Abstract
In prokaryotes, flagellar biogenesis is a complicated process involving over 40 genes. The phytopathogen Xanthomonas campestris pv. campestris possesses a single polar flagellum, which is essential for the swimming motility. A σ54 activator, FleQ, has been shown to be required for the transcriptional activation of the flagellar type III secretion system (F-T3SS), rod, and hook proteins. One of the two rpoN genes, rpoN2, encoding σ54, is essential for flagellation. RpoN2 and FleQ direct the expression of a second alternative sigma FliA (σ28) that is essential for the expression of the flagellin FliC. FlgM interacts with FliA and represses the FliA regulons. An flgM mutant overexpressing FliC generates a deformed flagellum and displays an abnormal motility. Mutation in the two structural genes of F-T3SS, flhA and flhB, suppresses the production of FliC. Furthermore, FliA protein levels are decreased in an flhB mutant. A mutant defective in flhA, but not flhB, exhibits a decreased infection rate. In conclusion, the flagellar biogenesis of Xanthomonas campestris requires alternative sigma factors RpoN2 and FliA and is temporally regulated by FlhA, FlhB, and FlgM.
The flagellar type III secretion system (F-T3SS) is essential for the export of extracytoplasmic components of the flagella (3, 10, 41, 43). In enteric bacteria, the assembly of F-T3SS is initiated by the formation of a membrane-embedded MS ring in which 26 monomers of FliF proteins form a pore at the appropriate position of the inner membrane (69). FliF interacting with the switch protein FliG (23, 40) and FlhAB (32, 44) is required for flagellar rotation and assembly. FlhA, together with FlhB and FliOPQR, residing in the MS ring is essential for flagellar export (44). FlhB controls the choice between secretion of flagellar rod-hook protein and flagellar filament protein (17, 18, 49, 51). FlhA and FlhB form a docking platform for soluble proteins FliI and FliH (50, 71). FliI is an ATPase (28). The energy of ATP hydrolysis by FliI initiates flagellar protein transport (48, 58).
Expression of flagellar genes is generally regulated by several regulators in a hierarchical manner (3, 8, 10, 43). In Enterobacteriaceae, which possess multiple peritrichous flagella, the flagellar genes can be classified into three classes: the class I proteins FlhDC expressed from a σ70-directed promoter are the master regulators that regulate the transcription of the class II genes comprising fliA (σ28) and the genes encoding F-T3SS, rod, and hook proteins (2, 39). Expression of the class III genes, including those for flagellin proteins, requires an active FliA. Several feedback pathways have been reported to coordinate the gene expression and flagellin assembly. An anti-sigma factor, FlgM, expressed from a class II promoter binds to FliA and inhibits its activity. This inhibition terminates with complete formation of the hook-basal body complex through which FlgM is expelled from the cytoplasm (35). The duration of expression of class III genes is limited due to increased proteolysis of FliA in the absence of FlgM (5). Chaperones, FliT and FlgN, also play roles in shifting from class II to class III promoters. FliT binds directly to FlhC and represses the expression of the class II genes (76). FlgN upregulates flgM and represses class III genes (1).
A second type of regulation is represented by a bacterial group, including vibrios and pseudomonads, that possesses single, polar flagella. Two alternative sigma factors (RpoN and FliA) are implicated in the regulation of the transcriptional hierarchy of flagellar genes. In coordination with RpoN, two NtrC-type activators, FlrA/FleQ and FlrBC/FleSR, regulate the transcription of the class II and III promoters (14, 60). The expression of class IV promoters is dependent on FliA under the negative regulation of FlgM (14).
A third type of regulation is found in Caulobacter crescentus, which also possesses a single polar flagellum. The production of flagellin (class III) is directly dependent on RpoN and the activator FlbD (a class II protein) (54). The expression of flbD is dependent on σ73 and the activator CtrA (class I). Regulator FliX binds to FlbD and inhibits its activity (16, 53, 54). After the assembling of an early structure of the basal body, FliX becomes an activator of FlbD, which initiates the transcription of late flagellar genes (52). Similar to the case for FlgM in the enteric bacteria, vibrios, and pseudomonads, FliX regulates a critical checkpoint coupling flagellar gene regulation to assembly.
Xanthomonas campestris pv. campestris, a member of the Pseudomonadaceae bearing a single polar flagellum, is a plant-pathogenic bacterium causing black rot in cruciferous plants (74). By examining the genome sequence of X. campestris pv. campestris ATCC 33913 (GenBank accession no. NC_003902), more than 40 genes have been predicted to be involved in flagellar biogenesis and motility. It is known that expression of the flagellin gene (fliC) is upregulated by Clp, a homolog of the cyclic AMP receptor protein (36). Our previous studies have demonstrated that fleQ, encoding a σ54-cognate activator, is essential for normal flagellation and for the transcription of the promoters upstream of fliE, fliL, fliQ, flgB, flgG, and flhF (27). In this work, we show that the expression of flagellar genes of X. campestris pv. campestris is regulated in a three-tier hierarchy. Alternative sigma factors, RpoN2 (σ54) and FliA (σ28), are required for the expression of class II and class III flagellar promoters, respectively. FlgM represses the activity of a FliA-dependent promoter via protein-protein interaction. The effects of mutations in two F-T3SS structural genes, flhA and flhB, on flagellar genes expression and virulence are also examined.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise specified, Luria-Bertani (LB) broth and LB agar plates (47) were employed as the media for the growth of X. campestris pv. campestris and Escherichia coli at 28 and 37°C, respectively. Saccharomyces cerevisiae Y187 was grown in a yeast-peptone-dextrose (YPD) medium, and the transformed Y187 strains were selected and maintained in synthetic dropout (SD/−) media as described in the Clontech Matchmaker III yeast two-hybrid system manual. Antibiotics added for selection were ampicillin (100 μg/ml), kanamycin (50 μg/ml), gentamicin (50 μg/ml), and tetracycline (10 μg/ml).
TABLE 1.
Bacteria and plasmids used in this work
| Bacterial strain or plasmid | Description | Reference or source |
|---|---|---|
| Xanthomonas campestris pv. campestris strains | ||
| Xc17 | Local purified virulent strain | 78 |
| XcflhA | flhA mutant derived from Xc17, flhA::Gm Apr Gmr | This work |
| XcflhA(c) | XcflhA harboring plasmid pBBADflhA, Apr Gmr Tcr | This work |
| XcflhB | flhB mutant derived from Xc17, flhBΔ612-564::Gm, Apr Gmr | This work |
| XcflhB(c) | XcflhB harboring plasmid pBBADflhB, Apr Gmr Tcr | This work |
| XcfliA | fliA mutant derived from Xc17, fliA::pOK12 Apr Kmr | This work |
| XcrpoN2 | rpoN2 mutant derived from Xc17, rpoN2Δ91-1081::Gm Apr Gmr | This work |
| XcfleQ | fleQ mutant derived from Xc17, fleQ::pOK12 Apr Kmr | 27 |
| XcflgM | flgM mutant derived from Xc17, flgM::Gm Apr Gmr | This work |
| XcflgM(c) | XcflgM harboring plasmid pRKflgM, Apr Gmr Tcr | This work |
| XcfliC | fliC mutant derived from Xc17, fliCΔ489-948::Gm Apr Gmr | This work |
| Escherichia coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 25 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm met(DE3) | Novagen |
| Saccharomyces cerevisiae strain Y187 | MATα ura3-52 his3-200 ade2-101 trp1-901 leu2-3,112 gal4Δ gal80Δ URA3::GAL1UAS-GAL1TATA-lacZ | Clontech |
| Plasmids | ||
| pOK12 | E. coli cloning vector, Kmr | 72 |
| pBB | E. coli cloning vector derived from pOK12, Kmr | This work |
| Pucgm | Gentamicin resistance gene cassettes donor, Apr Gmr | 66 |
| pFY13-9 | Promoter-probing vector derived from pRK415 using lacZ as the reporter, Tcr | 36 |
| pBBAD22K | Broad-host-range expression vector, Kmr | 68 |
| pRK415 | Broad-host-range expression vector, Tcr | 31 |
| pET30b | E. coli expression vector, Kmr | Novagen |
| PFYfliC | Transcriptional fusion derivative of pFY13-9 carrying fliC promoter (319 bp), Tcr | This work |
| PFYflhF | Transcriptional fusion derivative of pFY13-9 carrying flhF promoter (1391 bp), Tcr | 27 |
| pBBADflhA | pBBAD22K derivative with total CDS of flhA cloned downstream of an araBAD promoter | This work |
| pBBADflhB | pBBAD22K derivative with total CDS of flhB cloned downstream of an araBAD promoter | This work |
| PRKflgM | pRK415 derivative with entire CDS of flgM and its promoter region | This work |
| PETfliC | pET30b derivative containing entire fliC CDS fused with His tag at both ends | This work |
| PETfliA | pET30b derivative containing entire fliA CDS fused with His tag at both ends | This work |
| PETrpoN2 | pET30b derivative containing entire rpoN2 CDS fused with His tag at both ends | This work |
| pACT2 | GAL4 activation domain | Clontech |
| pACT-flgM | pACT2 derivative containing entire flgM CDS fused with GAL4 activation domain | This work |
| pAS2-1 | GAL4 DNA-binding domain | Clontech |
| pAS2-fliA | pAS2-1 derivative containing entire fliA CDS fused with GAL4 DNA-binding domain | This work |
Recombinant DNA techniques.
The standard methods for DNA manipulation were those described by Sambrook et al. (65). Restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were purchased from Takara (Kyoto, Japan). Chemicals and reagents were purchased from Sigma (St. Louis, MO) and E. Merck (Darmstadt, Germany).
Construction of flhA, flhB, rpoN2, fliC, flgM, and fliA mutants.
The flhA, flhB, rpoN2, flgM, and fliC mutants were constructed by insertional mutagenesis. A gentamicin resistance (Gmr) gene from pUCGM (66) was inserted into the target gene that had been cloned in suicide plasmid pOK12 (72) or pBB. The orientation of the inserted Gmr gene was the same as that of the target gene as confirmed by sequencing. The recombinant plasmids were then electroporated into a wild-type X. campestris pv. campestris strain, Xc17. Allelic exchange between the chromosomal gene and the mutagenized plasmidic copy was achieved by double crossover. The mutant strains were selected by antibiotic sensitivity and checked by PCR. A fliA mutant was constructed by inserting the plasmid pOK12 into the fliA gene via single crossover. The plasmids and oligonucleotide primers used in this work are listed in Tables 1 and 2. The details of the construction of recombinant suicide plasmids are provided as below.
TABLE 2.
Oligonucleotides used in this work
| Oligonucleotide | Sequence (5′→3′)a | Enzyme site added |
|---|---|---|
| flhAw-F | GGTACCagcgcatgagcgcccaacc | KpnI |
| flhAw-R | AAGCTTccggctcagctaatcgtgcc | HindIII |
| flhBw-F | GGTACCccaatgtccgagtccgaaga | KpnI |
| flhBw-R | AAGCTTgggcgctcatgcgctggccc | HindIII |
| flhB_PstI-F | cgaaggcggcCTGCAGcacc | PstI |
| flhB_PstI-R | ggaactggCTGCAGaagctg | PstI |
| flgM-F | tgttgtcggccaatgacc | |
| flgM-R | Cagtgcccagttcacctc | |
| fliA-F | AGATCTgacgagatccgtcgg | BglII |
| fliA-R | AAGCTTagcgacagcaccagc | HindIII |
| fliAw-F | CATATGatgagtaccgccacc | NdeI |
| fliAw-R | CTCGAGatcgtcaatttcgac | XhoI |
| fliCw-F | CATATGgcacaggtaatcaaca | NdeI |
| fliCw-R | GCGGCCGCctgcagcaggctcagcac | NotI |
| fliCp-F | CTCGAGttgcgcgcaaaaacccgt | XhoI |
| fliCp-R | TCTAGAttttgatatctcctctaa | XbaI |
| flgMw-F | AGATCTGatgagccagaaaatcgaaggc | BglII |
| flgMw-R | GGATCCttgcccgccagttgctggttc | BamHI |
| rpoN2w-F | CATATGaagacgaccatctctg | NdeI |
| rpoN2w-R | CTCGAG tcccgcccgggcaagcag | XhoI |
| fliA_YB-F | CATATGaagatcaaacgctttgttgccc | NdeI |
| fliA_YB-R | GGATCCtcttcaaggcgaagaacgagac | BamHI |
| flgM_YA-F | CATATGagtaccgccaccgcaacgaccg | NdeI |
| flgM_YA-R | GGATCCatcgtcaatttcgacaccggca | BamHI |
Uppercase letters indicate the restriction enzyme site.
(i) flhA.
A 2,105-bp DNA fragment containing the entire coding sequence (CDS) of flhA of Xc17 was amplified by PCR with primer pair flhAw-F/-R and cloned into pBB to obtain pBBflhAw. A Gmr gene was inserted into the BamHI site of pBBflhAw to form pBBflhA::Gm.
(ii) flhB.
A 1,140-bp DNA fragment containing the entire CDS of flhB of Xc17 was amplified by PCR with primer pair flhBw-F/-R and cloned into pBB to obtain pBBflhBw. Two PstI sites were introduced into the flhB gene in pBBflhBw by inverse PCR with flhB_PstI-F/-R. After digestion with PstI, the PCR fragment was self-ligated to form pBBflhB-PstI. A Gmr gene was inserted into the created PstI site to form pBBflhB::Gm.
(iii) rpoN2.
A 2,661-bp PstI-XhoI fragment containing the entire CDS of rpoN2 of Xc17 was obtained by chromosomal walking and cloned in pOK12. A Gmr was inserted into the cloned rpoN2 gene to replace an internal 996 bp of a SphI fragment. The derived plasmid was named pOKrpoN2::Gm.
(iv) flgM.
An 847-bp DNA fragment containing the entire CDS of flgM of Xc17 was amplified by PCR with primer pair flgM-F/-R and cloned into pBB to obtain pBBflgM. A Gmr gene was inserted into the sole PstI site of flgM to form pBBflgM::Gm.
(v) fliC.
A 1,194-bp DNA fragment containing the entire CDS of fliC of Xc17 was amplified by PCR with primer pair fliCw-F/-R and cloned into pBB to obtain pBBfliCw. A Gmr gene was inserted into pBBfliCw to replace an internal 459 bp of a SalI fragment of fliC to form pBBfliC::Gm.
(vi) fliA.
A 347-bp DNA fragment containing the internal region of fliA of Xc17 was amplified with primer pair fliA-F/-R and cloned into pOK12 to form pOKfliA.
Construction of flhA, flhB, and flgM complementary plasmids.
The entire CDSs of flhA and flhB were cut from pBBflhAw and pBBflhBw with KpnI and HindIII and then subcloned into the broad-host-range expression vector pBBAD22K (68) to obtain complementary plasmids pBBADflhA and pBBADflhB, respectively. Expression of the cloned genes was under the control of an arabinose-inducible promoter. An EcoRI/HindIII fragment of 548 bp containing the entire flgM gene and the upstream promoter region was cut from pBBflgM and subcloned into a broad-host-range vector pRK415 (31) to form pRKflgM.
Antibody preparation and Western blotting.
For antibody preparation, His-tagged antigen proteins were overexpressed from pET30b (Novagen, Madison, WI)-derived plasmids in E. coli strain BL21(DE3) after 4 h of induction with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The proteins were purified on an Ni-nitrilotriacetic acid resin column (Pharmacia), and ca. 1 mg of each protein was used for four injections to immunize a rabbit or five mice at intervals of 1 week. Overexpression plasmids were cloned as described below.
(i) fliA.
The entire CDS (765 bp) of fliA was amplified by PCR with primers fliAw-F/-R and cloned into pBB to obtain pBBfliAw. The CDS region was then subcloned into the NdeI/XhoI sites of pET30b to generate pETfliA.
(ii) fliC.
The entire CDS of fliC in pBBfliCw was subcloned into the NdeI/NotI sites of pET30b to generate pETfliC.
(iii) rpoN2.
The entire CDS (1,401 bp) of rpoN2 was amplified by PCR with primers rpoN2w-F/-R and cloned into pBB to obtain pBBrpoN2w. The CDS region was subcloned into the NdeI/XhoI sites of pET30b to generate pETrpoN2.
(iv) Western blotting.
Unless otherwise specified, 10 μg of crude extract prepared from test strains was separated on a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Perkin-Elmer Inc.). After hybridization with the specific polyclonal antisera and alkaline phosphatase-conjugated goat anti-rabbit antibodies (or alkaline phosphatase-conjugated anti-mouse antibodies), membranes were developed with appropriate substrates (Zymed Laboratories Inc.).
Determination of promoter activity.
The upstream promoter-containing regions of the fliC gene were PCR amplified with primers fliCp-F/R and cloned into pFY13-9, a promoter-probing vector using promoterless lacZ as the reporter (36), to form transcriptional fusion constructs. Promoter activity was monitored by measuring the β-galactosidase activities of the flagellar promoter-lacZ fusion constructs in the different strains as previously described (27).
Motility assay.
Motility assays were carried out by inoculating 3 μl (ca. 105 CFU) of mid-log-phase cultures onto the surfaces of a freshly prepared semisolid XOL medium (19) containing 0.3% agar. The subsurface distribution of bacteria, an indication of movement, was observed at 48 h postinoculation.
TEM.
Bacteria were grown in LB medium without agitation to avoid breakage of flagella. Cells were harvested by centrifugation at 4,000 × g for 5 min at 4°C. The cells were washed twice and resuspended in cold deionized water, which was then deposited onto grids coated with Formva (Standard Technology). The grids with the cells were floated on a drop of 2% uranyl acetate for 30 s for staining. After air drying for 15 min, transmission electron microscopy (TEM) characterization was performed in a JEOL JEM-1200CXII electron microscope.
Pathogenicity tests.
Pathogenicity tests were carried out by inoculating the X. campestris pv. campestris cells from overnight cultures onto leaves, which had been cut with a pair of scissors, of 6-week-old potted cabbage plants (77). Statistical results for 15 rounds of independent inoculation experiments are presented.
Yeast two-hybrid assay.
Protein-protein interaction assay was performed by using the Matchmaker III yeast two-hybrid system (Clontech) with a 312-bp fragment of flgM CDS (amplified with primers flgM_YA-F/-R) cloned into the NdeI/BamHI sites of pACT2 to form pACTflgM and a 768-bp fragment of fliA CDS (amplified with primers fliA_YB-F/-R) cloned into the NdeI/BamHI sites of pAS2-1 to form pAS2-fliA. The recombinant and parental plasmids were transformed into yeast strain Y187. Cotransformants were selected on a synthetic complete dropout medium (SD/−Trp/−Leu). Protein interaction was detected by β-galactosidase activity using the colony lift filter approach as described in the Clontech manual.
EPS assay.
Cells of X. campestris pv. campestris were grown in LB or XOLN medium (20) (40 ml in 250-ml flasks) for 72 h. The cells were removed by centrifugation (12,000 × g for 15 min at 4°C), and the exopolysaccharide (EPS) in the culture supernatants was precipitated in the presence of 0.5 M NaCl and 70% ethanol. The amounts of EPS were determined by the anthrone method as described previously (38).
Plate assays for extracellular hydrolytic enzymes.
The ability of the X. campestris pv. campestris strains to secrete extracellular enzymes was tested on LB plates containing skim milk (1%), starch (0.2%), polygalacturonic acid (PGA) (1%), or carboxymethylcellulose (0.5%). The cells (ca. 1× 107 CFU in 10 μl) were deposited on the surface of the plates. After 48 h of incubation, the plates were examined either by direct visualization or by being treated with appropriate reagents. Protease activity was judged by the appearance of clear zones surrounding the colonies on milk plates. In amylase assays, starch plates were stained with iodine (5% in ethanol). PGA plates were developed with 1% hexadecyl-trimethyl-ammonium bromide. The region exhausted of PGA by secreted pectinase displayed a clear zone. Carboxymethylcellulose plates were stained with 0.1% Congo red for 5 min, rinsed once with water, and washed twice with 20 ml of 1.0 M NaCl. Cellulase-positive colonies manifested pale-yellow clear zones against a red background.
Bioinformatic analysis.
The FliA-dependent promoters in X. campestris pv. campestris were predicted using the tools in RSAT (http://rsat.scmbb.ulb.ac.be/rsat/). A weight matrix was generated based on 25 published FliA-dependent promoter sequences (see Table S1 in the supplemental material). A genome-scale Patser search on X. campestris pv. campestris strain ATCC 33913 was performed using the FliA matrix (see Table S2 in the supplemental material), and the 21 best-hit sequences were selected (see Table S3 in the supplemental material).
RESULTS
The X. campestris pv. campestris flhA and flhB genes are essential for motility and flagellar biogenesis.
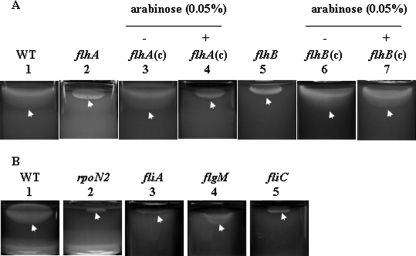
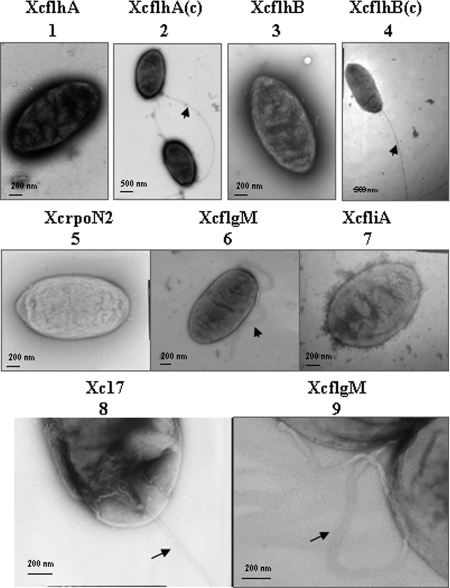
Putative F-T3SS genes flhA (XCC1909) and flhB (XCC1910) were identified in the genome of X. campestris pv. campestris ATCC 33913. Modeling of data using the program TMHMM 2.0 (33) suggested that the N-terminal parts of FlhA and FlhB contained seven and four transmembrane helices, respectively. To study the function of flhA and flhB in flagellum formation, flhA (XcflhA) and flhB (XcflhB) mutants of Xc17, a virulent X. campestris pv. campestris strain exhibiting high sequence homology with X. campestris pv. campestris ATCC 33913, were constructed by insertional mutagenesis. Motility assays were carried out in 0.3% agar medium. The results revealed that both XcflhA and XcflhB were nonmotile (Fig. 1A). Electron micrographs demonstrated that XcflhA and XcflhB lost the typical single, polar flagellum (Fig. 2).
FIG. 1.
Swimming motility assay of Xc17 and Xc17-derived mutant strains. WT, wild-type strain Xc17; flhA(c), flhA mutant complemented with pBBADflhA; flhB(c), flhB mutant complemented with pBBADflhB. For each tube, 3 μl (ca. 105 CFU) mid-log phase culture was inoculated onto the surface of freshly prepared semisolid (0.3% agar) XOL medium and observed after 48 h of incubation.
FIG. 2.
TEM of Xc17 and Xc17-derived mutant strains. 1, flhA mutant; 2, flhA mutant complemented with pBBADflhA; 3, flhB mutant; 4, flhB mutant complemented with pBBADflhB; 5, rpoN2 mutant; 7, fliA mutant; 6 and 9, flgM mutant; and 8, Xc17. The cells of Xc17 and complementary strains of flhA and flhB mutants exhibit a long, straight polar flagellum. Mutations in flhA, flhB, rpoN2, and fliA abolish the flagellation. An flgM mutant has a truncated flagellum with an abnormal structure (9).
To confirm the effect of mutation, plasmids pBBADflhA and pBBADflhB were used to complement XcflhA and XcflhB, respectively. The motility assay demonstrated that the complementary strain XcflhB(c) regained motility in the presence or absence of 0.05% arabinose (Fig. 1A). However, the motility of XcflhA(c) was restored only in a soft-agar medium without arabinose (Fig. 1A). This suggested that the addition of arabinose might induce overexpression of FlhA that impaired the motility. Based on this observation, all tested strains in the following experiments were cultured in media without arabinose. TEM observation showed that mutant strains complemented in trans had a normal flagellar morphology (Fig. 2).
Production of FliC protein is suppressed in flhA and flhB mutants.
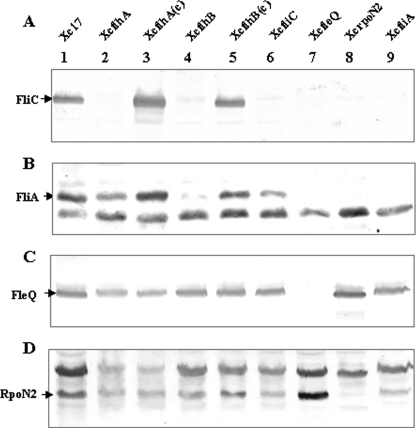
To elucidate the effects of flhA and flhB on the production of flagellin, the flagellin FliC was detected by Western blotting using polyclonal anti-FliC antibodies. A fliC (XCC1941) mutant, XcfliC, was used as a negative control. As shown in Fig. 1B, XcfliC was nonmotile, and it did not have a flagellum (data not shown). The Western blot results showed a band of approximately 40.4 kDa, which corresponded to the FliC protein, in the wild-type strain Xc17 but not in the flhA and flhB mutants (Fig. 3A). Production of FliC protein was restored in XcflhA(c) and XcflhB(c) (Fig. 3A). These results demonstrated that flhA and flhB were required for the synthesis of flagellin FliC.
FIG. 3.
Western blotting, using polyclonal antibodies raised against the flagellin (FliC) (A), σ28 (FliA) (B), activator (FleQ) (C), and σ54 (RpoN2) (D), of whole-cell extracts from X. campestris pv. campestris wild-type Xc17 and Xc17-derived mutant strains. Lane 1, Xc17; lane 2, flhA mutant; lane 3, flhA mutant complemented with pBBADflhA; lane 4, flhB mutant; lane 5, flhB mutant complemented with pBBADflhB; lane 6, fliC mutant; lane 7, fleQ mutant; lane 8, rpoN2 mutant; lane 9, fliA mutant. The positions of FliC, FliA, FleQ, and RpoN2 are indicated.
The rpoN2, fliA, and flgM genes are essential for motility and normal flagellar biogenesis.
Two rpoN homologs encoding σ54 were identified in the genome of X. campestris pv. campestris ATCC 33913. To distinguish these two genes, rpoN (XCC2802), located in a conserved region in most bacteria, was named rpoN1 and the other was named rpoN2 (XCC1935). Genes encoding the σ28 factor (fliA, XCC1906) and the anti-σ28 factor (flgM, XCC1955), as well as rpoN2, were located in a large flagellar gene cluster (27).
To elucidate the function of these genes in flagellation, rpoN2, fliA, and flgM mutants were constructed by insertional mutagenesis. Motility assay revealed that rpoN2 and fliA mutants were nonmotile on soft-agar medium (Fig. 1B). Electron micrographs revealed that rpoN2 and fliA mutants did not have any flagellum present (Fig. 2). A mutation of flgM severely reduced motility (Fig. 1B). TEM observation demonstrated that XcflgM had a short, immature flagellum (Fig. 2). Upon further investigation, we noticed that a normal flagellum had a smooth, long whip-like structure, while in XcflgM, the flagellum was short with an abnormal structural appearance (Fig. 2). These observations suggested that the two sigma factors, σ54 and σ28, were necessary for flagellar biogenesis and motility. The anti-sigma factor FlgM was essential for normal flagellar structure formation and full motility.
Synthesis of FliC is regulated by a cascade of three sigma factors: RpoD, RpoN2, and FliA.
To elucidate the hierarchy of regulation between RpoN2/FleQ and FliA, Western blotting was carried out using antibodies raised against FliA, RpoN2, FleQ, and FliC. FliC protein was absent in fliA, fleQ, and rpoN2 mutants (Fig. 3A). Consequently, FliA, FleQ, and RpoN2 were necessary for the production of FliC. A 28-kDa FliA protein could be detected in the total cell lysates of Xc17 and the fliC mutant XcfliC but was undetectable in the fliA, fleQ, and rpoN2 mutants (Fig. 3B). Therefore, RpoN2 and FleQ were required for the production of FliA. A 51.2-kDa RpoN2 protein and a 54.4 kDa FleQ protein were detected in the cell lysates of Xc17, XcfliC, and XcfliA, while they were absent in the respective mutants (Fig. 3C and D). This suggested that the regulation of the synthesis of both FleQ and RpoN2 was independent of FleQ, RpoN2, FliA, and FliC.
Results from a series of lacZ transcriptional fusion experiments indicated that the upstream regions of fliA and fleN did not have promoter activity (data not shown). Hence, the expression of fliA, fleN, and flhF was dependent on the promoter region upstream of flhF. The transcription levels of fliC and fliA were monitored by β-galactosidase activity of promoter-lacZ fusion constructs pFYfliC and pFYflhF (27) in Xc17 and flagellar mutants at 3-h intervals from an initial optical density at 550 nm of 0.35 (Table 3). The data demonstrated that the expression of fliC in rpoN2, fleQ, and fliA mutants was reduced to 28 to 33, 27 to 30, and 48 to 64%, respectively, of that in Xc17. The promoter activity of the upstream region of flhF decreased to 6 to 14 and 24 to 27% in rpoN2 and fleQ mutants, respectively, but increased to 140 to 150% in a fliA mutant (Table 3). These results suggested that RpoN2, FleQ, and FliA were required for the transcription of fliC. Moreover, the transcription of the flhF promoter was dependent on RpoN2/FleQ and was moderately autoregulated by FliA.
TABLE 3.
β-Galactosidase activities of the fliC promoter-lacZ and flhF promoter-lacZ fusions in Xc17 and flagellar mutants
| Promoter | Time (h) | Mean β-galactosidase activity (Miller units) ± SD in straina
|
||||
|---|---|---|---|---|---|---|
| Xc17 | rpoN2 mutant | fleQ mutant | fliA mutant | flgM mutant | ||
| fliC | 1 | 70.4 ± 2.6 | 19.3 ± 4.7 (28) | 18.8 ± 1.1 (27) | 34.1 ± 5.6 (48) | 168.7 ± 32.8 (240) |
| 4 | 50.1 ± 6.9 | 13.8 ± 2.8 (28) | 14.9 ± 4.6 (30) | 29.9 ± 6.4 (60) | 111.8 ± 8.7 (223) | |
| 7 | 30.6 ± 2.8 | 10.0 ± 1.4 (33) | 8.6 ± 1.7 (28) | 19.6 ± 3.8 (64) | 81.4 ± 6.2 (266) | |
| fliF | 1 | 68.4 ± 7.0 | 9.5 ± 2.9 (14) | 18.0 ± 0.4 (26)b | 102.0 ± 14.2 (149) | 32.4 ± 1.1 (47) |
| 4 | 49.9 ± 5.4 | 2.9 ± 4.4 (6) | 12.2 ± 5.6 (24)b | 69.8 ± 11.2 (140) | 24.5 ± 6.0 (49) | |
| 7 | 30.3 ± 2.9 | 3.4 ± 5.8 (11) | 8.3 ± 2.5 (27)b | 45.6 ± 3.9 (150) | 22.8 ± 3.7 (75) | |
Values in parentheses are percent activity in the mutant strains relative to that in Xc17.
Data are from reference 28.
In conclusion, the synthesis of FliC is under a hierarchical regulation of the two alternative sigma factors RpoN2/FleQ and FliA at the transcriptional level. An RpoN-dependent promoter was identified upstream of flhF-fleN-fliA (27), and a FliA-dependent promoter was identified upstream of fliC (see Table S3 in the supplemental material). According to the promoter type, flagellar genes could be classified into three classes: the first class consisted of RpoD-dependent genes, the second class included the RpoN2/FleQ regulons, and genes in the FliA regulons belonged to the third class.
The Western blot data also showed that expression of RpoN2 and FleQ was not significantly affected by the mutation of flhA or flhB (Fig. 3C and D). FliA production was slightly decreased in the flhA mutant and significantly reduced in the flhB mutant (Fig. 3B). The defect in expression was rescued after complementation in trans by plasmid-borne flhA and flhB genes, respectively (Fig. 3B). This indicated that flhB was involved not only in the regulation of class III genes but also in that of class II genes.
The anti-sigma factor FlgM negatively regulates the production of FliC via an interaction with FliA.
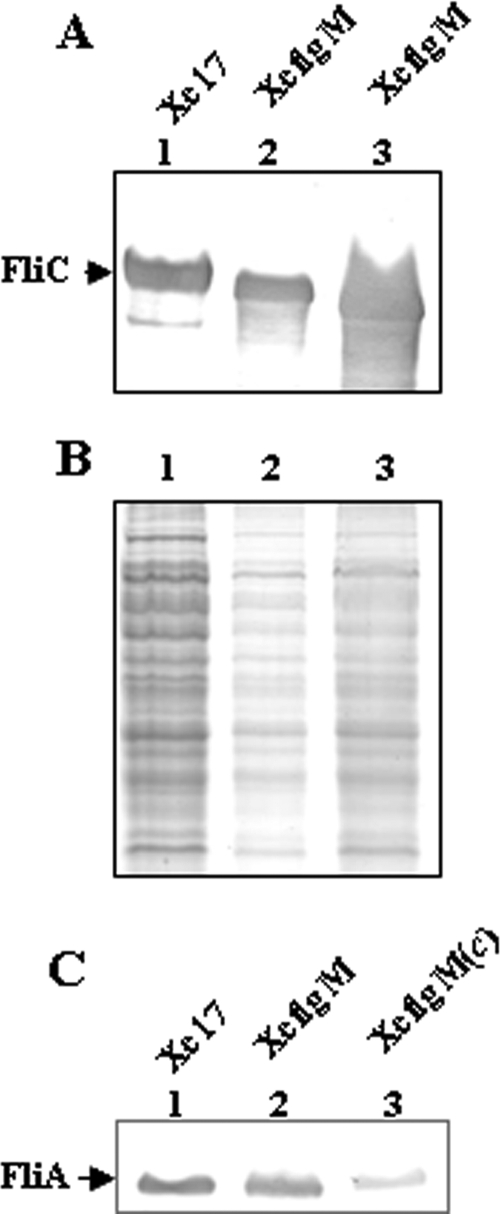
To detect the expression of FliC in XcflgM, different amounts of cell lysate of XcflgM were prepared, separated on an SDS-polyacrylamide gel (Fig. 4B), and then immunoblotted with anti-FliC antiserum (Fig. 4A). The blotting results showed that the FliC protein was overproduced about fivefold in an flgM mutant in comparison with Xc17 (Fig. 4A). The promoter activity of fliC was enhanced to 223 to 266% in XcflgM compared to the parental strain (Table 3), indicating the expression of fliC was negatively regulated by FlgM at the transcriptional level. The quantity of FliA did not change significantly in XcflgM (Fig. 4C), even though the transcription level of the flhF promoter underwent a 25% decrease at the late log phase, compared to Xc17 (Table 3).
FIG. 4.
(B) Different amounts of whole-cell extracts were separated on an SDS-polyacrylamide gel. Lane 1, 10 μg Xc17; lane 2, 2 μg flgM mutant; lane 3, 8 μg flgM mutant. (A) This was followed by Western blotting with polyclonal antibodies raised against the flagellin (FliC). (C) Western blotting, using polyclonal antibodies against σ28 (FliA), of the cell extract. Lane 1, Xc17; lane 2, flgM mutant; lane 3, flgM mutant complemented with pRKflgM.
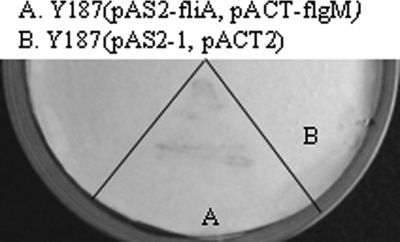
To detect the interaction between FliA and FlgM, the entire coding regions of fliA and flgM were cloned in an in-frame translational fusion with the DNA-binding domain and activating domain of the yeast activator GAL4 in plasmids pAS2-1 and pACT2, respectively, followed by transforming into yeast strain Y187. Cotransformants were selected on a synthetic complete dropout medium without leucine and tryptophan. Yeast clones harboring only one of the recombinant plasmids (pAS2-fliA or pACT-flgM) or the parental plasmids, pAS2-1 and pACT2, were also used as controls. Protein interaction was evaluated by β-galactosidase activity using the colony lift filter approach. As shown in sector A of Fig. 5, the yeast clone cotransformed by the two recombinant plasmids had β-galactosidase activity (blue color), while the clones harboring pAS2-fliA or pACT-flgM (data not shown) or the two parental plasmids (white color; Fig. 5, sector B) displayed insignificant activity. The results suggested possible cross talk between FliA and FlgM.
FIG. 5.
Colony filter lift assay to identify the specificity of interaction between FliA and FlgM. S. cerevisiae Y187 carrying recombinant plasmids pAS2-fliA and pACT-flgM (sector A) and parental plasmids pAS2-1 and pACT2 (sector B) were grown on yeast synthetic dropout medium (−Trp/−Leu) and transferred to a Whatman no. 5 paper filter. Cells were permeabilized by freeze-thaw treatment of the filters. The filters carrying the cells were then placed over filters presoaked with Z-buffer-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution. A blue color revealed within 30 min was considered positive. Assays were repeated three times with at least four independent transformants.
The FlhA mutant has an attenuated virulence.
FlhA and FlhB are F-T3SS proteins. To determine the effects of FlhAB on pathogenicity, a virulence assay was carried out in cabbage leaves by wound inoculation with Xc17 and flhA and flhB mutants. The results showed that XcflhB and the complementary strain XcflhB(c) exhibited the same virulence as the parental strain (15 successful infections/15 inoculations). Conversely, XcflhA produced a low infection rate, i.e., 33%, and the rate increased to 80% in the complementation strain XcflhA(c). The production of important virulence factors, i.e., extracellular enzymes and EPSs, was assayed to determine the cause of the low infection rate. The diameters of digested zones for amylases, cellulases, proteases, and pectinases were about 2.15 to 2.21, 1.67 to 1.84, 1.62 to 1.80, and 0.75 to 0.85 cm, respectively, for Xc17, XcflhA, XcflhB, and their complementary strains (Table 4). The EPS productivity was about 195 to 255 and 2,480 to 3,110 μg/ml for all tested strains grown in LB and XOLN media (Table 4). In conclusion, there was no significant difference in the activities of extracellular enzymes and the productivities of EPS for all tested strains.
TABLE 4.
Activities of extracellular enzymes and production of EPS in the flhA and flhB mutants, their complementary strains, and Xc17
| Strain | Mean diameter (cm) of digestive zone ± SD for extracellular enzymea
|
Mean EPS production (μg/ml) ± SD in medium
|
||||
|---|---|---|---|---|---|---|
| Amylase | Celluloses | Pectinase | Protease | LB | XOLN | |
| Xc17 | 2.21 ± 0.59 (100) | 1.84 ± 0.27 (100) | 0.85 ± 0.01 (100) | 1.62 ± 0.08 (100) | 252 ± 51 | 2,914 ± 24 |
| XcflhA | 2.17 ± 0.61 (98) | 1.76 ± 0.31 (95) | 0.77 ± 0.11 (91) | 1.80 ± 0.08 (110) | 230 ± 7 | 2,968 ± 34 |
| XcflhA(c) | 2.17 ± 0.57 (98) | 1.70 ± 0.31 (92) | 0.76 ± 0.15 (89) | 1.72 ± 0.13 (106) | 233 ± 36 | 3,110 ± 48 |
| XcflhB | 2.16 ± 0.62 (98) | 1.74 ± 0.33 (94) | 0.83 ± 0.13 (97) | 1.74 ± 0.08 (107) | 255 ± 15 | 2,480 ± 75 |
| XcflhB(c) | 2.15 ± 0.64 (97) | 1.74 ± 0.33 (94) | 0.84 ± 0.12 (99) | 1.64 ± 0.02 (101) | 195 ± 8 | 2,597 ± 51 |
Diameters of clearing zones were measured in plates containing the appropriate substrate. Values in parentheses are percent diameter for the mutant strains relative to that for Xc17.
DISCUSSION
The regulation of flagellar biogenesis in X. campestris pv. campestris is a simplified form of that in vibrios/pseudomonads.
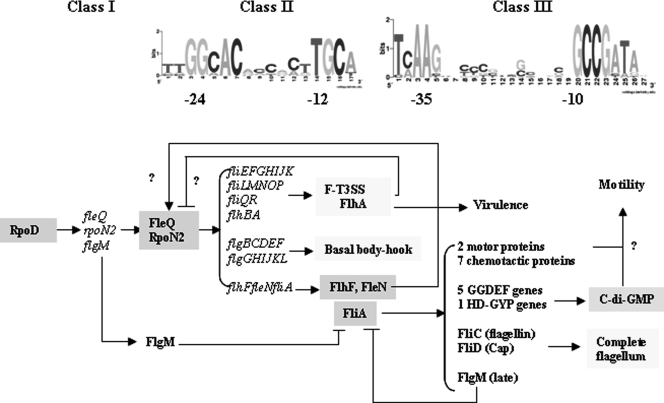
The biogenesis of the bacterial flagellum is a very complex process involving the temporal and spatial coordination of gene expression and protein assembly. As shown in this work, the expression of the three classes of flagellar promoters of X. campestris pv. campestris is dependent on σ70, σ54, and σ28, respectively, as commonly seen in vibrios and pseudomonads (14, 60). Nevertheless, two-tier RpoN-cognate activators, FlrA/FleQ and FlrBC/FleSR, are involved in flagellar gene expression in vibrios/pseudomonads (14, 60), whereas in X. campestris pv. campestris, FleQ appears to be the sole RpoN regulator involved in the flagellation (26). A similar model has been reported for Helicobacter pylori, except that only the FleSR-type regulator was identified (29, 57). A proposed model of the flagellar regulation pathway in X. campestris pv. campestris is illustrated in Fig. 6.
FIG. 6.
Model of the flagellar transcriptional cascade in X. campestris pv. campestris. Flagellar genes can be placed in three temporal classes according to the type of promoter recognized by the different sigma factors. Class I proteins RpoN2 (σ54) and the cognate activator FleQ are the master regulator directing the expression of class II genes. Early FlgM transcribed from a class I promoter inhibits class II sigma factor FliA until completion of the production of class II proteins and assembly of the F-T3SS-basal body-hook structure. Late (class III) genes belong to the FliA (σ28) regulons. Flagellar filament, cap proteins, and motility regulatory proteins are identified. Late FlgM acts as a final brake to the whole process. The promoter consensus based on the type II and type III flagellar genes of X. campestris pv. campestris is indicated at the top.
The early flagellar genes are RpoN2/FleQ dependent.
The rpoN gene, encoding the σ54 factor, is widely present in the microbial genome. RpoN is involved not only in nitrogen assimilation but also in many unrelated functions, such as utilization of carbon sources, nitrogen fixation, motility, alginate biosynthesis, and virulence, in different species (45, 61, 67). Most bacteria have only one rpoN gene, yet exceptions have been found in some nitrogen fixation bacteria and plant pathogens. The functions of the rpoN homologs in the same genome could be redundant or irrelevant (34, 46, 59). An allelic-exchange mutation in rpoN2 totally abolishes flagellar biogenesis and motility. This suggests that the two rpoN genes in X. campestris pv. campestris work independently. Further study is necessary to determine the mechanism of the specificity of the two RpoN proteins. Our previous data have characterized five RpoN/FleQ-dependent promoters encoding three types of proteins (Fig. 6): (i) regulators (FliA, FlhF, and FleN); (ii) secretion system (fliEFGHIJK, fliLMNOP, and fliQR); and (iii) rod and hook (flgBCDEF and flgGHIJKL). In Pseudomonas aeruginosa, FlhF and FleN play a role in the early stage of flagellation by regulating the localization and number of flagella (15, 55). A putative RpoN-dependent GG-N10-GC promoter (aGGaacaccacttGCa) is identified upstream of flhBA, indicating that it may also belong to the class II genes (Fig. 6).
Late flagellar genes and some motility/chemotaxis and c-di-GMP-signaling-related genes belong to the FliA regulons.
FliA is a σ70 family protein that recognizes a −35/−10-type promoter sequence. Twenty-one putative σ28-dependent genes are predicted in this work (see Table S3 in the supplemental material). Most are highly related to the late stage of flagellar biogenesis (such as flgM, fliC, and fliD) and to motility/chemotaxis (including XCC3653, XCC1891, XCC1727, XCC1870, XCC1871, XCC1883, XCC2315, and XCC3522). The most interesting finding is that five GGDEF domain-containing proteins (XCC0407, XCC1443, XCC1777, XCC3519, and XCC3546) and one HD-GYP domain-containing protein (XCC0350) are likely regulated by FliA. Both the GGDEF and HD-GYP domains are related to the synthesis and breakdown of an important bacterial intracellular secondary messenger, 3′,5′-cyclic diguanylic acid (c-di-GMP) (4, 62, 63). Recently, c-di-GMP has been shown to be involved in EPS production, biofilm formation, quorum sensing, virulence, and flagellar mobility (6, 37, 56, 75). c-di-GMP may be involved in the sigma factor regulatory pathways. In a pathogenic E. coli strain, FliA regulates adhesion and invasion via a c-di-GMP-dependent pathway (11). c-di-GMP is also involved in the signaling pathway of the σS network (73). In the X. campestris pv. campestris genome, 29 proteins with a GGDEF motif and three proteins containing an HD-GYP domain have been identified, and, more importantly, some are involved in motility (64). More experiments are required to evaluate whether the expression of c-di-GMP-related genes is FliA dependent and to elucidate its roles in flagellar biogenesis and motility.
F-T3SS structural proteins FlhB and FlhA are required for the production of FliA and for virulence.
FlhA and FlhB are two F-T3SS structural proteins. It is not surprising that the defect in F-T3SS formation results in an accumulation of FlgM and represses expression from a FliA-dependent promoter. The dramatic decrease of FliA protein in an flhB mutant suggests that FlhB might play a role in the expression and/or the stability of FliA. The effect of FlhA (and FlhB) on class one or two promoters has been reported for some bacteria. In Proteus mirabilis, an flhA mutant expresses 10-fold-lower levels of flhCD (class 2) transcripts (20). In H. pylori, FlhA acts as a master regulator and positively regulates transcription in the RpoN (class 2) and FliA (class 3) regulons (57). In Campylobacter jejuni, mutation in either flhA or flhB reduces the transcription from σ54-dependent flagellar genes (26).
Two classes of T3SS have been identified in gram-negative bacteria (13, 22). Besides F-T3SS, the nonflagellar T3SS (NF-T3SS) is involved in the delivery of protein effectors across eukaryotic cytoplasm to promote pathogenesis (12, 42). F- and NF-T3SS share many structural and functional features, and nine components of these two classes are homologous in sequence (13, 70). Furthermore, functional coordination between F- and NF-T3SS has been reported for some species. In Yersinia enterocolitica, the virulence-associated phospholipase YpIA can be secreted via F- and NF-T3SS (79, 80). In C. jejuni, FlhA coordinately regulates the bacterial motility and virulence (9, 30), and many virulence factors are secreted via F-T3SS (24). In Bacillus thuringiensis, an flhA mutation results in a defect in flagellar biogenesis, motility, virulence factors secretion, and cytotoxicity (7, 21). Our work demonstrated that the flhA mutant has an attenuated virulence, but the production of extracellular enzymes and EPS is not influenced. FlhA is highly homologous (30%) to HrcV (XCC1229), a protein of NF-T3SS. Since the nonmotile strain XcflhB exhibits a normal infection rate, F-T3SS and motility of X. campestris pv. campestris are not necessary for pathogenesis. Thus, further experiments will be necessary to determine whether FlhA participates in pathogenicity via NF-T3SS.
Supplementary Material
Acknowledgments
We thank Andy Hsieh for taking the time to carefully read and edit this paper.
This study was supported by grants NSC 96- and 97-2317-B-468-001 from the National Science Council, Republic of China, to R.-M.H.
Footnotes
Published ahead of print on 9 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aldridge, P., J. Karlinsey, and K. T. Hughes. 2003. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol. Microbiol. 491333-1345. [DOI] [PubMed] [Google Scholar]
- 2.Al Mamun, A. A., A. Tominaga, and M. Enomoto. 1996. Detection and characterization of the flagellar master operon in the four Shigella subgroups. J. Bacteriol. 1783722-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apel, D., and M. G. Surette. 2008. Bringing order to a complex molecular machine: the assembly of the bacterial flagella. Biochim. Biophys. Acta 17781851-1858. [DOI] [PubMed] [Google Scholar]
- 4.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204163-167. [DOI] [PubMed] [Google Scholar]
- 5.Barembruch, C., and R. Hengge. 2007. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 6576-89. [DOI] [PubMed] [Google Scholar]
- 6.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2007. Regulation of biofilm formation in Yersinia pestis. Adv. Exp. Med. Biol. 603201-210. [DOI] [PubMed] [Google Scholar]
- 7.Bouillaut, L., N. Ramarao, C. Buisson, N. Gilois, M. Gohar, D. Lereclus, and C. Nielsen-Leroux. 2005. FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl. Environ. Microbiol. 718903-8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brutinel, E. D., and T. L. Yahr. 2008. Control of gene expression by type III secretory activity. Curr. Opin. Microbiol. 11128-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo, C. D., E. Taboada, J. H. Nash, P. Lanthier, J. Kelly, P. C. Lau, R. Verhulp, O. Mykytczuk, J. Sy, W. A. Findlay, K. Amoako, S. Gomis, P. Willson, J. W. Austin, A. Potter, L. Babiuk, B. Allan, and C. M. Szymanski. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 27920327-20338. [DOI] [PubMed] [Google Scholar]
- 10.Chevance, F. F., and K. T. Hughes. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claret, L., S. Miquel, N. Vieille, D. A. Ryjenkov, M. Gomelsky, and A. Darfeuille-Michaud. 2007. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J. Biol. Chem. 28233275-33283. [DOI] [PubMed] [Google Scholar]
- 12.Coburn, B., I. Sekirov, and B. B. Finlay. 2007. Type III secretion systems and disease. Clin. Microbiol. Rev. 20535-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4811-825. [DOI] [PubMed] [Google Scholar]
- 14.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. [DOI] [PubMed] [Google Scholar]
- 15.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutton, R. J., Z. Xu, and J. W. Gober. 2005. Linking structural assembly to gene expression: a novel mechanism for regulating the activity of a sigma54 transcription factor. Mol. Microbiol. 58743-757. [DOI] [PubMed] [Google Scholar]
- 17.Ferris, H. U., and T. Minamino. 2006. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 14519-526. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, G. M., T. Hirano, H. U. Ferris, L. L. Devgan, M. Kihara, and R. M. Macnab. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 481043-1057. [DOI] [PubMed] [Google Scholar]
- 19.Fu, J. F., and Y. H. Tseng. 1990. Construction of lactose-utilizing Xanthomonas campestris and production of xanthan gum from whey. Appl. Environ. Microbiol. 56919-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furness, R. B., G. M. Fraser, N. A. Hay, and C. Hughes. 1997. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J. Bacteriol. 1795585-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghelardi, E., F. Celandroni, S. Salvetti, D. J. Beecher, M. Gominet, D. Lereclus, A. C. Wong, and S. Senesi. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 1846424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grunenfelder, B., S. Gehrig, and U. Jenal. 2003. Role of the cytoplasmic C terminus of the FliF motor protein in flagellar assembly and rotation. J. Bacteriol. 1851624-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerry, P. 2007. Campylobacter flagella: not just for motility. Trends Microbiol. 15456-461. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 26.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50687-702. [DOI] [PubMed] [Google Scholar]
- 27.Hu, R. M., T. C. Yang, S. H. Yang, and Y. H. Tseng. 2005. Deduction of upstream sequences of Xanthomonas campestris flagellar genes responding to transcription activation by FleQ. Biochem. Biophys. Res. Commun. 3351035-1043. [DOI] [PubMed] [Google Scholar]
- 28.Imada, K., T. Minamino, A. Tahara, and K. Namba. 2007. Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proc. Natl. Acad. Sci. USA 104485-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43307-322. [DOI] [PubMed] [Google Scholar]
- 30.Kalmokoff, M., P. Lanthier, T. L. Tremblay, M. Foss, P. C. Lau, G. Sanders, J. Austin, J. Kelly, and C. M. Szymanski. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 1884312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 32.Kihara, M., T. Minamino, S. Yamaguchi, and R. M. Macnab. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 1831655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 34.Kullik, I., S. Fritsche, H. Knobel, J. Sanjuan, H. Hennecke, and H. M. Fischer. 1991. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the sigma 54 gene (rpoN). J. Bacteriol. 1731125-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kutsukake, K., and T. Iino. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 1763598-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, M. C., S. F. Weng, and Y. H. Tseng. 2003. Flagellin gene fliC of Xanthomonas campestris is upregulated by transcription factor Clp. Biochem. Biophys. Res. Commun. 307647-652. [DOI] [PubMed] [Google Scholar]
- 37.Lee, V. T., J. M. Matewish, J. L. Kessler, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 651474-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, H. M., and Y. H. Tseng. 1979. Exopolysaccharide synthesis in Xanthomonas oryzae. Proc. Natl. Sci. Council ROC 3279-284. [Google Scholar]
- 39.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 1767345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowder, B. J., M. D. Duyvesteyn, and D. F. Blair. 2005. FliG subunit arrangement in the flagellar rotor probed by targeted cross-linking. J. Bacteriol. 1875640-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 5777-100. [DOI] [PubMed] [Google Scholar]
- 42.McCann, H. C., and D. S. Guttman. 2008. Evolution of the type III secretion system and its effectors in plant-microbe interactions. New Phytol. 17733-47. [DOI] [PubMed] [Google Scholar]
- 43.McCarter, L. L. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9180-186. [DOI] [PubMed] [Google Scholar]
- 44.McMurry, J. L., J. S. Van Arnam, M. Kihara, and R. M. Macnab. 2004. Analysis of the cytoplasmic domains of Salmonella FlhA and interactions with components of the flagellar export machinery. J. Bacteriol. 1867586-7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N). Mol. Microbiol. 10903-909. [DOI] [PubMed] [Google Scholar]
- 46.Michiels, J., M. Moris, B. Dombrecht, C. Verreth, and J. Vanderleyden. 1998. Differential regulation of Rhizobium etli rpoN2 gene expression during symbiosis and free-living growth. J. Bacteriol. 1803620-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Minamino, T., and K. Namba. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451485-488. [DOI] [PubMed] [Google Scholar]
- 49.Minamino, T., and R. M. Macnab. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 1824906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minamino, T., and R. M. Macnab. 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 371494-1503. [DOI] [PubMed] [Google Scholar]
- 51.Moriya, N., T. Minamino, K. T. Hughes, R. M. Macnab, and K. Namba. 2006. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J. Mol. Biol. 359466-477. [DOI] [PubMed] [Google Scholar]
- 52.Muir, R. E., and J. W. Gober. 2001. Regulation of late flagellar gene transcription and cell division by flagellum assembly in Caulobacter crescentus. Mol. Microbiol. 41117-130. [DOI] [PubMed] [Google Scholar]
- 53.Muir, R. E., and J. W. Gober. 2004. Regulation of FlbD activity by flagellum assembly is accomplished through direct interaction with the trans-acting factor, FliX. Mol. Microbiol. 54715-730. [DOI] [PubMed] [Google Scholar]
- 54.Muir, R. E., J. Easter, and J. W. Gober. 2005. The trans-acting flagellar regulatory proteins, FliX and FlbD, play a central role in linking flagellar biogenesis and cytokinesis in Caulobacter crescentus. Microbiology 1513699-3711. [DOI] [PubMed] [Google Scholar]
- 55.Murray, T. S., and B. I. Kazmierczak. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 1886995-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakhamchik, A., C. Wilde, and D. A. Rowe-Magnus. 2008. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl. Environ. Microbiol. 744199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52947-961. [DOI] [PubMed] [Google Scholar]
- 58.Paul, K., M. Erhardt, T. Hirano, D. F. Blair, and K. T. Hughes. 2008. Energy source of flagellar type III secretion. Nature 451489-492. [DOI] [PubMed] [Google Scholar]
- 59.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2002. The four different sigma(54) factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol. Microbiol. 4675-85. [DOI] [PubMed] [Google Scholar]
- 60.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 391595-1609. [DOI] [PubMed] [Google Scholar]
- 61.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9218-228. [DOI] [PubMed] [Google Scholar]
- 63.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Ryan, R. P., Y. Fouhy, J. F. Lucey, B. L. Jiang, Y. Q. He, J. X. Feng, J. L. Tang, and J. M. Dow. 2007. Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol. Microbiol. 63429-442. [DOI] [PubMed] [Google Scholar]
- 65.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 66.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 15831-834. [PubMed] [Google Scholar]
- 67.Studholme, D. J., and M. Buck. 2000. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol. Lett. 1861-9. [DOI] [PubMed] [Google Scholar]
- 68.Sukchawalit, R., P. Vattanaviboon, R. Sallabhan, and S. Mongkolsuk. 1999. Construction and characterization of regulated l-arabinose-inducible broad host range expression vectors in Xanthomonas. FEMS Microbiol. Lett. 181217-223. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki, H., K. Yonekura, K. Murata, T. Hirai, K. Oosawa, and K. Namba. 1998. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in type III protein export. J. Struct. Biol. 124104-114. [DOI] [PubMed] [Google Scholar]
- 70.Tampakaki, A. P., V. E. Fadouloglou, A. D. Gazi, N. J. Panopoulos, and M. Kokkinidis. 2004. Conserved features of type III secretion. Cell. Microbiol. 6805-816. [DOI] [PubMed] [Google Scholar]
- 71.Thomas, J., G. P. Stafford, and C. Hughes. 2004. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. USA 1013945-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100189-194. [DOI] [PubMed] [Google Scholar]
- 73.Weber. H., C. Pesavento, A. Possling, G. Tischendorf, and R. Hengge. 2006. Cyclic-di-GMP-mediated signalling within the sigma(S) network of Escherichia coli. Mol. Microbiol. 621014-1034. [DOI] [PubMed] [Google Scholar]
- 74.Williams, P. H. 1980. Black rot: a continuing threat to world crucifers. Plant Dis. 64736-742. [Google Scholar]
- 75.Wolfe, A. J., and K. L. Visick. 2008. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 190463-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamamoto, S., and K. Kutsukake. 2006. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1886703-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, B. Y., and Y. H. Tseng. 1988. Production of exopolysaccharide and levels of protease and pectinase activity in pathogenic and non-pathogenic strains of Xanthomonas campestris pv. campestris. Bot. Bull. Acad. Sin. 2993-99. [Google Scholar]
- 78.Yang, B. Y., H. F. Tsai, and Y. H. Tseng. 1988. Broad host range cosmid pLAFR1 and non-mucoid mutant XCP20 provide a suitable vector-host system for cloning genes in Xanthomonas campestris pv. campestris. Chin. J. Microbiol. Immunol. 2140-49. [PubMed] [Google Scholar]
- 79.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 1841324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 966456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.