cells which exhibited growth arrest but retained the full spectrum of biosynthetic functions required for transcription and translation of a codon-modified mRNA for up to 4 days.
The physiological roles of toxin proteins remain ill defined. Although various propositions have been advanced, two theories remain most prominent, with one contending that TA modules induce programmed cell death (PCD) and the other supporting the role of MazF in causing bacterial stasis. The evidence for the first model comes from studies of the mazEF TA genes in E. coli, located downstream of relA. The relA gene encodes a ppGpp synthase and is upregulated in response to uncharged tRNA at the ribosomal A site during amino acid starvation and other stressful conditions, including antibiotic exposure (1, 14, 21). In E. coli strain MC4100, overproduction of ppGpp by hyperexpressing RelA′, a truncated version of ppGpp synthase, represses mazEF promoter expression. Based on the above-described evidence, these authors suggested that stress-induced upregulation of ppGpp in E. coli strain MC4100 reduced synthesis of the labile MazE antitoxin and promoted its degradation in the presence of an activated ClpPA system and, hence, unleashed the toxic effect of MazF to advance to PCD (1).
Another model, proposed by Gerdes et al. (14), supported the role of TA systems in bacterial stasis under conditions of stress. In support of their hypothesis, Christensen et al. (6) found transcription of mazEF to increase during amino acid starvation induced by serine hydroxamate, accompanied by no cell killing. Pedersen et al. (21) also showed that the toxicity of MazF in E. coli is reversible and can be rescued by the antitoxin MazE within 6 h after MazF induction. Another TA module, HipAB, from E. coli was recently shown to induce a bacteriostatic state by the expression of the toxin HipA, which can also be reversed by the expression of its cognate antitoxin, HipB (18). Finally, the toxin of an orphan family of TA loci, evolutionarily unrelated to previously described TA families and found in plasmids of the Inc18 group, induces loss of cell proliferation in both Bacillus subtilis and E. coli, resulting in viable but nonreplicative cells within a defined time window beyond which cell death might ensue (19). Based on these studies, these authors proposed that the MazF toxin may not induce cell killing but rather may promote bacterial stasis.
In a recent study, we showed that the two open reading frames immediately upstream of the sigB operon in Staphylococcus aureus, herein designated mazESa and mazFSa, represent a TA system (11). We demonstrated that the toxin MazFSa is a sequence-specific endoribonuclease which cleaves model substrate ctpA mRNA at a consensus U-rich sequence of VUUV′ (where V and V′ are A, C, or G and may or may not be identical) both in vivo and in vitro. Overexpression of MazFSa was also found to reduce the CFU counts (11), while coexpression of MazEFSa had no effect on reducing CFUs. In this report, we demonstrate that ectopic overexpression of MazFSa induces S. aureus to undergo bacteriostasis rather than cell death. Importantly, MazFSa does not cleave all mRNAs containing VUUV′ sequences. The mRNAs of some essential genes and also a global regulator are found to be protected in vivo. We thus propose that this “protective effect” is mediated by RNA-binding proteins, since these mRNAs, while protected in vivo, can be cleaved by MazFSa in vitro. Thus, our report here suggests the selectivity of MazFSa for target mRNAs and helps explain why the expression of toxin is not immediately bactericidal and, hence, can be reversed by the expression of antitoxin within a defined time window. Additionally, our data also explain why cells undergoing MazF-induced growth arrest can still retain transcriptional and translational competence.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
We used E. coli strains DH5α and BL21(DE3)/pLysS and Staphylococcus aureus ALC6094 for these studies. S. aureus ALC6094 is a derivative of strain Newman that contains the lacUV5-regulated T7 polymerase gene integrated into the geh locus in the chromosome (8, 11). For our studies with MazFSa, ALC6094 was transformed with the plasmid pG164, which carries an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 promoter driving the expression of mazFSa (8, 11). In some studies, we also deployed a sarA deletion mutant of ALC6094. Cultures were routinely grown in LB for E. coli and in Trypticase soy broth or O3GL medium for S. aureus, with aeration at 37°C. For pulse-chase labeling experiments, S. aureus ALC6094 was grown in chemically defined medium (CDM) as described previously (15). The media were supplemented with either ampicillin (70 μg/ml) or chloramphenicol (10 μg/ml). Rifampin (rifampicin) at 200 μg/ml was also used to determine RNA decay in the culture at an optical density at 650 nm (OD650) of 0.4.
Bacterial viability assay.
Bacteria were stained with membrane-permeant Syto 9 and membrane-impermeant propidium iodide by using a Live/Dead BacLight bacterial viability kit (Molecular Probes, Eugene, OR). The kit is based on a combination of two probes; Syto 9 is a membrane-permeant nucleic acid stain (green fluorescence at 530 nm upon excitation at 488 nm) used to label all cells, whereas propidium iodide (red fluorescence at 620 nm upon excitation at 488 nm) enters only cells with compromised membranes. Bacteria with intact cell membranes indicative of live cells were stained fluorescent green, whereas bacteria with damaged membranes were stained fluorescent red due to acquisition of propidium iodide. The green/red fluorescence ratios were quantitated in an FL600 fluorescence reader (BioTek Instruments, Winooski, VT). The percentage of live cells was calculated from the standard curve according to the manufacturer's protocol. In brief, appropriate dilutions from the samples were prepared, stained with Live/Dead probes, and measured for fluorescence in triplicate. Culture that was heat-killed at 60°C for 1 h was the positive control and was defined as dead cells, whereas a fresh culture without any MazFSa induction was defined as 100% live cells. Suspensions containing various proportions of live and dead cells (0, 10, 50, 90, or 100%) were used to prepare the standard curves (relationship between the percentage of live bacteria and the green/red fluorescence ratio). The analysis of relative viability of the samples was derived from the standard curves.
Construction of plasmids.
Using S. aureus Newman chromosomal DNA as the template, the following genes or gene fragments were amplified by PCR: sarA, recA (600 bp of the C terminus), hla, and sigB (GenBank accession number NC_002745). The list of primers used for these and other studies described below can be obtained from the authors. The PCR products were digested with NcoI and BamHI and cloned into the NcoI and BamHI sites of pET14b (Novagen) to produce pET14b-sarA, pET14b-recA, pET14b-hla, and pET14b-sigB. The construction of plasmids pG164-MazF(His6) and pETDuet1-MazEF(His6), encoding MazFSa and MazEFSa, respectively, was described previously (11). We also constructed a derivative of pG164-MazF(His6) in which a fragment containing a modified pEPSA5 promoter (10) driving the sarA coding region was cloned downstream of the transcription terminator of mazF. The expression from the modified pEPSA5 promoter lacking the XylR repressor was moderate but constitutive, while the expression from the T7 promoter in pG164 remained IPTG-inducible (8, 10, 11). The recombinant plasmid, designated pG164-MazF(His6)-pEPSA5-sarA, was then introduced into S. aureus strain RN4220 by electroporation. The plasmid harvested from RN4220 was then transformed into S. aureus strain ALC6094 and its isogenic sarA mutant. All of the above DNA techniques were performed according to standard procedures (25).
Protein expression and purification.
To express MazFSa(His6), MazESa and MazFSa(His6) were coexpressed in E. coli BL21(DE3)/pLysS harboring the plasmid pETDuet1-MazEF(His6) after IPTG induction (1 mM) for 6 h. The cells were harvested and subjected to lysis by ultrasonification. The MazE-MazFSa(His6) complex was purified with a nickel-nitrilotriacetic acid resin affinity column (Novagen) according to the manufacturer's protocol. MazFSa(His6) was further purified from the denatured MazE-MazFSa(His6) complex (in 6 M guanidine-HCl) and refolded by stepwise dialysis as described previously (11).
Production of anti-MazFSa, anti-SarA, and anti-SigB antibodies and Western blotting.
The purified MazFSa(His6) protein (100 μg) was used to immunize three BALB/c × SJL/J (F1 cross) mice to obtain the anti-MazFSa antibodies as described previously (5). Anti-SarA and anti-SigB monoclonal antibodies were obtained as described previously (4, 5). Western blot analysis was carried out on cellular lysates as described previously (11, 12).
In vitro cleavage of mRNA by MazFSa.
The sarA and recA mRNAs were transcribed from BamHI-linearized pET14b-sarA and pET14b-recA plasmids by using a T7 large-scale transcription kit (Promega) as described previously (11). Five micrograms of mRNAs was digested with 15 pmol of MazFSa at 37°C for various time periods in 20-μl reaction mixtures containing 40 U RNase inhibitor, 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 1 mM dithiothreitol (DTT). The reaction mixture was then extracted with phenol-chloroform, ethanol precipitated to remove proteins, and subjected to agarose gel electrophoresis, followed by detection of the cleaved fragments by Northern blotting with [α-32P]dCTP-radiolabeled sarA and recA DNA probes (see below).
Northern blot hybridization.
Total RNA from S. aureus cells was prepared by using Trizol (Invitrogen, CA) and a reciprocating shaker with 0.1-mm silica-zirconia beads as previously described (3). For detection of specific transcripts, gel-purified DNA probes were radiolabeled with [α-32P]dCTP by use of a random-primed DNA-labeling kit (Roche Diagnostics GmbH) and hybridized under aqueous-phase conditions at 65°C. The blots were subsequently washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS) twice at room temperature and 1× SSC, 0.1% SDS twice at 65°C and autoradiographed as previously described (20).
Pulse-chase labeling of de novo protein synthesis with [35S]methionine.
S. aureus ALC6094 harboring pG164-MazF(His6) was inoculated for overnight growth in O3GL medium at 37°C, followed by 1:100 dilution into the CDM without methionine the next morning. When the culture reached an OD650 of 0.4, IPTG (1 mM) was added to induce the expression of MazFSa, while another aliquot without IPTG was used as the noninduced culture control. At selected time intervals (0 to 90 min) after induction, 1 ml of the induced or noninduced culture was taken and added to a test tube containing 15 μCi [35S]methionine. After isotopic labeling for 15 min at 37°C, 0.2 ml of regular methionine (40 mg/ml) was added to chase the culture for 5 min. The labeled cells were pelleted and washed with Tris-buffered saline (TBS), followed by resuspension in lysis buffer (TBS containing 2 mM phenylmethylsulfonyl fluoride proteinase inhibitor and 50 mM EDTA). The cells were then lysed with 0.1-mm silica-zirconia beads in a reciprocating shaker as previously described (3). The clarified supernatant was treated with human immunoglobulin G (200 μg/ml) for 1 h with gentle mixing at 4°C to facilitate binding to protein A. Anti-SarA or anti-SigB monoclonal antibody was then added to immunoprecipitate SarA or SigB protein by gentle mixing at 4°C overnight. A 100-μl (50% slurry) amount of goat polyclonal antibody to mouse conjugated to immunoglobulin G-agarose (Abcam) was then added to pull down the immunoprecipitate by mixing the agarose beads with the reaction mixture for 1 h at 4°C. The sample was pelleted by centrifugation at 200 × g for 1 min and washed with TBS. Fifty microliters of sample buffer was added to the agarose beads, followed by heating at 100°C for 5 min to release the proteins. Samples were then resolved by SDS-polyacrylamide gel electrophoresis (PAGE). For the detection of resolved radiolabeled proteins, the gel was fixed with a fixing solution (isopropanol/water/acetic acid [25:65:10]) for 30 min, followed by soaking in Amplify solution (Amersham) with agitation for 15 to 30 min. The gel was then dried for autoradiography.
Northwestern blotting.
Total-cell lysate from S. aureus was prepared by lysing cells in the lysis buffer (50 mM Tris-HCl, 5 mM EDTA, pH 8.0) with 0.1-mm silica-zirconia beads in a reciprocating shaker as previously described (3). 32P-labeled mRNA probes were transcribed from BamHI-linearized pET14b-sarA, pET14b-recA, pET14b-hla, and pET14b-sigB plasmids by using a T7 large-scale transcription kit (Promega) according to the manufacturer's protocol. Northwestern blotting was carried out with a modified protocol as previously described (24). In brief, cell lysates (20 μg) were resolved on 16% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membrane. Proteins on the membrane were denatured in denaturing buffer (50 mM Tris-HCl, pH 8.3, 50 mM DTT, 1 mM EDTA, 1 M guanidine-HCl) for 1 h at room temperature. The membrane was then washed twice with renaturing buffer (50 mM Tris-HCl, pH 7.5, 2 mM DTT, 2 mM EDTA, 0.5 M NaCl, 0.1% Tween 20, 10% glycerol) for 1 h at room temperature, and proteins were allowed to renature in the renaturing buffer at 4°C for 24 h. The membrane was then blocked with blocking solution (0.02% bovine serum albumin, 30 mM HEPES-NaOH, pH 8.0, 20 μg/ml yeast tRNA) for 1 h at room temperature and hybridized with 2 ng/ml of 32P-labeled RNA probes (5 × 106 cpm) in binding buffer (30 mM HEPES-NaOH, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 20 μg/ml yeast tRNA) for 3 h at room temperature. The membrane was washed three times with washing buffer (30 mM HEPES-NaOH, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 2 mM EDTA), air dried, and exposed to films to visualize bands that represented putative RNA-binding proteins.
RESULTS
Overproduction of MazFSa induced cell stasis rather than cell death.
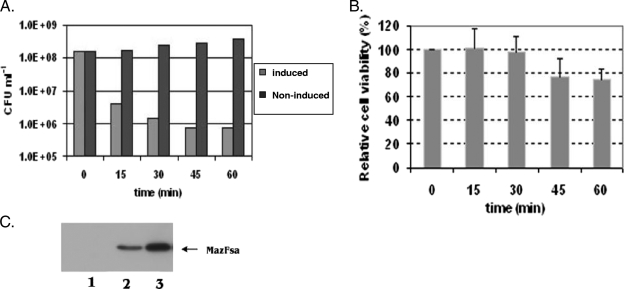
To determine the extent to which ectopic overexpression of MazFSa affects S. aureus, we examined its induction by IPTG in S. aureus ALC6094 harboring pG164-MazF(His6) at different growth phases and the effects on CFU counts and cell viability. Cultures showed normal growth under the uninduced conditions (Fig. 1A). When MazFSa was induced in early logarithmic phase (at an OD650 of 0.4), the CFU counts decreased rapidly, with 99.5% of the original CFU lost at 60 min postinduction (Fig. 1A). To ensure that MazFSa was indeed expressed in the induced culture, we monitored and detected successful expression of MazFSa on immunoblots with anti-MazFSa antibodies (Fig. 1C). To examine whether the cells associated with loss of CFU were indeed dead, the induced and noninduced cultures were further examined for viability with a BacLight Live/Dead bacterial viability assay kit (Molecular Probes), which stains membrane-compromised dead bacteria fluorescent red due to the uptake of propidium iodide. Although there was a ∼100-fold difference in CFU counts between the induced and noninduced cultures at an OD650 of 0.4 at 60 min postinduction, the difference between the cultures in cell death was less than 27% (Fig. 1B). These results suggest that cells exposed to ectopic overexpression of MazFSa are not killed in the first hour but instead are induced to undergo stasis.
FIG. 1.
Reduction of the CFU counts by ectopic overexpression of MazFSa. (A) Reduction of CFUs with S. aureus ALC6094 harboring pG164-MazF(His6) under IPTG induction (1 mM) at an OD650 of 0.4. To determine the CFUs, samples were withdrawn at various time points as indicated, diluted, spread on TSA plates supplemented with chloramphenicol (10 μg/ml), and incubated at 37°C overnight. The results are those from one typical experiment out of three similar independent experiments. (B) Relative cell viability. Cultures induced at an OD650 of 0.4 with or without IPTG (1 mM) were assessed for percentage of cell viability with a Live/Dead BacLight bacterial viability kit at indicated time points, as described in Materials and Methods. The percentage of viable cells from the induced culture at each time point was further compared to that of the noninduced culture to arrive at the relative cell viability. The results represent those of three similar independent experiments. Error bars show standard deviations. (C) Western blot analysis of MazFSa expression after IPTG (1 mM) induction at an OD650 of 0.4. Twenty micrograms of cell lysate proteins from each time point was resolved by SDS-PAGE, transferred to PVDF membrane, and probed with the anti-MazFSa antibodies as described in Materials and Methods. Lanes 1 to 3 show cell lysates prepared after induction with IPTG for 0, 30, or 60 min, respectively.
MazFSa cleaves cellular mRNAs selectively.
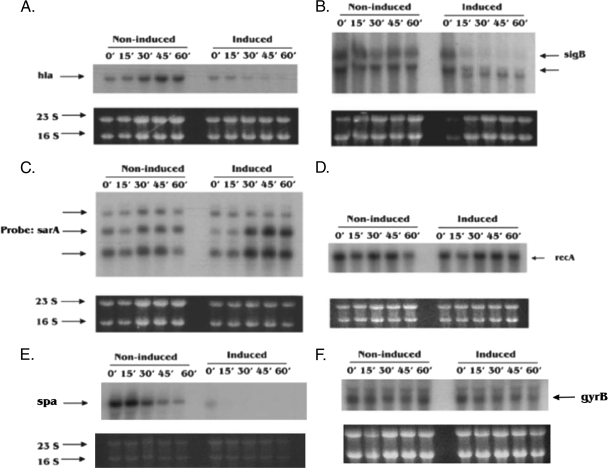
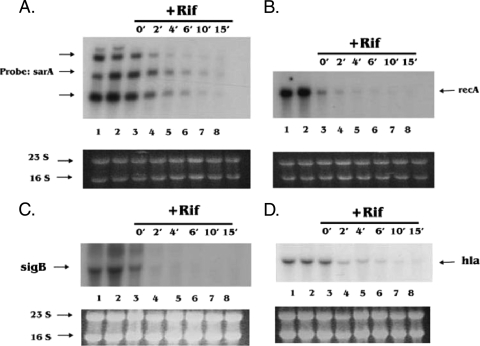
The overexpression of toxins from TA modules, such as relBE, mazEF, hipAB, and ɛζ, has been shown to be associated with a bacteriostatic state which can be fully reversed upon the expression of their cognate antitoxins (18, 19, 21). To examine this bacteriostatic state in S. aureus, we focused on the degradation of cellular mRNAs upon ectopic overexpression of MazFSa, which targets mRNA at the VUUV′ site. Northern blotting was carried out to analyze total cellular RNAs extracted at different time points after the induction of MazFSa as described in Materials and Methods. Six transcripts representing different gene categories (e.g., virulence, stress, regulator, and essential genes) that contain a number of VUUV′ sites (where V and V′ are A, C, or G and may or may not be identical) were chosen to evaluate the degradation of mRNAs after induction of MazFSa at an OD650 of 0.4. The transcript of the virulence gene hla, which has 45 VUUV′ sites, was found to degrade rapidly, being barely detectable at 60 min postinduction (Fig. 2A). The mRNA of another virulence gene, spa, was degraded in a manner similar to that of hla mRNA (Fig. 2E). The sigB transcript, which has 39 VUUV′ sites and encodes the stress-induced transcription factor SigB within an operon, was also cleaved after 30 min of induction with MazFSa (Fig. 2B). Remarkably, the transcript of the essential gene recA, which has 46 VUUV′ sites, was found to be stable against MazFSa- mediated degradation (Fig. 2D), as was the gyrB transcript (Fig. 2F), even at 60 min postinduction. Additionally, the three transcripts associated with the global regulator sarA, which has 24 VUUV′ sites, were preserved and possibly slightly upregulated after 30 min of induction (Fig. 2C). Notably, the 16S and 23S rRNAs were quite stable against MazFSa cleavage in vivo, with no appreciable changes in their band intensities on an agarose gel after exposure to MazFSa for 60 min, beginning at an initial OD650 of 0.4 (Fig. 2, bottom panels), thus indicating that rRNAs were protected from MazFSa cleavage in vivo, most likely by ribosomal proteins (30). When an S. aureus culture containing only the pG164 vector control (i.e., no MazFSa) was induced with IPTG, Northern blots with the above gene probes showed the same pattern as that of the uninduced control (data not shown), thus indicating that induction of the T7 polymerase gene with IPTG by itself did not preserve or upregulate sarA transcripts (e.g., by repressing a repressor or activating an activator), nor did it cause degradation of the mRNA transcripts we tested.
FIG. 2.
Degradation of mRNAs upon ectopic overexpression of MazFSa. A culture of S. aureus ALC6094 harboring pG164-MazF(His6) was induced at an OD650 of 0.4 with IPTG (1 mM), while the noninduced culture contained no IPTG. Total cellular RNAs were extracted at the indicated time points (minutes). Northern blot analysis was carried out with specific probes as described in Materials and Methods. (A to F) Transcripts were detected with hla, sigB, sarA, recA, spa, or gyrB probes. The data represent one typical experiment out of three similar independent experiments. The 23S and 16S bands served as the internal loading controls.
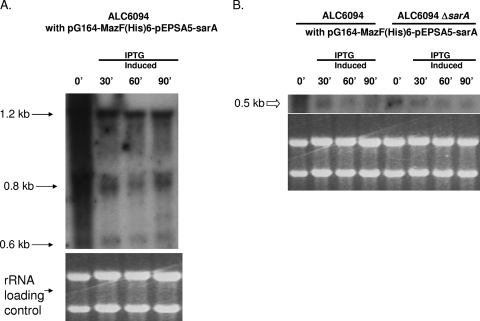
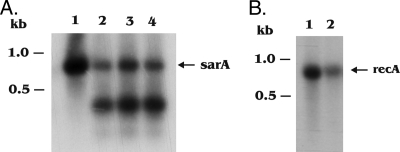
To further confirm that induced MazFSa did not cleave an mRNA encoding a repressor of the sarA promoter, we cloned in E. coli the sarA coding region downstream of a modified pEPSA5 promoter lacking the XylR repressor (10), thus resulting in mild constitutive expression of sarA from the modified pEPSA5 promoter. This fragment was then cloned downstream of the mazF transcription terminator in pG164-MazF(His6) to yield the recombinant plasmid pG164-MazF(His6)-pEPSA5-sarA containing an IPTG-inducible T7 promoter and a modified pEPSA5 promoter. The plasmid construct was then introduced into ALC6094 and its isogenic sarA mutant. Total cellular RNAs were then extracted from these cells before and after IPTG induction. Northern analyses of total cellular RNA with a sarA probe encompassing only the sarA coding region disclosed that the sarA transcripts, sized at 1.2, 0.8, and 0.6 kb and originating from the native triple promoters, were still preserved in the parental strain ALC6094 upon MazFSa induction (Fig. 3A). Notably, the sarA transcript originating from the modified pEPSA5 promoter in the recombinant plasmid, ∼0.5 kb in size (Fig. 3B), was preserved in the parental strain ALC6094 and in the isogenic sarA mutant. As this 0.5-kb transcript was transcribed from a heterologous promoter, this finding suggested that the preservation of this transcript upon MazFSa induction could not have arisen from MazF-mediated cleavage of a transcript encoding a repressor that targets the native sarA promoter. Taken together, the above results suggest that overexpression of MazFSa in S. aureus does not cleave all cellular mRNAs, since some mRNAs coding for selective virulence regulator or essential gene products were not targeted by MazFSa. This scenario where some gene transcripts are protected from MazFSa cleavage is more compatible with cell stasis than with cell death.
FIG. 3.
Northern blot analysis of sarA transcripts in Newman derivative ALC6094 and its isogenic sarA deletion mutant carrying pG164-MazF(His6)-pEPSA5-sarA. (A) The 1.2-, 0.8-, and 0.6-kb transcripts (long arrows) represent the native sarA transcripts from three distinct sarA promoters, which, as expected, were preserved upon MazFSa induction. (B) The ∼0.5-kb transcript (open arrow) represents the sarA transcript as driven from the modified pEPSA5 promoter in the plasmid pG164-MazFsa-pEPSA5-sarA. This transcript was preserved in the parental strain ALC6094 and the isogenic sarA mutant. Bottom, rRNA loading control. Total cellular RNAs were extracted at the indicated time points (minutes).
De novo synthesis of SarA and SigB upon ectopic overexpression of MazFSa.
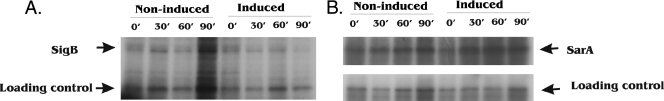
To confirm that proteins could still be synthesized in the face of MazFSa inhibition and to verify that the cleavage of selective mRNA truly leads to inhibition of de novo synthesis of the corresponding protein, we performed pulse-chase labeling of de novo-synthesized SarA and SigB with [35S]methionine upon ectopic overexpression of MazFSa. The SarA and SigB proteins were chosen for the preservation and destruction of their mRNAs, respectively. Briefly, total-cell lysates from both induced and noninduced cultures were immunoprecipitated with specific anti-SarA and anti-SigB monoclonal antibodies and resolved on SDS-polyacrylamide gels. The radiolabeled de novo-synthesized SarA and SigB were detected as described in Materials and Methods. The expression of SigB was normal in the noninduced culture, with no significant changes in the band intensities at 30 min (Fig. 4A); in contrast, inhibition of SigB synthesis was observed at 30 min after the induction of ectopic overexpression of MazFSa, and after 90 min, the band could barely be discerned (Fig. 4A). In the case of SarA, the protein continued to be synthesized in the noninduced culture but was upregulated after MazFSa induction (Fig. 4B). These data demonstrated that the sarA mRNA, being resistant to MazFSa cleavage, can be continuously translated to SarA protein in the face of inhibition by MazFSa. These results are consistent with the Northern blot analysis shown in Fig. 2, in which the sigB mRNA was degraded shortly after overexpression of MazFSa, while sarA mRNA was preserved or moderately upregulated in induced cultures. It is important to note that an unknown protein, which was pulled down nonspecifically by anti-SigB and anti-SarA antibodies, could be used as the loading control because it was constantly synthesized in noninduced and induced cultures.
FIG. 4.
Pulse-chase labeling of de novo-synthesized SarA and SigB with [35S]methionine upon ectopic overexpression of MazFSa. A culture of S. aureus ALC6094 harboring pG164-MazF(His6) was induced at an OD650 of 0.4 with IPTG (1 mM), while the noninduced culture received no IPTG. New protein synthesis was then monitored by isotopic labeling with [35S]methionine for 15 min under induced or noninduced conditions at indicated intervals (0 to 90 min) after IPTG induction. The cells were then lysed and immunoprecipitated with anti-SigB monoclonal antibody (A) or anti-SarA monoclonal antibody (B) as described in Materials and Methods. Equivalent amounts of cell lysate, derived from equal culture volumes, were subjected to SDS-PAGE, followed by autoradiography. The labeled unknown protein that was immunoprecipitated by anti-SigB and anti-SarA antibodies served as the internal loading control.
sarA and recA mRNAs were degraded following rifampin treatment.
To examine if decreased degradation of sarA and recA mRNAs or increased degradation of hla and sigB transcripts was attributable to mRNA stability or instability, respectively, we performed an RNA stability assay by adding rifampin to halt de novo mRNA synthesis, as described in Materials and Methods. The decay of the mRNAs was monitored by Northern blot analysis of total cellular RNA isolated from different time points after the addition of rifampin. As shown in Fig. 5B, the recA mRNA disappeared within 2 min after the addition of rifampin, while the sarA mRNA was slightly more stable, with a half-life of >2 min (Fig. 5A). Both mRNAs were barely detectable after 10 min (Fig. 5A and B). The half-lives and decay of hla and sigB mRNAs after rifampin treatment were similar to those of recA mRNA (Fig. 5C and D). Interestingly, the rRNA after rifampin treatment with the above blots appeared to be relatively stable (Fig. 5A to D, lower panels). This may be due to protection by ribosomal binding proteins or the possibility that the relatively high quantity of 16S and 23S ribosomal RNAs might have taken longer to degrade than particular transcripts in lower quantities. Collectively, our data indicate that the relative stability of the sarA and recA mRNAs in the presence of MazFSa is not likely due to the stability of their structure, since their degradation by RNase in the cells upon rifampin treatment is very similar to that of sigB and hla. As a sarA transcript arising from a modified pEPSA5 promoter in place of the native sarA promoter was also protected from MazFSa cleavage (Fig. 3B), we thus propose that the sarA and, possibly, recA mRNAs are likely protected by RNA-binding proteins in vivo. In contrast, the hla and sigB mRNAs were not protected from MazFSa-mediated cleavage in S. aureus.
FIG. 5.
Decay of RNAs upon blocking of de novo RNA synthesis with rifampin. Rifampin (200 μg/ml) was added 30 min after the culture containing S. aureus ALC6094 harboring pG164-MazF(His6) reached an OD650 of 0.4. RNA was extracted from control samples at time zero (time when culture reached an OD650 of 0.4) and at 15-min time points, shown in lanes 1 and 2, respectively, in panels A to D. Lanes 3 to 8 in panels A to D show the RNA extracted after the addition of rifampin for 0, 2, 4, 6, 10, and 15 min. (A to D) Transcripts detected with sarA, recA, hla, and sigB probes. 23S and 16S rRNA served as the internal loading controls.
In vitro cleavage of sarA and recA mRNAs by MazFSa.
Since both sarA and recA mRNAs contain the VUUV′ cleavage sites for MazFSa, we sought to determine if these two mRNAs can be cleaved by MazFSa in vitro. For this experiment, sarA and recA mRNAs generated with an in vitro transcription kit, as described in Materials and Methods, were digested by purified MazFSa. To enhance the sensitivity of the assay, we detected cleaved mRNA fragments by Northern blotting, using either sarA or recA DNA probes. Although there are multiple predicted VUUV′ cleavage sites in the sarA mRNA, only one major site was cleaved efficiently in a time-dependent manner (Fig. 6A). The remaining uncleaved sites are likely to be protected by the secondary structure of sarA mRNA, as MazFSa has been shown to cleave duplex RNA inefficiently (11). The C terminus of the truncated recA mRNA contains multiple VUUV′ sequences which were cleaved more completely by MazFSa, yielding smaller fragments that were not readily detectable with Northern blots (Fig. 6B). These results suggest that the sarA and recA mRNAs can be cleaved by MazFSa in vitro. In addition, their stability in vivo cannot be readily explained by multiple RNA duplexes or longer half-lives of the mRNA, but rather, these mRNAs may be protected from MazFSa by RNA-binding proteins, similar to what has been observed with protection of rRNA by ribosomal proteins (30).
FIG. 6.
Northern blot analysis of the in vitro cleavage of sarA and recA mRNAs by MazFSa. The sarA and recA mRNAs were transcribed from BamHI-linearized pET14b-sarA and pET14b-recA plasmids by using a T7 large-scale transcription kit (Promega) and digested with purified MazFSa as described in Materials and Methods. Amounts of 5 μg of sarA and recA mRNAs were digested with 15 pmol MazFSa at times indicated for panels A and B. (A) sarA mRNA cleavage by MazFSa. Lane 1, no MazFSa; lanes 2 to 4, digestion with MazFSa for 60, 90, and 120 min, respectively. (B) recA mRNA cleavage by MazFSa. Lane 1, no MazFSa; lane 2, digestion with MazFSa for 60 min. The cleaved fragments were detected with the 32P-radiolabeled sarA and recA DNA probes in Northern blots as described in Materials and Methods.
Northwestern analysis of the sarA and recA mRNA-binding proteins.
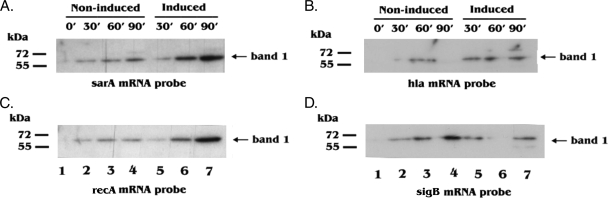
To investigate if RNA-binding protein(s) exists for sarA and recA mRNAs, we performed a Northwestern blot RNA-binding assay in which total-cell lysates from cultures both induced and uninduced for MazFSa were resolved on SDS-polyacrylamide gels, transferred to PVDF membranes, and probed with the radiolabeled sarA or recA mRNAs as described in Materials and Methods. The sarA mRNA probe detected a putative RNA-binding protein band of ∼60 kDa that increased in intensity upon induction of MazFSa for 60 to 90 min when compared with the amounts in noninduced controls (Fig. 7A, lanes 5 to 7). The recA mRNA probe also detected an mRNA-binding protein, with a pattern similar to that of the one probed with sarA mRNA (Fig. 7C). However, this increase in band intensity was not observed in the hla and sigB mRNA probes upon MazFSa induction (Fig. 7B and D). The identification of the putative 60-kDa RNA-binding protein is ongoing. Collectively, these results suggest that RNA-binding protein(s) may be responsible for the stability of sarA and recA mRNAs in the presence of MazFSa.
FIG. 7.
Northwestern blot analysis of cell lysates with sarA, recA, hla, and sigB mRNA probes. A culture of S. aureus ALC6094 harboring pG164-MazF(His6) was induced at an OD650 of 0.4 with IPTG (1 mM), while the control culture contained no IPTG. Samples were taken at 0, 30, 60, and 90 min. Amounts of 20 μg of the total-cell lysates from indicated time points (minutes) were resolved on 16% SDS-polyacrylamide gels and transferred to a PVDF membrane as described in Materials and Methods. The blots were then denatured, renatured, and hybridized with the 32P-labeled sarA mRNA (A), hla mRNA (B), recA mRNA (C), and sigB mRNA (D) probes. The major protein detected (band 1) is indicated on the right of each panel; the protein size markers are shown on the left.
DISCUSSION
The primary physiological role of TA systems has been the subject of much debate. One model proposes that TA loci induce bacterial PCD, while another favors the status of bacteriostasis upon toxin induction during periods of stress. Previously, we characterized mazEFSa, two genes immediately upstream of the sigB operon in S. aureus, as a TA system. The MazFSa toxin is an endoribonuclease that cleaves sequence-specific mRNA at VUUV′ sites. Ectopic overproduction of MazFSa inhibits cell growth (11), while the concomitant expression of MazESa can reverse this effect. In this study, we demonstrate that the growth arrest induced by MazFSa is attributable to bacterial stasis. Although there is a loss of CFU, most of the bacteria exposed to MazFSa for 1 h in our study are not dead, as demonstrated by the bacterial viability assay. We focused on the 1-h induction period because significant numbers of plasmid-free cells emerged after the first hour, which would affect the interpretation of the Live/Dead bacterial viability assay, as well as the total cellular RNA decay analysis.
The standard assay used to measure cell death (i.e., the inability to form colonies) cannot distinguish between cell death and bacterial stasis, wherein the cells are viable but cannot replicate to form colonies. For this reason, we deployed two assays to distinguish these two possibilities after MazFSa induction. In the CFU count assay, we found that, compared with the colony-forming activities of noninduced controls, ∼99.5% of the cells failed to replicate as colonies upon MazFSa induction during the early logarithmic phase (at an OD650 of 0.4) (Fig. 1A), but the difference between the induced and noninduced cultures in the viability assay with the Live/Dead BacLight kit was significantly less (<27%) (Fig. 1B).
It has been previously suggested that MazF of E. coli selectively degrades almost all cellular mRNAs in vivo, thus causing a precipitous drop in total protein synthesis and, hence, complete growth arrest (30). However, both 16S and 23S rRNAs of the E. coli cells were protected from such cleavage even though they contained a number of target cleavage sites for MazF of E. coli. Interestingly, both rRNAs were degraded in vitro by MazF of E. coli once the rRNA preparations were extracted with phenol, indicating that the resistance of rRNA to MazF cleavage in vivo may be due to protection by ribosomal proteins (30). Despite growth arrest attributable to the toxin, the cells still managed to maintain de novo synthesis of proteins from mRNAs resistant to MazFSa cleavage (Fig. 4). Suzuki et al. also reported that MazF-induced cells retain the full spectrum of biosynthetic functions necessary to support mRNA transcription and translation for up to 4 days (28). These results suggest that MazF-induced cells undergo stasis rather than cell death. However, if the cells are exposed to high levels of toxin for a prolonged period of time (i.e., over 6 h with MazF in E. coli), the effect can be irreversible. This result can be explained by a scenario where prolonged bacteriostasis might eventually lead to cell death due to pleiotropic effects (2).
In another TA system, Lioy et al. (19) showed that overexpression of the ζ toxin triggers bacteriostasis without gross inhibition of protein translation, which can be rescued by the expression of its cognate ɛ antitoxin within a time frame of 4 h. However, we were puzzled by the discrepancy between the MazF-mediated degradation of almost all cellular mRNA and the preservation of the biosynthetic competence in E. coli. In this study, we found that induced MazFSa cleaves cellular mRNAs selectively in S. aureus, with gene transcripts of essential genes (recA and gyrB) and also of the regulatory gene sarA not cleaved efficiently in vivo, while these transcripts can clearly be cleaved in vitro. The stability of the recA and sarA transcripts against MazFSa is not due to the stability of their structures or their longer half-lives, as these two mRNAs were degraded rapidly by intrinsic RNases upon the halting of de novo synthesis of mRNAs by rifampin (Fig. 5A and B). Based on the precedent that 16S rRNA is stable by virtue of its protection by ribosomal proteins (30), it is likely that recA, gyrB, and sarA mRNAs are protected by RNA-binding proteins.
One potential caveat about our studies is that the level of MazFSa expression was significantly higher than the level of MazFSa achieved as a result of the proteolytic destruction of the antitoxin MazESa. We took this approach because the native antitoxin MazESa was left undisturbed in our studies. Instead, we relied on the imbalance between the levels of MazFSa and the antitoxin MazESa to exert the toxic effect. Thus, it is difficult to assess if our observations here will be similar to those found under physiological conditions.
The stabilities of different mRNA species in bacteria vary widely. Thus, it is not surprising that the stability of individual transcripts is often the target of regulatory processes in bacterial cells as a means of target gene modulation in vivo. Different mRNAs are protected against intracellular ribonucleases to various degrees (13). The mRNA half-lives of some genes are influenced by interaction with various RNA-binding proteins, forming ribonucleoprotein complexes. One of the RNA-binding complexes, containing the chaperon GroE, has been shown to mediate mRNA protection against RNase E in E. coli (13). Based on this observation, we speculate that some gene transcripts might also be protected from MazF-mediated cleavage by RNA-binding proteins. Indeed, we found a putative protein binding to both labeled sarA and recA mRNA probes in our Northwestern blot analysis (Fig. 7). However, the binding specificity of this protein to sarA or recA mRNA is not yet clear; nonetheless, the expression of this protein band was upregulated upon induction of MazFSa for 60 to 90 min (Fig. 7), while this effect was much less with hla and sigB mRNA probes. Although there are other proteins that bound hla and sigB mRNA, as well as recA and sarA mRNA, it is conceivable that they might represent general RNA-binding proteins under physiological conditions or that they are nonspecific binding proteins.
Collectively, our findings suggest that MazFSa-induced S. aureus cells likely undergo stasis by protecting some essential and regulatory gene transcripts against MazFSa, probably by means of RNA-binding proteins. Accordingly, we predict that a strategy that blocks the activity of the RNA-binding proteins may usher S. aureus toward cell death rather than cell stasis.
Acknowledgments
These studies are supported in part by RO1 AI56114 from the NIH (to A.L.C.).
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 936059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 1868295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222511-514. [DOI] [PubMed] [Google Scholar]
- 4.Cheung, A. L., Y. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 671331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien, Y., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30991-1001. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332809-819. [DOI] [PubMed] [Google Scholar]
- 7.Condon, C. 2006. Shutdown decay of mRNA. Mol. Microbiol. 61573-583. [DOI] [PubMed] [Google Scholar]
- 8.D'Elia, M. A., M. P. Pereira, Y. S. Chung, W. Zhao, A. Chau, T. J. Kenney, M. C. Sulavik, T. A. Black, and E. D. Brown. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 1884183-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelberg-Kulka, H., R. Hazan, and S. Amitai. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 1184327-4332. [DOI] [PubMed] [Google Scholar]
- 10.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, C. G. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 431387-1400. [DOI] [PubMed] [Google Scholar]
- 11.Fu, Z., N. P. Donegan, G. Memmi, and A. L. Cheung. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 1898871-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, Z. B., K. L. Ng, T. L. Lam, and W. K. Wong. 2005. Cell death caused by hyper-expression of a secretory exoglucanase in Escherichia coli. Protein Expr. Purif. 4267-77. [DOI] [PubMed] [Google Scholar]
- 13.Georgellis, D., B. Sohlberg, F. U. Hartl, and A. von Gabain. 1995. Identification of GroEL as a constituent of an mRNA-protection complex in Escherichia coli. Mol. Microbiol. 161259-1268. [DOI] [PubMed] [Google Scholar]
- 14.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3371-382. [DOI] [PubMed] [Google Scholar]
- 15.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 26558-566. [DOI] [PubMed] [Google Scholar]
- 16.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 1863663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodkin-Gal, I., R. Hazan, A. Gaathon, S. Carmeli, and H. Engelberg-Kulka. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318652-655. [DOI] [PubMed] [Google Scholar]
- 18.Korch, S. B., and T. M. Hill. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 1883826-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lioy, V. S., M. T. Martin, A. G. Camacho, R. Lurz, H. Antelmann, M. Hecker, E. Hitchin, Y. Ridge, J. M. Wells, and J. C. Alonso. 2006. pSM19035-encoded zeta toxin induces stasis followed by death in a subpopulation of cells. Microbiology 1522365-2379. [DOI] [PubMed] [Google Scholar]
- 20.Manna, A. C., and A. L. Cheung. 2006. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 1884288-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45501-510. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112131-140. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrini, O., N. Mathy, A. Gogos, L. Shapiro, and C. Condon. 2005. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol. Microbiol. 561139-1148. [DOI] [PubMed] [Google Scholar]
- 24.Popovic, Z., and D. M. Templeton. 2005. A Northwestern blotting approach for studying iron regulatory element-binding proteins. Mol. Cell. Biochem. 26867-74. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 1832041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schukkink, R. F., and R. H. Plasterk. 1990. TcA, the putative transposase of the C. elegans Tc1 transposon, has an N-terminal DNA binding domain. Nucleic Acids Res. 18895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki, M., J. Zhang, M. Liu, N. A. Woychik, and M. Inouye. 2005. Single protein production in living cells facilitated by an mRNA interferase. Mol. Cell 18253-261. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, J., Y. Zhang, L. Zhu, M. Suzuki, and M. Inouye. 2004. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 27920678-20684. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12913-923. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Y., L. Zhu, J. Zhang, and M. Inouye. 2005. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 28026080-26088. [DOI] [PubMed] [Google Scholar]
- 32.Zhu, L., Y. Zhang, J. S. The, J. Zhang, N. Connell, H. Rubin, and M. Inouye. 2006. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 28118638-18643. [DOI] [PubMed] [Google Scholar]