Abstract
To sustain growth, the catabolic formation of the redox equivalent NADPH must be balanced with the anabolic demand. The mechanisms that ensure such network-wide balancing, however, are presently not understood. Based on 13C-detected intracellular fluxes, metabolite concentrations, and cofactor specificities for all relevant central metabolic enzymes, we have quantified catabolic NADPH production in Agrobacterium tumefaciens, Bacillus subtilis, Escherichia coli, Paracoccus versutus, Pseudomonas fluorescens, Rhodobacter sphaeroides, Sinorhizobium meliloti, and Zymomonas mobilis. For six species, the estimated NADPH production from glucose catabolism exceeded the requirements for biomass synthesis. Exceptions were P. fluorescens, with balanced rates, and E. coli, with insufficient catabolic production, in which about one-third of the NADPH is supplied via the membrane-bound transhydrogenase PntAB. P. versutus and B. subtilis were the only species that appear to rely on transhydrogenases for balancing NADPH overproduction during growth on glucose. In the other four species, the main but not exclusive redox-balancing mechanism appears to be the dual cofactor specificities of several catabolic enzymes and/or the existence of isoenzymes with distinct cofactor specificities, in particular glucose 6-phosphate dehydrogenase. An unexpected key finding for all species, except E. coli and B. subtilis, was the lack of cofactor specificity in the oxidative pentose phosphate pathway, which contrasts with the textbook view of the pentose phosphate pathway dehydrogenases as being NADP+ dependent.
For all combinations of prevalent carbon substrates, the approximately 60 reactions of heterotrophic central carbon metabolism provide the building blocks and energy at appropriate rates and stoichiometries to fuel about 300 anabolic reactions (43). Additionally, redox equivalents must be appropriately balanced between large numbers of producing and consuming reactions. Aerobically, the primary role of the redox cofactor NADH is respiratory ATP generation via oxidative phosphorylation. The chemically very similar redox cofactor NADPH, in contrast, drives anabolic reductions. To fulfill these rather distinct functions, the two redox couples are generally not in thermodynamical equilibrium and the NADPH-to-NADP+ ratio is in a more reduced state than the NADH-to-NAD+ ratio (25).
During heterotrophic growth on glucose, the major NADPH-generating reactions are considered to be the oxidative pentose phosphate (PP) pathway, the Entner-Doudoroff (ED) pathway, and the isocitrate dehydrogenase step in the tricarboxylic acid (TCA) cycle (Fig. 1A). The precise rate of formation of NADPH, however, depends on the actual carbon fluxes through these catabolic pathways, which can vary significantly with environmental conditions. The anabolic demand for NADPH, in contrast, is coupled to the rate of biomass formation (46) and, to various extents, to the reduction of thioredoxin for maintaining an appropriate redox state (3). Thus, in the absence of reactions that reoxidize NADPH at the expense of NAD+, the catabolic fluxes through the NADPH-generating reactions must be perfectly balanced with the requirements for NADPH in about 100 anabolic reactions. This situation is indeed found in yeasts (4, 6). Since bacteria often exhibit extensive catabolic fluxes through NADPH-producing reactions (22), they inevitably encounter situations in which the NADPH formation in stoichiometric coupling with catabolic fluxes will exceed the anabolic demand or, at high biomass yields, will be insufficient (55). Hence, a key question of metabolic network operation is how bacteria balance catabolic NADPH formation with anabolic consumption.
FIG. 1.
(A) Central metabolic network for heterotrophic growth on glucose. Experimentally investigated reactions that are potentially relevant for NADPH metabolism are indicated by white boxes. (B) Possible NADPH-balancing mechanisms with particular examples. Abbreviations: G6PDH, glucose-6P dehydrogenase; 6PGDH, 6P-gluconate dehydrogenase; GAPDH, glyceraldehyde-3P dehydrogenase; PEP, phosphoenolpyruvate; MAE, malic enzyme; MDH, malate dehydrogenase; acetyl-CoA, acetyl coenzyme A; ICTDH, isocitrate dehydrogenase; and EMP pathway, Emden-Meyerhof-Parnas pathway.
Potential biochemical mechanisms that may provide such NADPH balancing can be divided into those that avoid imbalances in the first place and those that decouple NADPH formation from catabolism (Fig. 1B). Imbalance-avoiding mechanisms include the appropriate choice of catabolic pathways, as in yeasts (6), and the differential expression of isoenzymes with different cofactor specificities (13, 14, 69).
The decoupling of catabolic NADPH formation from anabolism can potentially be achieved through three distinct mechanisms (Fig. 1B). First, this decoupling may be catalyzed by either a membrane-bound, proton-translocating transhydrogenase or a soluble, energy-independent transhydrogenase (5, 28). With both isoforms, Escherichia coli can counterbalance situations of catabolic under- and overproduction of NADPH (7, 9). The membrane-bound transhydrogenase PntAB provides about 40% of the total NADPH during growth on glucose in batch cultures (55). In contrast, glucose-limited chemostat growth with extensive TCA cycle fluxes results in significant overproduction of NADPH (16, 68), which is presumably balanced through the soluble transhydrogenase UdhA, shown previously to carry out this function in E. coli cells growing on acetate and in mutants with high PP pathway fluxes (55). This counterbalancing of NADPH overproduction can be readily transferred to other microbes through heterologous expression of the soluble transhydrogenase (17). Second, many bacteria (34, 35, 54, 60, 71) and yeast (49) contain NAD(H) kinases that can directly convert NAD(H) into NAD(P)H at the expense of ATP. By itself, this mechanism has not yet been implicated in NADPH balancing, but in combination with carbon flux rerouting, it may contribute (60). Third, biochemical redox cycles—combinations of reactions or isoenzymes with different cofactor specificities—can catalyze effective transhydrogenation without affecting net catabolic fluxes. Examples of such redox cycles include the simultaneous isoform operation of isocitrate dehydrogenase in animal mitochondria (57) and glyceraldehyde-3-phosphate (glyceraldehyde-3P) and alcohol dehydrogenases in Kluyveromyces lactis (50, 65). The NAD+- and NADP+-accepting glyceraldehyde-3P dehydrogenase from K. lactis can restore the growth of a Saccharomyces cerevisiae phosphoglucoisomerase mutant on glucose. Similarly, increased activities of cytosolic and mitochondrial alcohol dehydrogenases with distinct cofactor specificities have demonstrated the operation of such redox cycles in equilibrating NADPH overproduction in a K. lactis phosphoglucoisomerase mutant.
Here, we address the question of how eight metabolically diverse bacterial species balance their catabolic NADPH formation with their anabolic demand during growth on glucose as the sole carbon source. The analysis relies on inferring catabolic in vivo NAD(P)H formation rates from previously published (22) and newly 13C-detected intracellular fluxes, comprehensive in vitro determination of redox cofactor specificities in all major NAD(P)H producing reactions, quantitative metabolomics, and network-wide cofactor balancing.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The following bacterial species and strains were analyzed: Agrobacterium tumefaciens C58 (obtained from F. Narberhaus), Pseudomonas fluorescens 52-1C (obtained from B. Witholt), Rhodobacter sphaeroides ATH 2.4.1 (German Collection of Microorganisms and Cell Cultures; DSMZ 158), R. sphaeroides GA wild-type and pntAB knockout strains (obtained from T. Donohue), Paracoccus versutus A2 (DSMZ 582), Sinorhizobium meliloti (DSMZ 1981), Zymomonas mobilis NRRL B-806 (DSMZ 424), Bacillus subtilis 168 trpC2 (Bacillus Genetic Stock Center), and E. coli MG1655 (E. coli Genetic Stock Center; no. 6300). All physiological experiments were conducted with minimal medium supplemented with 3 g/liter glucose as the sole carbon source. For most species, M9 medium containing the following ingredients per liter of deionized water was used: 7.52 g of Na2HPO4·2H2O, 3.0 g of KH2PO4, 0.5 g of NaCl, and 2.5 g of (NH4)2SO4. The following components (per liter of final medium) were sterilized separately and then added: 1 ml of 0.1 M CaCl2, 1 ml of 1 M MgSO4, 0.6 ml of 100 mM FeCl3, 2 ml of vitamin solution (filter sterilized), and 10 ml of M9 trace salts solution. The vitamin solution contained (per 50 ml) 25 mg each of biotin, cyanocobalamin, niacin, calcium pantothenate, pyridoxine HCl, and thiamine HCl. The M9 trace salts solution contained (per liter) 0.18 g of ZnSO4·7H2O, 0.12 g of CuCl2·2H2O, 0.12 g of MnSO4·H2O, and 0.18 g of CoCl2·6H2O. For R. sphaeroides, a special trace salts solution that contained the following (per liter) was used: 1.5 g of nitrilotriacetic acid, 3.0 g of MgSO4·7H2O, 0.5 g of MnSO4·H2O, 1.0 g of NaCl, 0.1 g of FeSO4·7H2O, 0.1 g of CoCl2·6H2O, 0.135 g of CaCl2·2H2O, 0.1 g of ZnSO4·7H2O, 0.01 g of CuSO4·5H2O, 0.01 g of H3BO3, 0.01 g of Na2MoO4·2H2O, 0.015 g of NiCl2, and 0.02 g of Na2SeO3. The pH of the solution was adjusted to 6.5 with KOH. Aerobic batch cultures were grown at 30°C in 500-ml baffled flasks with 50 ml of M9 medium (P. versutus and Z. mobilis were grown in special minimal media [22]) on a gyratory shaker at 225 rpm (P. fluorescens at 250 rpm). Anaerobic cultures of Z. mobilis were grown at 30°C in 125-ml sealed glass flasks with 50 ml of minimal medium on magnetic stirrers at 225 rpm. The sterile medium was gassed with sterile filtered N2 for 15 min.
Construction of mutants.
A. tumefaciens soluble transhydrogenase (sth) and membrane-bound transhydrogenase (pntAB) knockout strains and the corresponding double-knockout strain were constructed using the pK18mobsacB suicide vector (47, 58). The two homologous regions flanking the sth and pntAB genes were amplified by PCR using the following oligonucleotide primer pairs: 5′-GCATCTAGACCGCATGGGTGAGTTGAAGAAGGA-3′ and 5′-GCAGGATCCAAGGCGGGGCAGGTGGTGGATACG-3′ and 5′-GCAGTCGACATGAAAAGATCGATGTGAAGGTG-3′ and 5′-GCATCTAGAATTTGGACGATGGGGAGTG-3′ for sth and 5′-GCAGGATCCGACTGCGGCTCCGGGCGACTG-3′ and 5′-GCATCTAGATCCTTAGGCGTTCCGATTCTCAAA-3′ and 5′-GCAGTCGACATGGCGATGCGAAGAAGT-3′ and 5′-GCAAAGCTTCGCGCTAGGGCTGGATAC-3′ for pntAB. The oligonucleotides were digested with the restriction enzymes BamHI/XbaI and XbaI/SalI, respectively, for sth and BamHI/XbaI and SalI/HindIII, respectively, for pntAB and ligated into the pK18mobsacB vector. The resulting plasmid was transferred into A. tumefaciens by electroporation, and kanamycin-resistant clones which had the plasmid inserted into the chromosome were selected. Those clones were grown on Luria-Bertani medium without kanamycin and then streaked onto plates containing 10% sucrose to select for a second homologous recombination event as described previously (47). Positive knockouts were analyzed by PCR using primers corresponding to a sequence inside sth or pntAB and one outside the flanking sequences used for the recombinations: 5′-CACCCGCGCCTTCGATCCCTACCC-3′ and 5′-GCTTGCCGTGCCGCGAATGTGCT-3′ for sth and 5′-GCCGCCGCCGCCGAAGATT-3′ and 5′-AGCCGCCAAGGATGACCGAGATGA-3′ for pntAB.
Determination of intracellular metabolites by liquid chromatography-tandem mass spectrometry.
Cells grown on glucose minimal medium were harvested at mid-exponential growth phase by rapid centrifugation of 1-ml cell broth aliquots in a tabletop centrifuge at 15,000 rpm and room temperature for 15 s. Cell pellets were immediately frozen in liquid N2 and subsequently extracted three times with 0.5 ml of 10 mM ammonium acetate buffer (pH 7.2) in 60% ethanol at 70°C for 1 min. The pooled supernatants of extraction were completely dried in a speed vac at 30°C and 0.12 × 105 kPa and resuspended in 50 μl of 10 mM ammonium acetate buffer (pH 7.2). Metabolite concentrations were measured on an Agilent 1100 series high-performance liquid chromatography system coupled with an Applied Biosystems/MDS SCIEX 4000 Q TRAP system. Metabolites extracted from 100% 13C-labeled yeast biomass were used as internal standards, except for NADPH and NADH, which were analyzed without internal standards. Data were recorded and analyzed with Analyst software version 1.4.2, build 1228. Chromatographic separation was achieved with a Waters Atlantis T3 150- by 2.1-mm by 3-μm column at 40°C by using an adapted version of a previously published protocol (40). Briefly, the injected volume was 8 μl, and the mobile phase at a flow rate of 250 μl/min was directly introduced into the mass spectrometer via electrospray ionization. The gradient profile was linear with two phases, where solution A was 10 mM tributylamine and 15 mM acetate in 5% methanol (pH 4.95) and solution B was 100% methanol: 0 min, 0% B; 5 min, 37% B; 12 min, 41% B; 24 min, 100% B; 28 min, 100% B; 29 min, 0% B; and 38 min, 0% B. Multiple-reaction monitoring settings were optimized individually for each metabolite (40).
In vitro enzyme activities.
Cells were harvested by centrifugation at 4°C and washed twice in 0.9% NaCl and 10 mM MgSO4. Upon 10-fold concentration in cell lysis buffer (100 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol, and Complete EDTA-free protease inhibitor cocktail [Roche]), cells were disrupted by passage through a French press at 4°C. Cell extracts were obtained by centrifugation at 23,100 × g for 30 min at 4°C. The supernatant was used for the enzymatic assays in 1 ml of reaction buffer at 25°C. The reduction of NAD(P)+ or the oxidation of NAD(P)H was monitored spectrophotometrically at 340 nm. The protein concentrations of the extracts were determined by the biuret assay (24).
The in vitro enzyme activities of glucose-6-phosphate (glucose-6P), 6-phosphate-gluconate (6P-gluconate), and isocitrate dehydrogenases were determined by using assay mixtures of 100 mM Tris-HCl, pH 7.5, 2.5 mM MnCl2, 50 μl of extract, 1 mM NAD(P)+, and 2 mM of glucose-6P, 6P-gluconate, or isocitrate (2). Alternatively, experimentally determined intracellular concentrations of cofactors and substrates were used for glucose-6P and 6P-gluconate dehydrogenases. The assay mixtures for glyceraldehyde-3P dehydrogenase contained 125 mM triethanolamine, 5 mM l-cysteine, 20 mM potassium arsenate, 50 mM K2HPO4, pH 9.2, 50 μl of extract, 1 mM NAD(P)+, and 3 mM glyceraldehyde-3P (18). The reverse reaction of malate dehydrogenase was evaluated with a mixture of 100 mM Tris-HCl, pH 8.8, 20 μl of extract, 0.2 mM NAD(P)H, and 1 mM oxaloacetate (2). The NADP+-dependent malic enzyme was assessed with a mixture of 100 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 50 mM KCl, 100 μl of extract, 1 mM NADP+, and 30 mM l-malate, pH 7.8 (2, 14). Malic enzyme was assayed for NADP+-dependent activity only since interference of the NAD+-dependent activity with malate dehydrogenase could not be excluded. Alcohol dehydrogenase in Z. mobilis was analyzed with a mixture of 100 mM Tris-HCl, pH 7.5, 2.5 mM MnCl2, 10 μl of extract, 0.2 mM NAD(P)H, and 1 mM acetaldehyde. All reactions were started by adding the substrate, and the specific activities were obtained by dividing the measured slope of NAD(P)H formation or consumption by the total cell protein concentration. For in vitro enzyme activities under near-in vivo conditions, the substrate concentrations were chosen from the determined in vivo values and the reaction was monitored for the short initial period before it slowed down (typically a few seconds).
The membrane fraction and the membrane-free soluble fraction were separated by centrifuging the previously obtained supernatant at 159,000 × g and 4°C for 3 h. Reverse transhydrogenase activity (optimized for E. coli membrane-bound transhydrogenase) in the membrane fraction was then assayed with a mixture of 50 mM sodium phosphate, pH 7.0, 100 μl of the membrane fraction (resuspended in the same amount of sodium phosphate buffer as the total volume during centrifugation), 0.2 mM NADPH, and 1 mM 3-acetylpyridine adenine dinucleotide (APAD+) (67). Forward soluble transhydrogenase activity (optimized for E. coli soluble transhydrogenase) in the membrane-free soluble fraction was assayed in a mixture of 50 mM Tris-HCl, pH 7.6, 2 mM MgCl2, 100 μl of the membrane-free soluble fraction, 0.2 mM NADPH, and 1 mM APAD+ (55). The reduction of APAD+ and the oxidation of NADPH were monitored spectrophotometrically at 400 and 310 nm simultaneously to exclude interferences in the adsorption of both NADPH and APADH. The extinction coefficients were experimentally determined by standard curves to be 1.75 mM−1 cm−1 at 310 nm and 3.51 mM−1 cm−1 at 400 nm for APADH and 3.24 mM−1 cm−1 at 310 nm and 0.16 mM−1 cm−1 at 400 nm for NADPH.
NADPH balancing.
Previously published net carbon fluxes (22) were used as the basis for all the calculations except for the E. coli wild type, the R. sphaeroides wild type and pntAB mutant (T. Donohue), and the newly constructed mutants of A. tumefaciens. The new flux results were obtained by the same method used for the other species. Briefly, ratios of converging fluxes were calculated from gas chromatography-mass spectrometry-detected 13C patterns in protein-bound amino acids from cells grown on [13C]glucose minimal medium (19). Absolute intracellular fluxes were calculated by 13C-constrained flux balancing that combines experimentally determined fluxes in and out of the cell and the abovementioned flux ratios (20, 70) (see Table SA1 in the supplemental material). NADPH formation was determined from the carbon fluxes through the cofactor-dependent reactions multiplied by the experimentally determined relative cofactor specificities for NADP+. NADPH consumption was calculated from the NADPH requirements for biomass production (11, 46). For malic enzyme, the detection of only NADP+-dependent activity was possible and the cofactor specificities were therefore assumed on the basis of literature data for each organism (Table 1). For P. versutus and E. coli, the malic enzyme was chosen to be NAD+-dependent under these conditions.
TABLE 1.
Qualitative cofactor specificities based on genetic evidence and reported enzyme activities
| Enzymeb | Cofactor or form | Activity ina:
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
A. tumefaciens
|
S. meliloti
|
P. fluorescens
|
R. sphaeroides
|
P. versutusd
|
Z. mobilis
|
E. coli
|
B. subtilis
|
|||||||||||||||||
| G | E | Reference | G | E | Reference(s) | G | E | Reference | G | E | Reference | G | E | G | E | Reference | G | E | Reference(s) | G | E | Reference | ||
| G6PDH | NADP+ | + | + | (+) | + | + | + | (+) | + | (+) | + | (+) | + | (+) | + | 10 | (+) | + | ||||||
| NAD+ | + | + | + | + | + | + | + | + | − | + | ||||||||||||||
| 6PGDH | NADP+ | + | + | + | + | 41 | + | + | 63 | (+) | − | (+) | + | (+) | − | (+) | + | 10 | + | + | 69 | |||
| NAD+ | + | + | + | − | + | − | − | + | + | |||||||||||||||
| GAPDH | NADP+ | − | − | (+) | + | − | − | − | − | 10 | + | − | 18 | |||||||||||
| NAD+ | (+) | + | (+) | + | (+) | + | (+) | + | (+) | + | (+) | + | (+) | + | + | + | ||||||||
| ICTDH | NADP+ | (+) | + | + | + | 62 | (+) | + | (+) | + | (+) | + | (+) | − | + | + | 10, 62 | + | + | 62 | ||||
| NAD+ | − | − | − | − | − | − | − | − | ||||||||||||||||
| MAE | NADP+ | + | + | 31 | + | + | 14, 15 | (+) | + | 31 | (+) | − | (+) | + | + | + | + | + | 44 | + | + | 13 | ||
| NAD+ | + | + | + | + | ||||||||||||||||||||
| MDH | NADP+ | − | − | − | − | − | − | − | 48 | − | − | |||||||||||||
| NAD+ | (+) | + | (+) | + | (+) | + | (+) | + | (+) | + | − | − | (+) | + | (+) | + | ||||||||
| THc | Membr. | + | + | + | − | + | − | 21 | + | + | 27 | + | − | − | 66 | + | + | 7, 9 | − | |||||
| Soluble | + | − | (+) | − | + | − | −− | + | − | − | + | − | + | |||||||||||
Enzyme activity measured directly is indicated by +, activity assumed because of homology is indicated by (+), the known absence of activity is indicated by −, and the lack of information on enzyme activity is indicated by no symbol. Genetic evidence (G) is based on annotation and/or prediction from amino acid sequences; enzymatic evidence (E) is based on measured enzyme activities with both NAD+ and NADP+ (Table 2; see also Table SA2 in the supplemental material).
Enzyme abbreviations: G6PDH, glucose-6P dehydrogenase; 6PGDH, 6P-gluconate dehydrogenase; GAPDH, glyceraldehyde-3P dehydrogenase; ICTDH, isocitrate dehydrogenase; MAE, malic enzyme; MDH, malate dehydrogenase; and TH, transhydrogenase.
Activities of soluble and membrane-bound (membr.) transhydrogenases were measured.
Data on P. versutus enzyme activities are not available in the literature.
Variations in biomass composition.
For the standard biomass composition of E. coli (16, 46), portions of NADPH of 66, 21, and 6% are required for protein, lipid, and RNA biosynthesis, respectively, depending on the growth rate. The remaining 7% covers the reducing power needed for DNA, lipopolysaccharides, peptidoglycans, glycogens, and polyamines. The dependency of NADPH requirements for protein and RNA on the growth rate was already included in the previously published biomass composition model (11, 16) (see Fig. SA1 in the supplemental material). However, the third major anabolic sink of NADPH, lipids, was not considered to be growth rate dependent; hence, its contribution to cellular biomass was varied between 5 and 15% to account for variations in biomass compositions among species. The differences in the relative lipid content were equally balanced by altering both RNA and protein contents, and the consecutive changes in the NADPH requirements for biomass production were calculated as described previously (11, 16).
Reversibility analysis of transhydrogenases.
For the energy-dependent transhydrogenase reaction that involves the translocation of protons over the membrane, the Gibbs energy of reaction was determined as follows:
 |
where ΔrG′ is the range of Gibbs energy, ΔfG′i0 is the transformed standard Gibbs energy of formation for each reactant, i is the considered reaction, v is velocity, R is the universal gas constant, T is the absolute temperature in kelvins, ΔpH is the pH difference between the periplasm and cytoplasm, z is the number of the translocated protons per reaction, F is the Faraday constant, and ΔΦ is the membrane potential. Transformed standard Gibbs energies of reactions were taken from Kümmel et al. (38), and different membrane potentials as well as ΔpH values were considered to calculate the proton motive force (1, 23, 32, 33, 36, 39, 53, 61). The Gibbs energy of the energy-independent transhydrogenase reaction was calculated with the same equation lacking the third term (proton motive force).
RESULTS
Cofactor specificities of the main NADPH-producing enzymes.
To elucidate how microbes manage NADPH metabolism, we chose eight metabolically diverse bacterial species whose intracellular fluxes during growth on glucose were reported previously (22). These are the anaerobic Z. mobilis, the Rhizobiaceae A. tumefaciens and S. meliloti, the metabolically versatile facultative phototroph R. sphaeroides, the facultative autotroph P. versutus, the versatile P. fluorescens, and the model bacteria E. coli and B. subtilis. For some of these species, i.e., P. fluorescens and Z. mobilis, with exclusive catabolism through the ED pathway, and P. versutus, with high TCA cycle flux (22), significant NADPH overproduction is expected from the glucose flux distribution. This NADPH overproduction conclusion was based on the typically made assumption in stoichiometric modeling for 13C flux analysis (16, 29, 42, 56) or analysis at the genome level (51) that dehydrogenases have exclusive cofactor specificities.
Since any conclusion on NADPH metabolism critically depends on the validity of this assumption, we first searched the literature for enzymatic evidence and public databases for genetic evidence of redox cofactor specificities in all reactions that could principally generate or consume NADPH in the central metabolisms of the eight species (Table 1). Direct biochemical evidence for cofactor specificity was available for only a few enzyme-organism combinations for which Km values for NAD+ and NADP+ were determined separately. In a few cases, distinct binding sites for NADH and NADPH in the deduced protein sequences suggested that these enzymes might indeed exhibit dual cofactor specificities (45). Such cofactor specificity typically depends on conserved domains where distinct amino acids stabilize or interfere with the additional phosphate group of NADPH (8, 30, 45). Hence, available protein sequences were analyzed to obtain indications of cofactor specificities (Table 1). Overall, this survey indicated that cofactor specificity was at least questionable in many cases.
For more direct biochemical evidence, we then systematically determined in vitro enzyme activities with both NAD+ and NADP+ for all reactions that could principally provide or consume NADPH in the central metabolisms of all eight species (specific and relative activities are in Table SA2 in the supplemental material and Table 2, respectively). In contrast to the common belief, both dehydrogenases in the oxidative PP pathway of most species exhibited significant activity with NADH as well as NADPH, ranging from 20 to 50% of the total activity under saturated in vitro conditions (Table 2). The sole exception with complete specificity for NADPH was E. coli, as was also shown earlier (10). Species like Z. mobilis and P. fluorescens, in contrast, contain glucose-6P and 6P-gluconate dehydrogenases without any apparent preference for either cofactor or two isoenzymes with different cofactor specificities.
TABLE 2.
Relative cofactor specificities of catabolic enzymes
| Context | Enzymeb | Cofactor | Relative cofactor specificityc (%) ± SD in:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A. tumefaciens | S. meliloti | P. fluorescens | R. sphaeroides | P. versutus | Z. mobilis | E. coli | B. subtilis | |||
| Saturated in vitro conditionsa | G6PDH | NADP+ | 66 ± 6 | 63 ± 3 | 56 ± 11 | 70 ± 2 | 78 ± 4 | 51 ± 3 | 100 ± 0 | 88 ± 1 |
| NAD+ | 34 ± 6 | 37 ± 3 | 44 ± 11 | 30 ± 2 | 22 ± 4 | 49 ± 3 | 0 ± 0 | 12 ± 1 | ||
| 6PGDH | NADP+ | 87 ± 0 | 93 ± 0 | 47 ± 10 | − | 94 ± 8 | − | 100 ± 0 | 89 ± 1 | |
| NAD+ | 13 ± 0 | 7 ± 0 | 53 ± 10 | − | 6 ± 8 | − | 0 ± 0 | 11 ± 1 | ||
| GAPDH | NADP+ | 0 ± 0 | 0 ± 0 | 13 ± 6 | 2 ± 0 | 1 ± 1 | 1 ± 0 | 2 ± 2 | 3 ± 3 | |
| NAD+ | 100 ± 0 | 100 ± 0 | 87 ± 6 | 98 ± 0 | 99 ± 1 | 99 ± 0 | 98 ± 2 | 97 ± 3 | ||
| ICTDH | NADP+ | 98 ± 1 | 98 ± 0 | 99 ± 1 | 96 ± 3 | 99 ± 0 | − | 100 ± 0 | 100 ± 0 | |
| NAD+ | 2 ± 1 | 2 ± 0 | 1 ± 1 | 4 ± 3 | 1 ± 0 | − | 0 ± 0 | 0 ± 0 | ||
| MDH | NADP+ | 0 ± 0 | 0 ± 0 | − | 1 ± 1 | 0 ± 0 | − | 1 ± 2 | 1 ± 1 | |
| NAD+ | 100 ± 0 | 100 ± 0 | − | 99 ± 1 | 100 ± 0 | − | 99 ± 2 | 99 ± 1 | ||
| Nonsaturated, quasi in vivo conditions | G6PDH | NADP+ | 74 | 71 | 92 | 84 | 83 | ND | ND | 100 |
| NAD+ | 26 | 29 | 8 | 16 | 17 | ND | ND | 0 | ||
| 6PGDH | NADP+ | 92 | 92 | 80 | − | 73 | ND | ND | 96 | |
| NAD+ | 8 | 8 | 20 | − | 27 | ND | ND | 4 | ||
Relative cofactor specificities were derived from absolute specific units separately determined for NAD+ and NADP+ at saturated in vitro concentrations (units per gram of cell protein) (see Table SA2 in the supplemental material). Standard deviations were derived from duplicate determinations in at least two independent growth experiments.
Enzyme abbreviations: G6PDH, glucose-6P dehydrogenase; 6PGDH, 6P-gluconate dehydrogenase; GAPDH, glyceraldehyde-3P dehydrogenase; ICTDH, isocitrate dehydrogenase; and MDH, malate dehydrogenase.
−, absolute activities were close to zero and relative cofactor specificities were not determined; ND, enzyme activity was not determined.
While glyceraldehyde-3P dehydrogenases were specific mostly for NAD+, the P. fluorescens enzyme exhibited a small but significant amount of activity with NADP+ also. P. fluorescens contains one putative NADP+-dependent and two NAD+-dependent glyceraldehyde-3P dehydrogenase-encoding genes (Table 1) and may be similar to B. subtilis, which contains a NADP+-dependent isoform that catalyzes the gluconeogenic direction and is repressed during growth on glucose (18). Throughout all species investigated, the two TCA cycle enzymes isocitrate dehydrogenase and malate dehydrogenase were highly specific for the cofactors NADP+ and NAD+, respectively (Tables 1 and 2). Although malic enzyme activity is not required during growth on glucose, significant malic enzyme fluxes through the so-called pyruvate shunt in P. fluorescens were described previously (22). The operation of the pyruvate shunt in this organism was confirmed here by basically absent and exceptionally high in vitro activities of malate dehydrogenase and malic enzyme, respectively (see Table SA2 in the supplemental material).
Potential further sources of NAD(P)H are the pyruvate dehydrogenase complex and the 2-oxo-glutarate dehydrogenase. Since both contain the NAD+-specific dihydrolipoamide dehydrogenase, they are highly specific for NAD+ (12, 59). Succinate dehydrogenase delivers electrons directly to the respiratory chain via reduced flavin adenine dinucleotide and hence does not react with NADP+. The cofactor dehydrogenases in the bacterial respiratory chains were assumed to be specific for NADH, since specificity for NADPH has been described so far only for mitochondria (26, 64). The active alcohol dehydrogenase in Z. mobilis was determined to be specific exclusively for NADH (mean activity ± standard deviation, 896 ± 52 U g cell protein−1 for NADH and 1 ± 6 U g cell protein−1for NADPH), which is in agreement with previously reported values (37, 52).
In vivo cofactor specificities of the PP pathway dehydrogenases.
Next, we focused on the in vivo cofactor specificities of the two oxidative PP pathway dehydrogenases because their high catabolic fluxes in most species (22) may significantly influence the expected NADPH overproduction. In standard in vitro enzyme assays, oxidized cofactors NADP+ and NAD+ are typically used at the same concentration of 1 mM, which does not reflect the in vivo conditions. In order to obtain activities for the oxidative PP pathway dehydrogenases that were closer to those in the in vivo situation, we quantified intracellular cofactor and substrate concentrations for all species except Z. mobilis by targeted metabolomics (Table 3). The glucose-6P pool size was in the range of 0.5 to 1 mM and generally at least five times larger than the 6P-gluconate pool, while the NADP+ pool was about two to five times smaller than the NAD+ pool. In accordance with the findings reported by Harold (25), the NADP(H) pool was more reduced than the NAD(H) pool except in R. sphaeroides and P. versutus, where both pools were equally reduced.
TABLE 3.
Intracellular metabolite concentrations and Gibbs energies of transhydrogenase reactions
| Organism | Concna (μM) of:
|
NAD+/NADP+ ratioa | ΔrG′b (kJ mol−1) for:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Glucose-6P | 6P-gluconate | NADP+ | NAD+ | NADPH | NADH | mTH | sTH | ||
| A. tumefaciens | 522 ± 89 | 57 ± 6 | 203 ± 26 | 475 ± 20 | 138 ± 37 | 224 ± 26 | 2.34 ± 0.32 | −8.01-−32.20 | 0.57-−2.21 |
| S. meliloti | 548 ± 58 | 42 ± 1 | 142 ± 25 | 392 ± 16 | 316 ± 238 | 193 ± 49 | 2.77 ± 0.50 | −3.80-−32.44 | 0.82-−6.42 |
| P. fluorescens | 821 ± 161 | 150 ± 7 | 184 ± 22 | 343 ± 12 | 152 ± 22 | 152 ± 122 | 1.86 ± 0.23 | −3.97-−32.33 | 0.71-−6.25 |
| R. sphaeroides | 553 ± 35 | 86 ± 2 | 82 ± 20 | 447 ± 24 | 33 ± 56 | 180 ± 25 | 5.43 ± 1.73 | −6.60-−41.34 | 9.71-−3.62 |
| P. versutus | 1048 ± 85 | 147 ± 19 | 96 ± 8 | 421 ± 75 | 43 ± 13 | 190 ± 21 | 4.41 ± 0.87 | −8.68-−33.45 | 1.83-−1.54 |
| B. subtilis | 515 ± 117 | 281 ± 10 | 154 ± 9 | 615 ± 44 | 284 ± 89 | 245 ± 64 | 3.98 ± 0.37 | −4.71-−29.68 | −1.94-−5.51 |
| E. coli | 252 ± 46 | 32 ± 3 | 204 ± 7 | 854 ± 59 | 202 ± 55 | 320 ± 69 | 4.18 ± 0.33 | −6.40-−30.78 | −0.84-−3.82 |
Data are means ± standard deviations of results from triplicate experiments.
ΔrG′, estimated range of Gibbs energies of reaction for membrane-bound transhydrogenase (mTH; from NADH toward NADPH with H+ transfer) and soluble transhydrogenase (sTH; from NADPH toward NADH, energy independent).
Using these in vivo concentrations, we then determined in vitro enzyme activities under quasi in vivo conditions and compared them to activities obtained under saturated conditions (Fig. 2). Assuming an irreversible, two-substrate mechanism for the two PP pathway dehydrogenases, the formation of reduced cofactors was linear at saturating conditions but decreased after a few seconds with the in vivo reactant concentrations. The difference between quasi in vivo and saturated concentrations (2 mM for the substrate and 1 mM for both cofactors) resulted in lower enzyme activities generally shifted to a higher specificity for NADP+ (Fig. 2 and Table 2), despite the higher NAD+/NADP+ ratio in vivo (Table 3). These relative specificities from quasi in vivo conditions allowed for a biologically more relevant estimation of the NADPH formation rate in the PP pathway and were used for the NADPH balancing in the next part of the study.
FIG. 2.
(A) In vitro formation of NADPH (circles) or NADH (triangles) by glucose-6P dehydrogenase of A. tumefaciens under saturated conditions (closed symbols) and quasi in vivo conditions (open symbols). (B and C) Specific activities of glucose-6P dehydrogenases and 6P-gluconate dehydrogenases from six different species were deduced using only the linear phase of reduced cofactor formation under quasi in vivo conditions (open bars) and were compared to the specific activities under saturated conditions (closed bars). Activities with standard deviations are from at least duplicate experiments. N.D., not determined.
NADPH balancing.
13C-determined intracellular carbon fluxes specify the total amount of redox cofactors that are produced in each dehydrogenase reaction. For cofactor-specific enzymes, carbon flux data thus directly quantify the concomitant formation/consumption rate of the cofactor. For all redox enzymes, we estimated cofactor conversion rates by multiplying the absolute carbon fluxes (22) by the experimentally determined specificities for either cofactor (Table 2). For the PP pathway dehydrogenases, we used the cofactor specificities determined under quasi in vivo conditions.
E. coli was the only species with a negative NADPH balance (Fig. 3), which is fully consistent with the previously reported NADPH formation through the membrane-bound transhydrogenase PntAB under the tested condition (55). P. fluorescens exhibited most likely balanced NADPH metabolism (Fig. 3) because of two glucose-6P dehydrogenase isoenzymes with different cofactor specificities (based on amino acid sequences). The other six species exhibited positive NADPH balances on glucose (Fig. 3). P. versutus was the most extreme case, where even the unlikely extreme NADPH specificities of 0 and 100% for all nonspecific reactions resulted in significant catabolic overproduction of NADPH. Potential variances in the biomass composition, in particular the lipid content, that would affect the anabolic NADPH requirement were comparatively small (Fig. 3). Thus, independent of the above-mentioned uncertainties, all six species with catabolic NADPH overproduction must operate mechanisms that regenerate NADP+ or avoid the overproduction of NADPH. For A. tumefaciens, B. subtilis, and P. fluorescens, known isoenzymes with alternative cofactors may potentially suffice to avoid NADPH overproduction (Fig. 3). For A. tumefaciens and B. subtilis, however, an even balance could be achieved only if the appropriate isoenzyme was used exclusively. Hence, an additional NADPH-balancing mechanism is likely to be necessary for these two species. Since P. versutus, S. meliloti, R. sphaeroides, and Z. mobilis do not contain such dual cofactor isoenyzmes, they require some type of balancing mechanism.
FIG. 3.
Metabolism-wide NADPH balancing based on 13C flux distributions and cofactor specificities for glucose-6P and 6P-gluconate dehydrogenases (black bars) determined in vitro under quasi in vivo conditions. The black error bars indicate variances of NADPH requirements for biomass if the lipid proportion is allowed to vary between 5 and 15%. Dark gray bars indicate the theoretical range of the NADPH balance due to isoenzymes with distinct cofactor specificities. Light gray bars indicate the hypothetical range when 100% specificity was assumed for each cofactor for all enzymes with nonspecific in vitro activities. qGlc, specific glucose uptake rate.
The function of transhydrogenases.
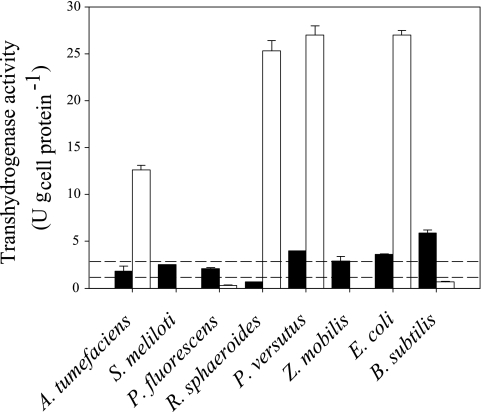
An obvious mechanism to maintain the NADPH balance may be catalyzed by transhydrogenases. Therefore, we determined in vitro transhydrogenase activities in both the soluble and membrane fractions of all species (Fig. 4). Significant activities above 1 U g cell protein−1 in the membrane fraction were detected only for A. tumefaciens, E. coli, P. versutus, and R. sphaeroides. Significant activity in the soluble fraction, in contrast, was detected only for B. subtilis under the glucose batch conditions used, with the possible exception of very low activities for E. coli and P. versutus. The background activities were determined to be 2.8 ± 0.2 U g cell protein−1 for an E. coli pntAB udhA double mutant (55) and 1.2 ± 0.3 U g cell protein−1 for a self-constructed A. tumefaciens pntAB sth mutant. In both mutants, both known forms of transhydrogenases were deleted and the remaining background activity arises from the nonspecific reduction of APAD+ in the soluble fraction that contains also other dehydrogenases. The functionality of the assay was verified by soluble transhydrogenase activities of 84.1 ± 7.3 U g cell protein−1 in an analysis of diluted crude cell extracts from the E. coli pntAB udhA double mutant with plasmid-based overexpression of E. coli UdhA.
FIG. 4.
Transhydrogenase activities in the soluble fractions (closed bars) and the membrane fractions (open bars) of cell extracts. Activities with standard deviations are from at least duplicate experiments. The upper and lower dashed lines indicate average background activities, which were determined with the soluble fractions of E. coli pntAB udhA and A. tumefaciens pntAB sth double mutants, respectively.
The soluble transhydrogenase is generally accepted as a redox equilibration enzyme with Gibbs energies of reaction in the range of +9 to −6 kJ mol−1 (Table 3), which indicate a fully reversible transhydrogenation reaction under all conditions. However, the absent activities demonstrate that a soluble transhydrogenase cannot constitute the postulated balancing mechanism in the species studied, with the possible exception of B. subtilis and P. versutus. Could a reverse flux from NADPH to NADH through the membrane-bound transhydrogenase be responsible? Depending on the assumed proton motive force (between 10 and 30 kJ mol−1 based on published values for ΔpH and Δφ) and the determined intracellular concentrations with their standard deviations, the Gibbs energy would be in the range of −4 to −41 kJ mol−1 (Table 3). Thus, the reaction is irreversible under physiological conditions and cannot balance the NADPH overproduction in A. tumefaciens, P. versutus, and R. sphaeroides. This conclusion was corroborated by 13C flux data for the R. sphaeroides pntAB mutant (27) and self-constructed A. tumefaciens pntAB, sth, and pntAB sth mutants, which exhibited fluxes and NADPH balances essentially unaltered from those of their parents (see Table SA1 in the supplemental material). Thus, A. tumefaciens, R. sphaeroides, S. meliloti, and Z. mobilis certainly use no transhydrogenase for redox balancing, while for B. subtilis and P. versutus, the soluble transhydrogenase could potentially fulfill this function.
DISCUSSION
How bacteria balance catabolic formation with the anabolic consumption of the redox cofactor NADPH has not yet been addressed seriously, presumably because this balancing is a complicated property that concerns the entire metabolic network. Thus, techniques of systems biology that became available only recently are necessary. By combining 13C flux data, metabolomics, and in vitro characterization of cofactor specificities for key enzymes under quasi in vivo conditions, we here addressed this question for eight bacterial species by taking a network approach. With the possible exception of P. fluorescens, catabolic and anabolic NADPH rates were not balanced during batch growth on glucose as the sole carbon source. E. coli was the only investigated species that produced insufficient NADPH from glucose catabolism and thus required significant additional NADPH formation via the energy-dependent transhydrogenase PntAB, as was shown previously (55). The other six species, in contrast, produced significantly more NADPH during catabolism than was required for anabolism. Thus, they must feature NADPH imbalance-avoiding or reoxidation mechanisms for NADPH to prevent an imbalance that would preclude sustained growth.
An obvious but previously neglected possibility is alternative cofactor specificities for catabolic enzymes, and this issue was investigated in detail. Although we found several catabolic reactions to accept both cofactors, most prominently the dehydrogenases of the oxidative PP pathway, even the unlikely exclusive use of NAD+ in the oxidative PP pathway would not prevent the significant overproduction of NADPH in several species. Moreover, when we characterized these cofactor specificities under quasi in vivo conditions based on intracellular metabolite concentrations in the different species, it became clear that other balancing mechanisms, at least in R. sphaeroides, S. meliloti, Z. mobilis, and in particular P. versutus, must exist. The most well-known valve-like mechanism is the soluble transhydrogenase, which has been shown to fulfill a valve function in E. coli (55) and, upon heterologous expression, also in S. cerevisiae (17). Of the four above-named species, however, only P. versutus contains such a soluble transhydrogenase.
How do the six overproducing species then maintain their redox balance? All evidence indicates that P. versutus and B. subtilis rely on their soluble transhydrogenase activities, but the latter may additionally exploit its alternative, NAD+-dependent 6P-gluconate dehydrogenase (69) and malic enzyme (13). For A. tumefaciens, R. sphaeroides, S. meliloti, and Z. mobilis, the overproduction of NADPH may potentially be avoided by assuming almost exclusive specificities for NADH in the glucose-6P dehydrogenase step of the ED pathway, although the quasi in vivo enzyme data suggest that this scenario is unlikely to occur to this extent. Both A. tumefaciens and R. sphaeroides exhibited significant in vitro transhydrogenase activities in their membrane fractions. With rather negative Gibbs energies of NADPH formation—given a large range of proton motive forces and the determined intracellular metabolite concentrations—this reaction is physiologically unidirectional and therefore not involved in balancing the catabolic overproduction of NADPH. The irrelevance of the membrane-bound transhydrogenases for redox balancing is fully consistent with the here-determined unaltered catabolic fluxes in PntAB-type transhydrogenase mutants of A. tumefaciens and R. sphaeroides. Since transhydrogenases can be excluded as key mechanisms, the most likely explanation for redox balancing in these four species is a combination of different mechanisms. The main one appears to be the cofactor nonspecificity of several catabolic enzymes or the use of isoenzymes, possibly in combination with putative redox cycles, NADH kinases, and/or a higher than anticipated NADPH demand for thioredoxin reduction or biomass formation.
A highly unexpected key finding was that, except for E. coli, none of the investigated species contained highly NADPH-specific dehydrogenases in the oxidative PP pathway. This finding contrasts with the textbook view of the PP pathway as being NADP+ dependent. The lack of high-level NADPH specificity, in particular for the glucose-6P dehydrogenase, may be related to the fact that this enzyme is also required for the ED pathway, the main or even exclusive glucose catabolic pathway in most species investigated here. In Z. mobilis, the very low biomass yield of 3% and high ethanol production directly link the demand of NADH for the alcohol dehydrogenase to the regeneration of NADH by glucose-6P and glyceraldehyde-3P dehydrogenases. Since the genes for transhydrogenases are absent, both of these dehydrogenases operate primarily or even exclusively with NADH in vivo (66). The nonspecificity of the key enzyme glucose-6P dehydrogenase appears to be a major mechanism to avoid catabolic NADPH overproduction in a number of species. Moreover, NAD+-dependent oxidative PP pathway flux may confer flexibility on glucose metabolism that enables the organism to cope with dynamic fluctuations in NADP+ and NADPH availability. For the modeling of species other than E. coli (51), the general role of the PP pathway has to be reconsidered to obtain realistic functionality and redox balances.
Supplementary Material
Footnotes
Published ahead of print on 30 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abee, T., K. J. Hellingwerf, and W. N. Konings. 1988. Effects of potassium ions on proton motive force in Rhodobacter sphaeroides. J. Bacteriol. 1705647-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al Zaid Siddiquee, K., M. J. Arauzo-Bravo, and K. Shimizu. 2004. Metabolic flux analysis of pykF gene knockout Escherichia coli based on 13C-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Appl. Microbiol. Biotechnol. 63407-417. [DOI] [PubMed] [Google Scholar]
- 3.Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2676102-6109. [DOI] [PubMed] [Google Scholar]
- 4.Bakker, B. M., K. M. Overkamp, A. J. van Maris, P. Kotter, M. A. Luttik, J. P. van Dijken, and J. T. Pronk. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2515-37. [DOI] [PubMed] [Google Scholar]
- 5.Bizouarn, T., M. Althage, A. Pedersen, A. Tigerstrom, J. Karlsson, C. Johansson, and J. Rydstrom. 2002. The organization of the membrane domain and its interaction with the NADP(H)-binding site in proton-translocating transhydrogenase from Escherichia coli. Biochim. Biophys. Acta 1555122-127. [DOI] [PubMed] [Google Scholar]
- 6.Blank, L. M., F. Lehmbeck, and U. Sauer. 2005. Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res. 5545-558. [DOI] [PubMed] [Google Scholar]
- 7.Boonstra, B., C. E. French, I. Wainwright, and N. C. Bruce. 1999. The udhA gene of Escherichia coli encodes a soluble pyridine nucleotide transhydrogenase. J. Bacteriol. 1811030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carugo, O., and P. Argos. 1997. NADP-dependent enzymes. I. Conserved stereochemistry of cofactor binding. Proteins 2810-28. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, D. M., T. W. Loo, S. Gillam, and P. D. Bragg. 1986. Nucleotide sequence of the pntA and pntB genes encoding the pyridine nucleotide transhydrogenase of Escherichia coli. Eur. J. Biochem. 158647-653. [DOI] [PubMed] [Google Scholar]
- 10.Csonka, L. N., and D. G. Fraenkel. 1977. Pathways of NADPH formation in Escherichia coli. J. Biol. Chem. 2523382-3391. [PubMed] [Google Scholar]
- 11.Dauner, M., and U. Sauer. 2001. Stoichiometric growth model for riboflavin-producing Bacillus subtilis. Biotechnol. Bioeng. 76132-143. [DOI] [PubMed] [Google Scholar]
- 12.Dietrichs, D., and J. R. Andreesen. 1990. Purification and comparative studies of dihydrolipoamide dehydrogenases from the anaerobic, glycine-utilizing bacteria Peptostreptococcus glycinophilus, Clostridium cylindrosporum, and Clostridium sporogenes. J. Bacteriol. 172243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doan, T., P. Servant, S. Tojo, H. Yamaguchi, G. Lerondel, K. Yoshida, Y. Fujita, and S. Aymerich. 2003. The Bacillus subtilis ywkA gene encodes a malic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate. Microbiology 1492331-2343. [DOI] [PubMed] [Google Scholar]
- 14.Driscoll, B. T., and T. M. Finan. 1997. Properties of NAD+- and NADP+-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti and differential expression of their genes in nitrogen-fixing bacteroids. Microbiology 143489-498. [DOI] [PubMed] [Google Scholar]
- 15.Dunn, M. F. 1998. Tricarboxylic acid cycle and anaplerotic enzymes in rhizobia. FEMS Microbiol. Rev. 22105-123. [DOI] [PubMed] [Google Scholar]
- 16.Emmerling, M., M. Dauner, A. Ponti, J. Fiaux, M. Hochuli, T. Szyperski, K. Wüthrich, J. E. Bailey, and U. Sauer. 2002. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 184152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiaux, J., Z. P. Cakar, M. Sonderegger, K. Wüthrich, T. Szyperski, and U. Sauer. 2003. Metabolic-flux profiling of the yeasts Saccharomyces cerevisiae and Pichia stipitis. Eukaryot. Cell 2170-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fillinger, S., S. Boschi-Muller, S. Azza, E. Dervyn, G. Branlant, and S. Aymerich. 2000. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 27514031-14037. [DOI] [PubMed] [Google Scholar]
- 19.Fischer, E., and U. Sauer. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270880-891. [DOI] [PubMed] [Google Scholar]
- 20.Fischer, E., N. Zamboni, and U. Sauer. 2004. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Anal. Biochem. 325308-316. [DOI] [PubMed] [Google Scholar]
- 21.French, C. E., B. Boonstra, K. A. Bufton, and N. C. Bruce. 1997. Cloning, sequence, and properties of the soluble pyridine nucleotide transhydrogenase of Pseudomonas fluorescens. J. Bacteriol. 1792761-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuhrer, T., E. Fischer, and U. Sauer. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 1871581-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gober, J. W., and E. R. Kashket. 1985. Measurement of the proton motive force in Rhizobium meliloti with the Escherichia coli lacY gene product. J. Bacteriol. 164929-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gornall, A., W. Bardawill, and M. David. 1949. Determinations of serum proteins by means of the Biuret reaction. J. Biol. Chem. 177751-766. [PubMed] [Google Scholar]
- 25.Harold, F. 1986. The vital force: a study of bioenergetics. Freeman and Company, New York, NY.
- 26.Harris, D. M., J. A. Diderich, Z. A. van der Krogt, M. A. Luttik, L. M. Raamsdonk, R. A. Bovenberg, W. M. van Gulik, J. P. van Dijken, and J. T. Pronk. 2006. Enzymic analysis of NADPH metabolism in beta-lactam-producing Penicillium chrysogenum: presence of a mitochondrial NADPH dehydrogenase. Metab. Eng. 891-101. [DOI] [PubMed] [Google Scholar]
- 27.Hickman, J. W., R. D. Barber, E. P. Skaar, and T. J. Donohue. 2002. Link between the membrane-bound pyridine nucleotide transhydrogenase and glutathione-dependent processes in Rhodobacter sphaeroides. J. Bacteriol. 184400-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoek, J. B., and J. Rydstrom. 1988. Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem. J. 2541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua, Q., C. Yang, T. Baba, H. Mori, and K. Shimizu. 2003. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J. Bacteriol. 1857053-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurley, J. H., R. Chen, and A. M. Dean. 1996. Determinants of cofactor specificity in isocitrate dehydrogenase: structure of an engineered NADP+ → NAD+ specificity-reversal mutant. Biochemistry 355670-5678. [DOI] [PubMed] [Google Scholar]
- 31.Iwakura, M., M. Tokushige, H. Katsuki, and S. Muramatsu. 1978. Studies on regulatory functions of malic enzymes. V. Comparative studies of malic enzymes in bacteria. J. Biochem. (Tokyo) 831387-1394. [DOI] [PubMed] [Google Scholar]
- 32.Kashket, E. R. 1981. Proton motive force in growing Streptococcus lactis and Staphylococcus aureus cells under aerobic and anaerobic conditions. J. Bacteriol. 146369-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katsu, T., H. Nakagawa, T. Kanamori, N. Kamo, and T. Tsuchiya. 2001. Ion-selective electrode for transmembrane pH difference measurements. Anal. Chem. 731849-1854. [DOI] [PubMed] [Google Scholar]
- 34.Kawai, S., C. Fukuda, T. Mukai, and K. Murata. 2005. MJ0917 in archaeon Methanococcus jannaschii is a novel NADP+ phosphatase/NAD+ kinase. J. Biol. Chem. 28039200-39207. [DOI] [PubMed] [Google Scholar]
- 35.Kawai, S., S. Mori, T. Mukai, W. Hashimoto, and K. Murata. 2001. Molecular characterization of Escherichia coli NAD+ kinase. Eur. J. Biochem. 2684359-4365. [DOI] [PubMed] [Google Scholar]
- 36.Kell, D. B., P. John, and S. J. Ferguson. 1978. The protonmotive force in phosphorylating membrane vesicles from Paracoccus denitrificans. Magnitude, sites of generation and comparison with the phosphorylation potential. Biochem. J. 174257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinoshita, S., T. Kakizono, K. Kadota, K. Das, and H. Taguchi. 1985. Purification of two alcohol dehydrogenases from Zymomonas mobilis and their properties. Appl. Microbiol. Biotechnol. 22249-254. [Google Scholar]
- 38.Kummel, A., S. Panke, and M. Heinemann. 2006. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 22006.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludovico, P., F. Sansonetty, and M. Corte-Real. 2001. Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry. Microbiology 1473335-3343. [DOI] [PubMed] [Google Scholar]
- 40.Luo, B., K. Groenke, R. Takors, C. Wandrey, and M. Oldiges. 2007. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J. Chromatogr. A 1147153-164. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Drets, G., A. Gardiol, and A. Arias. 1977. 6-Phospho-d-gluconate: NAD+ 2-oxidoreductase (decarboxylating) from slow-growing rhizobia. J. Bacteriol. 1301139-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marx, A., A. A. deGraaf, W. Wiechert, L. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 49111-129. [DOI] [PubMed] [Google Scholar]
- 43.Michal, G. 1999. Biochemical pathways. Spektrum Akademischer Verlag GmbH, Heidelberg, Germany .
- 44.Murai, T., M. Tokushige, J. Nagai, and H. Katsuki. 1971. Physiological functions of NAD+- and NADP+-linked malic enzymes in Escherichia coli. Biochem. Biophys. Res. Commun. 43875-881. [DOI] [PubMed] [Google Scholar]
- 45.Naylor, C. E., S. Gover, A. K. Basak, M. S. Cosgrove, H. R. Levy, and M. J. Adams. 2001. NADP+ and NAD+ binding to the dual coenzyme specific enzyme Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase: different interdomain hinge angles are seen in different binary and ternary complexes. Acta Crystallogr. D 57635-648. [DOI] [PubMed] [Google Scholar]
- 46.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates Inc., Sunderland, MA.
- 47.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175282-294. [DOI] [PubMed] [Google Scholar]
- 48.Nipkow, A., B. Sonnleitner, and A. Fiechter. 1985. Effect of carbon-dioxide on growth of Zymomonas mobilis in continuous culture. Appl. Microbiol. Biotechnol. 21287-291. [Google Scholar]
- 49.Outten, C. E., and V. C. Culotta. 2003. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 222015-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Overkamp, K. M., B. M. Bakker, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 2002. Two mechanisms for oxidation of cytosolic NADPH by Kluyveromyces lactis mitochondria. Yeast 19813-824. [DOI] [PubMed] [Google Scholar]
- 51.Price, N. D., J. L. Reed, and B. O. Palsson. 2004. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat. Rev. Microbiol. 2886-897. [DOI] [PubMed] [Google Scholar]
- 52.Reid, M. F., and C. A. Fewson. 1994. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 2013-56. [DOI] [PubMed] [Google Scholar]
- 53.Riondet, C., R. Cachon, Y. Wache, G. Alcaraz, and C. Divies. 1999. Changes in the proton-motive force in Escherichia coli in response to external oxidoreduction potential. Eur. J. Biochem. 262595-599. [DOI] [PubMed] [Google Scholar]
- 54.Sakuraba, H., R. Kawakami, and T. Ohshima. 2005. First archaeal inorganic polyphosphate/ATP-dependent NAD+ kinase, from hyperthermophilic archaeon Pyrococcus horikoshii: cloning, expression, and characterization. Appl. Environ. Microbiol. 714352-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauer, U., F. Canonaco, S. Heri, A. Perrenoud, and E. Fischer. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 2796613-6619. [DOI] [PubMed] [Google Scholar]
- 56.Sauer, U., V. Hatzimanikatis, J. E. Bailey, M. Hochuli, T. Szyperski, and K. Wüthrich. 1997. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat. Biotechnol. 15448-452. [DOI] [PubMed] [Google Scholar]
- 57.Sazanov, L. A., and J. B. Jackson. 1994. Proton-translocating transhydrogenase and NAD+- and NADP+-linked isocitrate dehydrogenases operate in a substrate cycle, which contributes to fine regulation of the tricarboxylic acid cycle activity in mitochondria. FEBS Lett. 344109-116. [DOI] [PubMed] [Google Scholar]
- 58.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 59.Scrutton, N. S., A. Berry, and R. N. Perham. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 34338-43. [DOI] [PubMed] [Google Scholar]
- 60.Singh, R., J. Lemire, R. J. Mailloux, and V. D. Appanna. 2008. A novel strategy involved in anti-oxidative defense: the conversion of NADH into NADPH by a metabolic network. PLoS ONE 3e2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slonczewski, J. L., B. P. Rosen, J. R. Alger, and R. M. Macnab. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. USA 786271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steen, I. H., D. Madern, M. Karlstrom, T. Lien, R. Ladenstein, and N. K. Birkeland. 2001. Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state, and phylogenetic affiliation. J. Biol. Chem. 27643924-43931. [DOI] [PubMed] [Google Scholar]
- 63.Stournaras, C., P. Maurer, and G. Kurz. 1983. 6-Phospho-d-gluconate dehydrogenase from Pseudomonas fluorescens. Properties and subunit structure. Eur. J. Biochem. 130391-396. [DOI] [PubMed] [Google Scholar]
- 64.Tarrio, N., M. Becerra, M. E. Cerdan, and M. I. Gonzalez Siso. 2006. Reoxidation of cytosolic NADPH in Kluyveromyces lactis. FEMS Yeast Res. 6371-380. [DOI] [PubMed] [Google Scholar]
- 65.Verho, R., P. Richard, P. H. Jonson, L. Sundqvist, J. Londesborough, and M. Penttila. 2002. Identification of the first fungal NADP+-GAPDH from Kluyveromyces lactis. Biochemistry 4113833-13838. [DOI] [PubMed] [Google Scholar]
- 66.Viikari, L. 1988. Carbohydrate metabolism in Z. mobilis. Crit. Rev. Biotechnol. 7237-261. [Google Scholar]
- 67.Yamaguchi, M., and C. D. Stout. 2003. Essential glycine in the proton channel of Escherichia coli transhydrogenase. J. Biol. Chem. 27845333-45339. [DOI] [PubMed] [Google Scholar]
- 68.Yang, C., Q. Hua, T. Baba, H. Mori, and K. Shimizu. 2003. Analysis of Escherichia coli anaplerotic metabolism and its regulation mechanisms from the metabolic responses to altered dilution rates and phosphoenolpyruvate carboxykinase knockout. Biotechnol. Bioeng. 84129-144. [DOI] [PubMed] [Google Scholar]
- 69.Zamboni, N., E. Fischer, D. Laudert, S. Aymerich, H. P. Hohmann, and U. Sauer. 2004. The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway. J. Bacteriol. 1864528-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zamboni, N., E. Fischer, and U. Sauer. 2005. FiatFlux—a software for metabolic flux analysis from 13C-glucose experiments. BMC Bioinformatics 6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zerez, C. R., D. E. Moul, E. G. Gomez, V. M. Lopez, and A. J. Andreoli. 1987. Negative modulation of Escherichia coli NAD+ kinase by NADPH and NADH. J. Bacteriol. 169184-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.