Abstract
Mammalian reoviruses are nonenveloped particles containing a genome of 10 double-stranded RNA (dsRNA) gene segments. Reovirus replication occurs within viral inclusions, which are specialized nonmembranous cytoplasmic organelles formed by viral nonstructural and structural proteins. Although these structures serve as sites for several major events in the reovirus life cycle, including dsRNA synthesis, gene segment assortment, and genome encapsidation, biochemical mechanisms of virion morphogenesis within inclusions have not been elucidated because much remains unknown about inclusion anatomy and functional organization. To better understand how inclusions support viral replication, we have used RNA interference (RNAi) and reverse genetics to define functional domains in two inclusion-associated proteins, μNS and μ2, which are interacting partners essential for inclusion development and viral replication. Removal of μNS N-terminal sequences required for association with μ2 or another μNS-binding protein, σNS, prevented the capacity of μNS to support viral replication without affecting inclusion formation, indicating that μNS-μ2 and μNS-σNS interactions are necessary for inclusion function but not establishment. In contrast, introduction of changes into the μNS C-terminal region, including sequences that form a putative oligomerization domain, precluded inclusion formation as well as viral replication. Mutational analysis of μ2 revealed a critical dependence of viral replication on an intact nucleotide/RNA triphosphatase domain and an N-terminal cluster of basic amino acid residues conforming to a nuclear localization motif. Another domain in μ2 governs the capacity of viral inclusions to affiliate with microtubules and thereby modulates inclusion morphology, either globular or filamentous. However, viral variants altered in inclusion morphology displayed equivalent replication efficiency. These studies reveal a modular functional organization of inclusion proteins μNS and μ2, define the importance of specific amino acid sequences and motifs in these proteins for viral replication, and demonstrate the utility of complementary RNAi-based and reverse genetic approaches for studies of reovirus replication proteins.
Mammalian reoviruses are the type species of the genus Orthoreovirus, within the Reoviridae family, which includes important human and veterinary pathogens, such as rotavirus and bluetongue virus, respectively. Reoviruses serve as highly tractable experimental models for studies of viral replication and pathogenesis (51). Reovirus virions are nonenveloped, double-shelled particles that display icosahedral symmetry and contain 10 segments of double-stranded RNA (dsRNA). Following internalization of virions by receptor-mediated endocytosis, the viral outer capsid undergoes acid-dependent proteolysis within endosomes to generate core particles containing all components of the viral transcriptional machinery (3, 18, 58). Transcriptionally active core particles released into the cytoplasm synthesize full-length, message-sense, single-stranded RNAs (ssRNAs) corresponding to each viral gene segment (4, 14). These ssRNAs are competent for translation and serve as templates for minus-strand synthesis to generate nascent genomic dsRNA (33, 52, 53). Synthesis of the complementary strand appears to be concomitant with assortment of the 10 genome segments into progeny particles (1). The viral replication cycle is completed by condensation of outer capsid proteins onto newly formed dsRNA-containing particles, producing fully assembled infectious progeny (70).
Reovirus replication and assembly are thought to occur within viral inclusions that form in the cytoplasm of infected cells (19). Viral inclusions contain dsRNA (56), viral proteins (19), and both complete and incomplete virion particles (19). The viral nonstructural proteins μNS and σNS and minor core protein μ2 are collectively required for the genesis and maturation of viral inclusions in reovirus-infected cells (2, 28). Expression of these proteins from cloned cDNAs in the absence of infection results in spontaneous assembly of intracytoplasmic structures exhibiting inclusion morphology (6, 39). Virus-like inclusions recruit constituent proteins of the viral core (10), indicating that viral inclusion-forming proteins create an organizing center for the ultimate development of a functional replication matrix. However, the precise role of viral inclusions in the reovirus replication pathway, including dsRNA synthesis, gene segment assortment, genome packaging, and virion assembly, is poorly understood, owing largely to limited mechanistic insight into the individual and corporate functions of inclusion-forming proteins.
The ∼80-kDa μNS protein, encoded by the M3 genome segment, is 721 amino acid residues in length (38, 41). The μNS protein associates with viral mRNA (1), core protein μ2 (12), and nonstructural protein σNS (6, 39). Transiently expressed μNS, in the absence of other viral proteins, forms inclusion-like structures that are similar in appearance and localization to the globular inclusions observed in cells infected with prototype strain type 3 Dearing (T3D) (12). In addition, μNS associates with intact viral cores and recruits core proteins λ1, λ2, λ3, and σ2 into viral inclusion-like structures in transient-transfection assays (10, 12). Interactions of μNS with cores prevent assembly of the virion outer capsid and prolong transcription (11), which may benefit viral replication at earlier stages of infection by maximizing RNA and protein production. Thus, μNS protein plays a fundamental role in viral inclusion formation and provides a protected environment in which RNA replication, assortment, and packaging converge in particle assembly.
A truncated isoform of μNS naturally produced during reovirus infection, known as μNSC (32), lacks the 40 N-terminal residues and has been proposed to result from initiation of translation at an alternative in-frame AUG codon in M3 mRNA (64). The N-terminal 40 residues of μNS contain interacting domains for μ2 and σNS (12, 39), neither of which associates with μNSC (12, 39). Using RNA interference (RNAi)-based trans-complementation approaches, μNSC was found to be incapable of supporting viral growth in μNS-silenced cells (2, 28), pointing to the significance of interactions between μNS, μ2, and σNS for viral replication. However, the concerted activities of μNS and other replication proteins within inclusions have not been defined in structural or functional terms.
The approximately 83-kDa μ2 protein, encoded by the M1 genome segment, is 736 amino acid residues in length and forms a structurally minor component of the reovirus core (17, 38, 41). The μ2 protein binds ssRNA and dsRNA (8) and demonstrates both nucleoside triphosphatase (NTPase) and RNA 5′-triphosphatase (RTPase) activities (26). Reassortant studies suggest that μ2 determines viral strain differences in transcriptional efficiencies of core particles (66). Phenotypes associated with a temperature-sensitive lesion in μ2 indicate that it also participates in RNA replication and particle assembly (16).
The μ2 protein binds to cellular microtubules and determines virus strain-specific differences in inclusion morphology, either globular or filamentous, in infected cells (45, 67). Sequence analysis of M1 segments belonging to independent viral isolates has revealed a correlation between inclusion morphology and a single amino acid polymorphism at position 208 (45). Virus strains that produce filamentous inclusions, such as prototype strains type 1 Lang (T1L) and type 2 Jones (T2J), contain Pro208, whereas strain T3D, which produces globular inclusions, contains Ser208 (45, 67). Inclusion-like structures arising from coexpression of plasmid-encoded μNS and μ2 in the absence of viral infection adopt a morphology predicted by the amino acid residue at position 208, independent of the μ2 strain origin (45). The presence of Pro at position 208 in μ2 is genetically linked to more efficient μ2-microtubule association and increased microtubule stabilization in reovirus-infected cells (45). In contrast, the μ2 proteins of strains encoding a Ser at position 208 are more prone to temperature-dependent misfolding that correlates with reduced efficiency of microtubule binding (39). Taken together, these findings indicate that amino acid position 208 of μ2 determines inclusion morphology by influencing its capacity to assume a conformation commensurate with microtubule binding and stabilization.
In studies reported here, we extended application of RNAi-based replication complementation and plasmid-based reverse genetics to define sequences and functional domains in μNS and μ2 proteins involved in viral replication. Our findings demonstrate the importance of extreme amino-terminal sequences in μNS that interact with μ2 and σNS proteins and sequences in the C-terminal one-half of μNS that mediate inclusion formation. Furthermore, these studies reveal the significance of μ2 sequences that specify or predict NTPase and RTPase activities, constitute a predicted nuclear localization motif, and determine inclusion morphology. An understanding of μNS and μ2 structure-function relationships gained through these studies will provide new insights into the genesis of viral inclusions and their role in reovirus replication.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Murine L929 (L) cells were grown in Joklik's modified Eagle's minimal essential medium (Irvine Scientific, Santa Ana, CA) supplemented to contain 5% fetal calf serum (Gibco-BRL, Gaithersburg, MD), 2 mM l-glutamine, 100 U of penicillin G/ml, 100 μg of streptomycin/ml, and 0.25 μg of amphotericin B/ml (Gibco-BRL). Human embryonic kidney 293T cells stably expressing reovirus-specific short hairpin RNAs (shRNAs) were maintained as described previously (28).
Reovirus prototype strains T1L and T3D are laboratory stocks. Recombinant strain (rs) T3D is a stock rescued by plasmid-based reverse genetics from cloned T3D cDNAs (27). Viral titers were determined by plaque assay using L-cell monolayers (60). Attenuated vaccinia virus strain rDIs-T7pol expressing T7 RNA polymerase was propagated in chicken embryo fibroblasts (25).
The μ1/μ1C-specific monoclonal antibody 8H6 (61) and antisera to μNS (28) and μ2 (68) proteins have been described previously.
Plasmid construction.
The construction of μNS expression vectors pM3 (encoding full-length μNS) and pM3(41-721) (encoding μNS amino acids 41 to 721), previously referred to as pcM31861m and pcM3dN138, respectively, has been described elsewhere (28). The expression vectors pM3(14-721) (encoding μNS amino acids 14 to 721), pM3(Δ14-40) (encoding μNS amino acids 1 to 13 and 41 to 721), pM3(1-675) (encoding μNS amino acids 1 to 675), and pM3(1-716) (encoding μNS amino acids 1 to 716) were generated by inserting PCR amplicons derived from pM3 into the KpnI-XhoI site of pcDNA3 (Invitrogen, Carlsbad, CA). The μNS expression vector pH570Q, H572S, containing His-to-Gln and His-to-Ser amino acid substitutions at positions 570 and 572, respectively, was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with specific primers and pM3 as template. Plasmid vector pT7-M3T3DM41I, which contains an ATG-to-ATC modification of the μNSC translation initiation codon, was generated using QuikChange with pT7-M3T3D template (27) and specific primers. This plasmid was used to recover the μNSC-negative virus rsT3D-μNSC-null.
The μ2 expression vector, pM1T1L, in which the complete T1L μ2 open reading frame is cloned into pcDNA3, was generated using a T1L M1 cDNA originally obtained from Earl Brown (68). The expression vectors containing single, double, or triple substitutions in the T1L μ2 protein—pK415A, pG416A, pK419A, pD446A, pG449A, pR100G,R101G, pRKR103-5GQG, pL106QL108Q, and pK109Q K110Q—were generated using QuikChange, specific primers, and pM1T1L template. The rescue vector pT7-M1T1L, encoding the entire T1L M1 gene, was constructed by inserting an M1 cDNA amplicon fused at its native 5′ terminus to the T7 RNA polymerase promoter into the SmaI-RsrII site of p3E5EGFP (62), resulting in complete replacement of sequences encoding green fluorescent protein (GFP) and the Ebola virus leader and trailer, thereby ligating the native M1 3′ terminus to the HDV ribozyme sequence. Two amino acid differences in the cDNA obtained from E. G. Brown (68), Phe302 and Leu383, compared to the published T1L M1 sequence (45) were changed to match the published sequence (Phe302 to Leu and Leu383 to Pro) using QuikChange. QuikChange also was used to generate constructs for rescue of the following T1L and T3D μ2 mutant viruses (summarized in Table 1): pT7-M1T1LP208S (pT7-M1T1L template), pT7-M1T1LL383P (pT7-M1T1L template), pT7-M1T1LF302L (pT7-M1T1L template), pT7-M1T3DS208P (pT7-M1T3D template) (27), and pT7-M1T3DS208P+L383P (pT7-M1T3D template) (27).
TABLE 1.
Reovirus strains used for studies of viral inclusion morphology
| Virus strain | M1 gene derivation | Amino acid at μ2 position:
|
||
|---|---|---|---|---|
| 208 | 302 | 383 | ||
| T1L | T1L | Pro | Phe | Leu |
| T3D | T3D | Ser | Phe | Leu |
| rsT3D | T3D | Ser | Phe | Leu |
| rsT3D-μ2S208P | T3D | Pro | Phe | Leu |
| rsT3D-μ2S208P+L383P | T3D | Pro | Phe | Pro |
| rsT3D-T1Lμ2 | T1L | Pro | Phe | Leu |
| rsT3D-T1Lμ2P208S | T1L | Ser | Phe | Leu |
| rsT3D-T1Lμ2F302L | T1L | Pro | Leu | Leu |
| rsT3D-T1Lμ2L383P | T1L | Pro | Phe | Pro |
Expression constructs encoding μ2 protein mutagenized within the N-terminal region were generated using pM1T1L template, specific primers, and either QuikChange (substitution mutants) or PCR (N-terminal deletion mutant).
Expression plasmid pGFP-μ2(99-110), encoding GFP fused C-terminally to μ2 amino acid residues 99 to 110, was engineered by cloning oligonucleotides specifying D99RRLRKRLMLKK110 into the XhoI-EcoRI site of pEGFP-C2 (Clontech, Mountain View, CA). Plasmid pGFP-μ2(99-110m), containing Ala substitutions of μ2 basic amino acids in pGFP-μ2(99-110), was generated using oligonucleotides encoding D99AALAAALMLAA110.
All mutations were confirmed by nucleotide sequence analysis. Primer sequences used for plasmid construction are available upon request.
Transient protein expression.
293T cells and L cells were transfected using Lipofectamine 2000 (Invitrogen) and TransIT-LT1 (Mirus, Madison, WI) transfection reagents, respectively, according to the manufacturers' instructions. To detect expressed proteins by immunofluorescence, cells grown on glass coverslips (Fisher Scientific, Pittsburgh, PA) were sequentially transfected with plasmid vectors, incubated for 24 h, fixed in 10% formalin, permeabilized with 1% Triton X-100, and incubated with antisera specific for viral proteins, a 1:1,000 dilution of Alexa Fluor anti-immunoglobulin G secondary antibody (Molecular Probes, Eugene, OR), and TO-PRO3 (Molecular Probes). GFP fusion proteins were detected by intrinsic GFP fluorescence. Cells were visualized using an inverted LSM510 confocal microscope (Carl Zeiss, New York, NY).
To detect expressed proteins by immunoblotting, lysates from transfected cells were prepared as described previously (28). Proteins were detected using enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ) following incubation with μNS- and μ2-specific antisera and appropriate secondary antibodies.
Infection with native and recombinant reovirus strains.
Monolayers of L cells were infected with various reovirus strains at a multiplicity of infection (MOI) of 2 PFU/cell. Following 1 h of adsorption, cells were washed with phosphate-buffered saline (PBS) to remove the inoculum, and fresh medium was added. Cultures were harvested at various intervals for virus titration, immunofluorescence assay, and immunoblotting.
LMB treatment.
Monolayers of L cells were adsorbed with rsT1L at an MOI of 10 (culture plates) or 20 (glass coverslips) PFU/cell. Following 1 h of adsorption, cells were washed with PBS to remove the inoculum, and fresh medium was added. At 8 h postinfection, growth medium was replaced with fresh medium containing 20 μg/ml leptomycin B (LMB), and cultures were processed 4 h later for plaque assay or confocal immunofluorescence microscopy.
Viability of uninfected and reovirus-infected cells in the presence of LMB was determined using the CellTiter 96 AQueous assay (Promega, Madison, WI), which is based on the capacity of viable cells to metabolically reduce 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS). L cells were distributed into 96-well plates at a density of 5.5 × 104 cells/well and infected with rsT1L at an MOI of 10 PFU/cell. Following 1 h of adsorption, cells were washed with PBS to remove the inoculum, and fresh medium was added. LMB was added 8 h postinfection at a final concentration of 20 μg/ml, and the MTS assay was performed according to the manufacturer's instructions after 4 h of LMB exposure. Absorbance in reaction wells was recorded at 485 λ following 1 h of incubation.
The subcellular localization of shuttling reporter proteins was determined in the presence and absence of LMB using the GFP-based vectors Rev68-90-GFP2-M9 (24) and pXRGG (34). Rev68-90-GFP2-M9 contains sequences derived from the human immunodeficiency virus type 1 (HIV-1) Rev protein nuclear export signal (NES; amino acid residues 68 to 90) and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) nuclear localization signal (NLS; M9). The pXRGG vector contains full-length HIV-1 Rev protein fused C-terminally to the hormone-responsive region of the rat glucocorticoid receptor, followed by GFP. Rev and the glucocorticoid receptor furnish pXRGG with one NLS and two NLSs, respectively, while Rev supplies an NES. L cells were transfected with plasmid vectors, and cultures were supplemented 20 h posttransfection with 20 μg/ml LMB. Cells were imaged 4 h later by confocal microscopy.
Complementation of reovirus replication in cells expressing reovirus shRNAs.
trans-complementation of viral replication by transient expression of viral proteins was performed as described previously (28). 293T cells (106) stably expressing M1 or M3 shRNA were seeded into six-well plates (Costar, Cambridge, MA) approximately 24 h prior to transfection. Cells were transfected with expression plasmids encoding μ2 or μNS protein, incubated for 4 or 6 h, respectively, and adsorbed with T3D at an MOI of 10 PFU/cell. Following 1 h of adsorption, cells were washed with PBS to remove the inoculum, and fresh medium was added. Cultures were harvested 24 h after infection, and viral titers in cell lysates were determined by plaque assay.
Rescue of reovirus mutants from cloned cDNAs.
Viruses containing engineered changes in μNS and μ2 proteins were generated from cloned cDNAs using plasmid-based reverse genetics (27). Recombinant viruses were isolated from cotransfection lysates at 5 days posttransfection by plaque assay using L cell monolayers.
RESULTS
Complementation of reovirus replication in cells stably expressing T3D M3 shRNA by transient expression of μNS.
Previous studies revealed that the N-terminal 40 residues of μNS are required for association with viral replication proteins μ2 and σNS and that C-terminal sequences in μNS are required for inclusion formation (9, 12, 39). To determine whether these μNS domains are required for viral replication, expression plasmids encoding wild-type (wt) and mutant forms of μNS were generated for use in RNAi-based replication-complementation assays. We first assessed the intracellular distribution of viral proteins in transiently transfected 293T cells. At 24 h posttransfection, wt μNS and an N-terminally truncated μNS isoform corresponding to μNSC (amino acids 41 to 721) were present in globular inclusion-like structures, similar morphologically to inclusions present in strain T3D-infected cells (Fig. 1A). Likewise, truncation mutants lacking amino acids 1 to 13 or 14 to 40, which are required for interaction of μNS with σNS and μ2, respectively (12, 39), formed distinctive globular structures. This finding contrasts with the diffuse cytoplasmic distribution of μNS protein lacking the C-terminal 46 or 5 amino acid residues. Substitution of two histidine residues (His570 to Gln and His572 to Ser) possessing Zn2+-binding potential and contained within a short sequence motif conserved among μNS proteins of orthoreoviruses and aquareoviruses (9) also resulted in a diffuse cytoplasmic pattern of protein localization. These results show that His570, His572, and C-terminal sequences, but not μ2- or σNS-interacting N-terminal sequences, of μNS are required for autoassembly into inclusion-like structures.
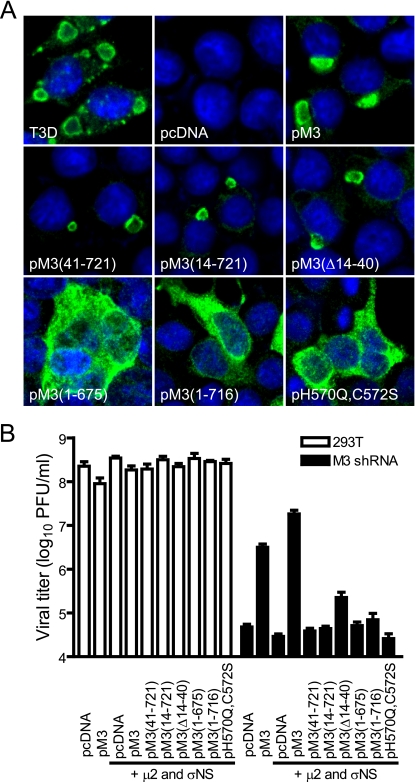
FIG. 1.
trans-complementation of reovirus replication in cells expressing M3 (μNS-specific) shRNA. (A) Subcellular localization of mutant μNS proteins. 293T cells were infected with T3D at an MOI of 2 PFU/cell or transfected with the indicated plasmid constructs, and μNS expression was examined 24 h later by confocal immunofluorescence microscopy using μNS-specific antiserum (green). Nuclei were stained with TO-PRO3 (blue). pcDNA, nonrecombinant vector control; pM3, encodes full-length μNS; pM3(41-721), encodes μNS amino acids 41 to 721; pM3(14-721), encodes μNS amino acids 14 to 721; pM3(Δ14-40), encodes μNS amino acids 1 to 13 and 41 to 721; pM3(1-675), encodes μNS amino acids 1 to 675; pM3(1-716), encodes μNS amino acids 1 to 716; pH570Q, H572S, encodes full-length μNS containing His-to-Gln and His-to-Ser amino acid substitutions at positions 570 and 572, respectively. (B) Viral growth in cells expressing vector-encoded μNS proteins. Parental or M3 shRNA-expressing 293T cells were transfected with plasmids expressing wt or mutant μNS proteins, wt μ2, or wt σNS as indicated, followed by infection with T3D at an MOI of 10 PFU/cell. At 24 h postinfection, viral titers were determined by plaque assay. Results are mean viral titers from three independent experiments. Error bars denote standard deviations.
Cultures of 293T cells stably expressing M3 shRNA (28) were transiently transfected with wt T3D μNS expression plasmid pM3, which contains three silent point mutations within the shRNA target sequence, followed by infection 4 h posttransfection with T3D at an MOI of 10 PFU/cell. Viral titers in culture lysates were determined 24 h postinfection by plaque assay. Yields of T3D were diminished ∼5,000-fold in shRNA-expressing cells transfected with nonrecombinant plasmid compared to an ∼70-fold reduction in cells transfected with pM3 (Fig. 1B). Thus, μNS protein provided in trans restored ∼98% of viral growth suppressed by treatment with M3 shRNA. An additional ∼6-fold enhancement in yield was achieved by coexpression of μ2 and σNS proteins with μNS, suggesting that the abundance or accessibility of μNS-interacting partners is an important factor in complementation efficiency using this RNAi-based system.
We next assessed the capacity of mutant forms of ectopically supplied μNS protein to complement reovirus replication in μNS-silenced cells. Expression plasmids encoding mutant μNS proteins and wt μ2 and σNS proteins were cotransfected into M3 shRNA-expressing cells, followed by infection with T3D. Mutant μNS proteins lacking amino acids 1 to 13, 1 to 40, 676 to 721, or 717 to 721, or containing the double-point mutations His570→Gln and His572→Ser, were incapable of restoring reovirus yields above control vector, although expression of a deletion mutant lacking amino acids 14 to 40 resulted in a very modest (∼5-fold) increase in viral titer (Fig. 1B). These results indicate that viral replication depends on the capacity of μNS to form inclusions and interact with μ2 and σNS proteins.
Importance of μNSC for viral replication in infected cells.
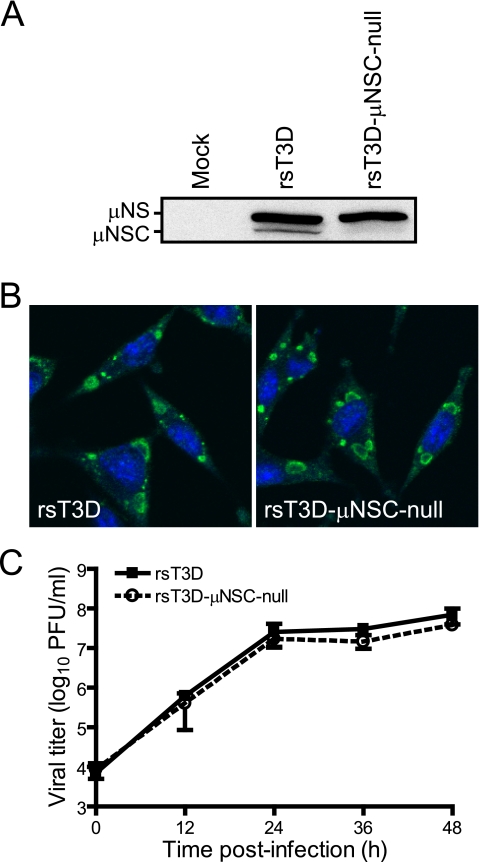
We demonstrated previously that heterologous expression of a naturally occurring, internally initiated form of μNS lacking amino acids 1 to 40, known as μNSC, is incapable of supporting reovirus replication in an RNAi-based complementation system (28). However, the precise function of μNSC in the viral life cycle is unknown. To better understand the biologic role of μNSC, we used reverse genetics (27) to generate a virus incapable of μNSC expression, rsT3D-μNSC-null, by altering the in-frame μNSC initiation codon (corresponding to nucleotides 139 to 141 of the μNS open reading frame) to the isoleucine-encoding sequence, ATC. All remaining genes were derived from wt T3D. Immunoblot analysis of L cells infected with plaque-purified rsT3D and rsT3D-μNSC-null viruses revealed that μNS and μNSC were expressed by rsT3D, but only μNS was expressed in rsT3D-μNSC-null-infected cells (Fig. 2A). Inclusion formation (Fig. 2B) and growth kinetics (Fig. 2C) for the two viruses were virtually identical. These results indicate that μNSC is neither required for viral inclusion formation nor viral replication in reovirus-infected cells.
FIG. 2.
Replication of a μNSC-null virus. (A) Immunoblot detection of μNS and μNSC expression in virus-infected cells. L cells were infected with rsT3D or rsT3D-μNSC-null at an MOI of 2 PFU/cell. At 24 h postinfection, cells were lysed and lysates were immunoblotted using μNS-specific antiserum. (B) Immunofluorescence detection of μNS and μNSC expression in virus-infected cells. L cells were infected as described for panel A and processed for confocal microscopy at 24 h postinfection using μNS-specific antiserum (green). Nuclei were stained with TO-PRO3 (blue). (C) Growth of rsT3D-μNSC-null. L cells were infected with rsT3D or rsT3D-μNSC-null at an MOI of 2 PFU/cell, and viral titers in culture lysates were determined by plaque assay at the intervals shown. Results are mean viral titers from three independent experiments. Error bars denote standard deviations.
Complementation of reovirus replication in cells stably expressing T3D M1 shRNA.
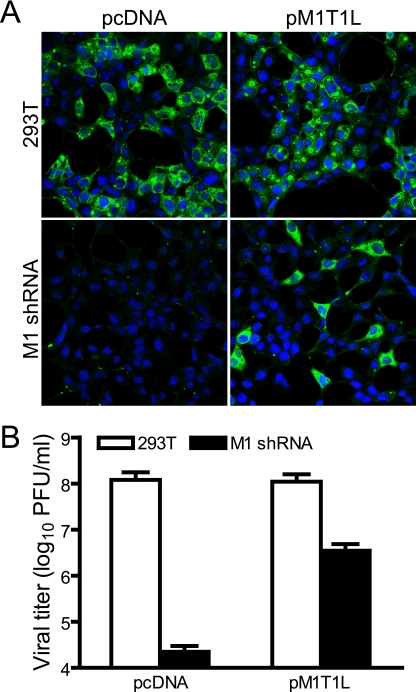
To develop a complementation system for studies of μ2 functions in viral replication, we established L cells constitutively expressing T3D M1-specific shRNA. A T1L μ2 expression plasmid containing a single silent nucleotide substitution within the shRNA target sequence (pM1T1L) was transfected into M1 shRNA-expressing cells, followed by infection 6 h posttransfection with T3D at an MOI of 10 PFU/cell. At 20 h postinfection, infected cells were subjected to immunofluorescence microscopy to detect outer capsid protein μ1/μ1C as a marker of viral replication. The μ1/μ1C protein was detected in shRNA-expressing cells transfected with pM1T1L but not in cells transfected with empty vector (Fig. 3A). Concordantly, transfection of cells with pM1T1L resulted in ∼150-fold-increased viral yields compared to cells transfected with control vector (Fig. 3B), confirming that reovirus replication in μ2-silenced cells is rescued by ectopic μ2 expression.
FIG. 3.
trans-complementation of reovirus replication in cells expressing M1 (μ2-specific) shRNA. Parental or M1 shRNA-expressing 293T cells were transfected with empty vector (pcDNA) or T1L μ2-expressing vector (pM1T1L), followed by infection with T3D at an MOI of 10 PFU/cell. (A) Immunofluorescence analysis of viral replication. At 20 h postinfection, cells were visualized by confocal immunofluorescence microscopy using a monoclonal antibody specific for viral structural protein μ1/μ1C (green). Nuclei were stained with TO-PRO3 (blue). (B) Quantitation of viral growth. At 24 h postinfection, viral titers in culture lysates were determined by plaque assay. Results are mean viral titers from two independent experiments. Error bars denote standard deviations.
Importance of an intact μ2 NTPase/RTPase domain for reovirus replication.
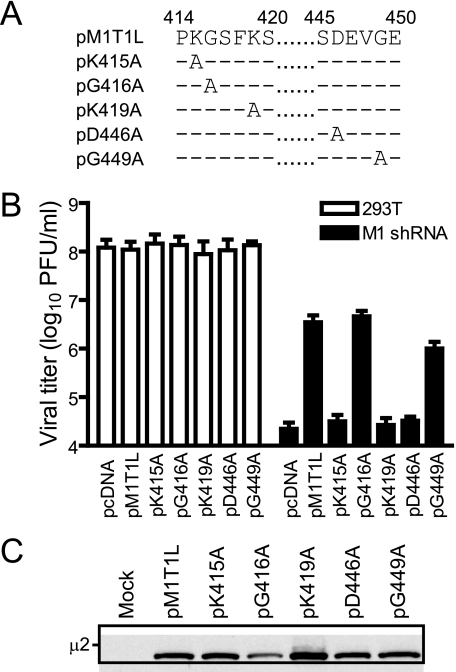
The μ2 protein contains conserved regions of primary sequence, A411VLPKGSFKS420 and D446EVG449, with similarity to the nucleotide-binding A and B motifs, respectively, of ATPases (26, 42). Previous studies showed that baculovirus-expressed μ2 exhibits NTPase and RTPase activities and that both functions are abolished by dual Ala substitutions of conserved Lys residues at positions 415 and 419 (26). To ascertain the importance of μ2 putative nucleotide-binding motifs in virus replication, we replaced Lys415, Gly416, Lys419, Asp446, and Gly449 individually with Ala (Fig. 4A). Mutant μ2 proteins with Gly416 to Ala or Gly449 to Ala mutations retained the capacity to restore viral growth in μ2-deficient cells, whereas mutants with Lys415 to Ala, Lys419 to Ala, or Asp446 to Ala substitutions failed to support growth above background levels (Fig. 4B). Differing complementation efficiencies among wt and mutant μ2 proteins were not explained by variations in expression levels, based on results of immunoblot assays (Fig. 4C). These findings indicate that both putative nucleotide-binding motifs in μ2 are vital to its function in viral replication, consistent with an essential role for μ2 NTPase/RTPase activity.
FIG. 4.
trans-complementation of M1 (μ2-specific) RNAi with μ2 proteins containing mutations in a domain required for NTPase and RTPase activities. (A) Substitutions in predicted NTP-binding motifs of T1L μ2 protein. Changes are shown relative to the wt sequence encoded by the parental vector, pM1T1L. (B) Viral growth in cells expressing vector-encoded μ2 proteins. Parental or M1 shRNA-expressing 293T cells were transfected with empty vector (pcDNA) or T1L μ2-expressing vectors, followed by infection with T3D at an MOI of 10 PFU/cell. At 24 h postinfection, viral titers in culture lysates were determined by plaque assay. Results are mean viral titers from three independent experiments. Error bars denote standard deviations. (C) Confirmation of μ2 expression in transfected cells. Expression of μ2 proteins from trans-complementation vectors was verified by immunoblotting of protein extracts from transiently transfected cells using μ2-specific antiserum.
Importance of a μ2 N-terminal region basic cluster for reovirus replication.
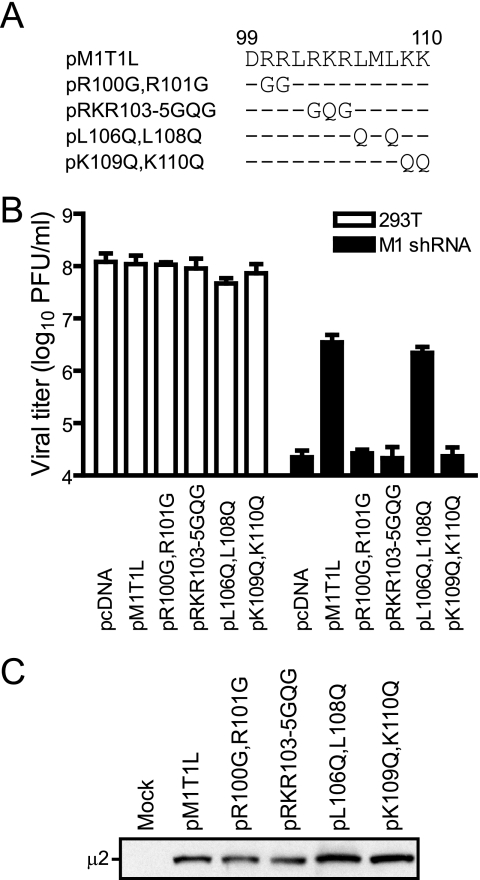
Alignment of μ2 sequences from reovirus strains representing serotypes 1, 2, and 3 reveals a highly conserved short stretch of predominantly basic amino acids that occupies positions 100 to 110 in the primary sequence, R100RLRKRLMLKK110 (67). Such motifs are associated with nuclear targeting activities (15, 21). Therefore, we investigated the importance of R100RLRKRLMLKK110 in viral replication by using complementation methods (Fig. 5A). Viral yields in cells expressing a mutant μ2 protein substituted at two apolar positions, Leu106/Leu108 to Gln106/Gln108, were equivalent to those expressing wt protein (Fig. 5B). In contrast, disruption of basic positions via double or triple substitution mutants—R100R101 to G100G101, R103 K104 R105 to G103Q104G105, and K109K110 to Q109Q110—eliminated the capacity of vector-derived μ2 to restore viral growth in μ2-silenced cells despite equivalent expression levels of wt and mutant μ2 proteins (Fig. 5C). These results indicate that a cluster of basic residues near the N terminus of μ2 is critical for viral replication and support the idea that these conserved sequences are contained within a discrete functional domain.
FIG. 5.
trans-complementation of M1 (μ2-specific) RNAi with μ2 proteins containing mutations in a conserved N-terminal polybasic region. (A) Substitutions in a basic amino acid domain. Changes are shown relative to the wt sequence encoded by the parental vector, pM1T1L. (B) Viral growth in cells expressing vector-encoded μ2 proteins. Parental or M1 shRNA-expressing 293T cells were transfected with empty vector (pcDNA) or T1L μ2-expressing vectors, followed by infection with T3D at an MOI of 10 PFU/cell. At 24 h postinfection, viral titers in culture lysates were determined by plaque assay. Results are mean viral titers from three independent experiments. Error bars denote standard deviations. (C) Confirmation of μ2 expression in transfected cells. Expression of μ2 proteins from trans-complementation vectors was verified by immunoblotting of protein extracts from transiently transfected cells using μ2-specific antiserum.
Influence of μ2 N-terminal region basic sequences on protein subcellular distribution.
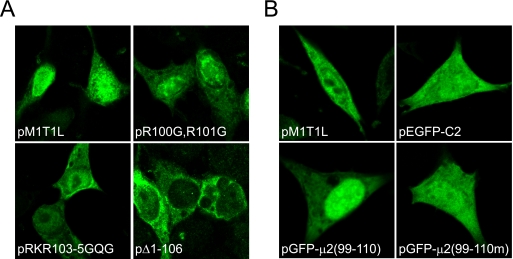
To determine whether perturbations in protein nucleocytoplasmic compartmentalization contribute to the lethal phenotype of mutations introduced at positions occupied by basic residues between amino acids 100 and 110, we examined the distribution of μ2 in transiently transfected cells using confocal microscopy. Native μ2 protein was found in both the nucleus and cytoplasm of 293T cells with similar staining intensities (Fig. 6A). As the molecular mass of μ2 exceeds the exclusion limit for passive diffusion of molecules into the nucleus (∼40 kDa) (7, 44), this finding is consistent with the presence of a functional NLS. Amino acid substitutions that abrogate the capacity of μ2 to trans-complement viral replication—R100R101 to G100G101 and R103 K104 R105 to G103Q104G105—elicited a modest to moderate shift in the distribution of μ2 from the nucleus to cytoplasm. Moreover, an N-terminally truncated μ2 protein lacking the first 106 amino acid residues displayed almost exclusive cytoplasmic localization. These results provide evidence that N-terminal region sequences in μ2 harbor an NLS.
FIG. 6.
Requirement for conserved N-terminal basic residues in μ2 subcellular localization. (A) Intracellular distribution of wt and mutant μ2 proteins. 293T cells were transfected with plasmid constructs encoding the indicated T1L-derived μ2 proteins, which were detected by confocal immunofluorescence microscopy using μ2-specific antiserum (green). pM1T1L, wt T1L μ2; pR100G,R101G, μ2 containing Arg100-to-Gly and Arg101-to-Gly substitutions; pRKR103-5GQG, μ2 containing Arg103-to-Gly, Lys104-to-Gln, and Arg105-to-Gly substitutions; pΔ1-106, truncated μ2 protein lacking the N-terminal 106 amino acid residues. (B) Intracellular distribution of GFP fused to μ2 protein sequences. L cells were transfected with the indicated plasmid constructs. Expression of wt T1L μ2 protein was detected by confocal immunofluorescence microscopy using μ2-specific antiserum (green). Intrinsic GFP fluorescence was detected using confocal microscopy (green). pEGFP-C2, enhanced GFP; pGFP-μ2(99-110), EGFP appended at the C terminus with the μ2 putative nuclear localization sequence, D99RRLRKRLMLKK110; pGFP-μ2(99-110m), pGFP-μ2(99-110) containing Ala substitutions for μ2 basic residues.
To directly determine whether the μ2 polybasic region and predicted NLS possess autonomous nuclear targeting activity, we examined the intracellular distribution of GFP extended at its C terminus with μ2 sequences, D99RRLRKRLMLKK110 (Fig. 6B). In contrast to the homogeneous dispersion of unmodified GFP in L cells, there was marked nuclear accumulation of GFP fused to the μ2 predicted NLS. Ala substitution of basic residues in the appended μ2 sequences nullified their impact on GFP nuclear translocation, resulting in a uniform distribution of the GFP-μ2 fusion protein indistinguishable from wt GFP. These findings indicate that the N-terminal polybasic region in μ2 can independently function as an NLS. Furthermore, because mutations in this same region profoundly interfere with viral replication (Fig. 5B), results of the μ2 localization experiments suggest that nuclear targeting of μ2 is required for completion of the reovirus infectious cycle.
Effect of LMB on μ2 localization and reovirus replication.
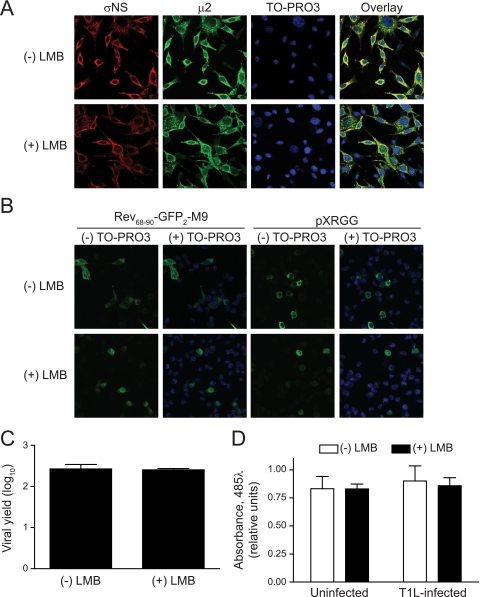
Because μ2 protein distributes to both the nucleus and cytoplasm (Fig. 6) (10, 12, 37, 39, 45), it is possible that μ2 shuttles between these compartments. The best-characterized and most commonly used nuclear export pathway is mediated by Crm1 (exportin 1), which facilitates translocation of cargos containing a Leu-rich NES, the Crm1 ligand (20, 30). LMB covalently associates with Crm1 and blocks binding to this NES (29). To determine whether Crm1-mediated nuclear export functions in μ2 subcellular distribution and viral replication, we examined the effect of LMB treatment on μ2 nuclear accumulation and viral replication in T1L-infected L-cell cultures. Addition of 20 μg/ml LMB at 8 h postinfection did not appreciably alter the nucleocytoplasmic distribution of μ2 at 12 h postinfection (Fig. 7A). Furthermore, viral yields in LMB-treated and control cells were virtually identical (Fig. 7C). The treatment conditions employed were adequate to induce nuclear sequestration of two GFP-based shuttling proteins, Rev68-90-GFP2-M9 and pXRGG, containing the Crm1-dependent NES of HIV-1 Rev (24, 34) (Fig. 7B). As an additional control, neither uninfected nor reovirus-infected cells displayed evidence of substantial LMB-induced cytotoxicity as judged by the capacity of treated cells to reductively metabolize MTS (Fig. 7D). Thus, experimental parameters commensurate with inhibition of Crm1-mediated nuclear export did not reveal spatial or functional evidence of Crm1-dependent μ2 nucleocytoplasmic shuttling.
FIG. 7.
Treatment of reovirus-infected cells with LMB. (A) Subcellular distribution of μ2. L cells were infected with T1L at an MOI of 20 PFU/cell. At 8 h postinfection, the culture medium was supplemented to contain 20 μg/ml LMB (+LMB) or left unsupplemented (-LMB). Cells were imaged 12 h postinfection using confocal immunofluorescence microscopy with σNS (red)- and μ2 (green)-specific antisera. Nuclei were stained with TO-PRO3 (blue). (B) Subcellular distribution of nuclear shuttling proteins. Modified GFPs enabled for nucleocytoplasmic shuttling were transfected into L cells. At 20 h posttransfection, the culture medium was supplemented to contain 20 μg/ml LMB (+ LMB) or left unsupplemented (-LMB). GFP was visualized 24 h posttransfection using confocal microscopy. Nuclei were left unstained (- TO-PRO3) or stained (blue) with TO-PRO3 (+ TO-PRO3). Rev68-90-GFP2-M9 contains sequences derived from the HIV-1 Rev protein nuclear export signal and the heterogeneous nuclear ribonucleoprotein A1 nuclear localization signal. The nuclear export signal for pXRGG is supplied by full-length Rev, and nuclear localization signals are furnished by both Rev and the rat glucocorticoid receptor hormone-responsive region. (C) Reovirus growth. L cells were infected with rsT1L at an MOI of 10 PFU/cell, and the culture medium was supplemented 8 h postinfection to contain 20 μg/ml LMB (+ LMB) or left unsupplemented (- LMB). Viral titers in culture lysates were determined at 0 and 12 h postinfection by plaque assay. Results are mean viral yields (relative to time zero) from three independent experiments. Error bars denote standard deviations. (D) Cell viability. L cells were infected with rsT1L at an MOI of 10 PFU/cell. At 8 h postinfection, culture medium was supplemented to contain 20 μg/ml LMB (+ LMB) or left unsupplemented (- LMB). Cell viability was determined 12 h postinfection with an MTS assay. Results are mean absorbances from three independent experiments. Error bars denote standard deviations. Reaction absorbance is directly proportional to cell viability.
Sequence polymorphisms in μ2 and viral inclusion morphology.
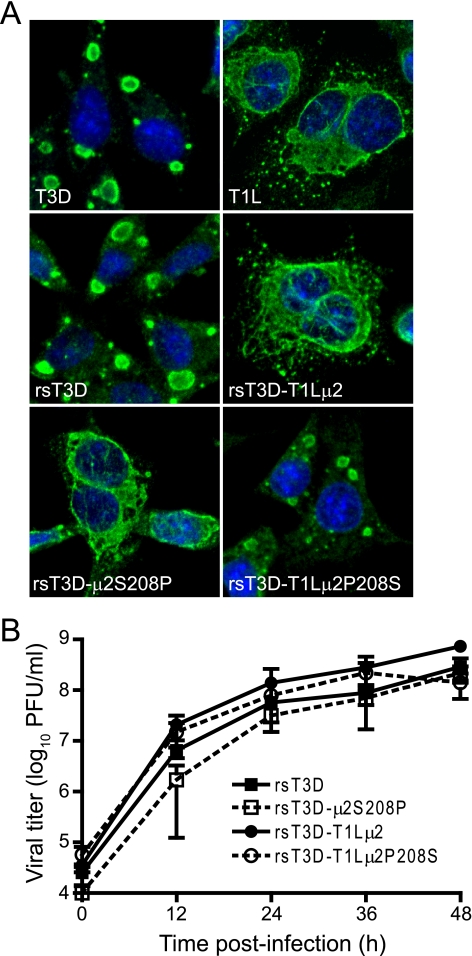
The μ2-encoding M1 gene is a determinant of strain-specific differences in viral inclusion morphology (45), which has been correlated with the efficiency of μ2-microtubule interactions and a single amino acid polymorphism in μ2 at position 208 (45, 67). To determine the significance of the Pro208/Ser208 polymorphism in viral replication, L cells were infected with wt and μ2-mutant viruses recovered by reverse genetics (Table 1) and subjected to immunofluorescence analysis at 24 h post infection. Consistent with patterns displayed by native viruses, rsT3D, which contains a Ser residue at μ2 position 208, produced exclusively globular inclusions, whereas recombinant virus containing the T1L-derived μ2 protein (rsT3D-T1Lμ2), in which Pro occurs at position 208, produced only filamentous inclusions (Fig. 8A). Moreover, this pattern was reversed upon reciprocal Ser208-to-Pro and Pro208-to-Ser substitutions in T3D (rsT3D-μ2S208P) and T1L (rsT3D-T1Lμ2P208S) μ2 proteins, respectively. These results confirm that the Pro/Ser polymorphism at position 208 in μ2 protein serves as an independent determinant of viral inclusion morphology in L cells.
FIG. 8.
Modulation of viral inclusion morphology by engineered changes at μ2 amino acid position 208. (A) Inclusion morphology of μ2 mutant viruses. L cells were infected at an MOI of 2 PFU/cell with wt viruses or viral mutants with substitutions at amino acid position 208 in the T1L and T3D μ2 proteins. At 24 h postinfection, cells were imaged using confocal immunofluorescence microscopy after staining with μNS-specific antiserum (green). Nuclei were stained with TO-PRO3 (blue). T3D and T1L, native viruses; rsT3D, wt strain; rsT3D-T1Lμ2, rsT3D containing T1L-derived μ2; rsT3D-μ2S208P, rsT3D containing a Ser208-to-Pro substitution in μ2; rsT3D-T1Lμ2P208S, rsT3D-T1Lμ2 with a Pro208-to-Ser substitution in μ2. (B) Growth of μ2 mutant viruses. L cells were infected with the indicated viruses at an MOI of 2 PFU/cell, and titers in culture lysates were determined by plaque assay at the intervals shown. Results are mean viral titers from three independent experiments. Error bars denote standard deviations.
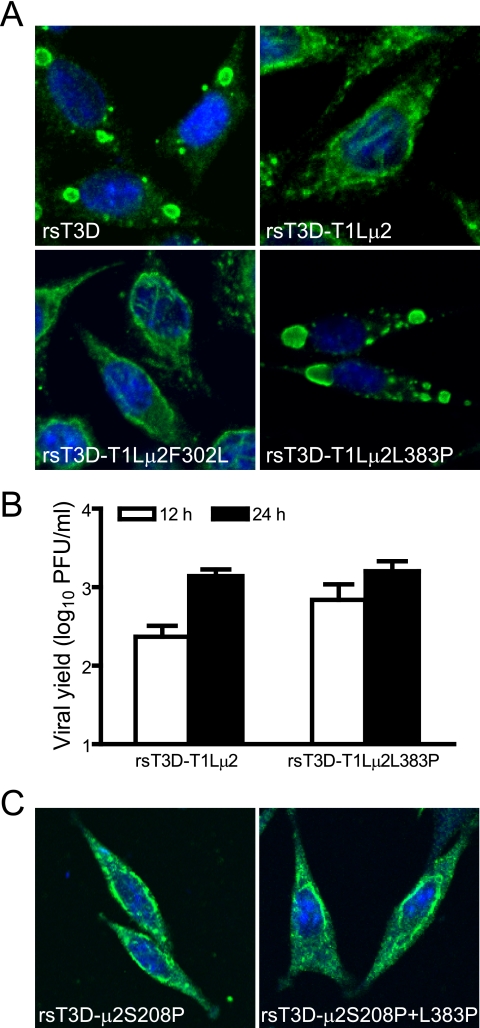
Stocks of reovirus maintained in different laboratories exhibit polymorphisms in the μ2 protein (67). The T1L and T3D stocks used in our laboratory were acquired from the laboratory of Bernard Fields and contain Phe and Leu residues at μ2 positions 302 and 383, respectively. However, a cloned T1L μ2 cDNA derived from a reovirus stock maintained in the laboratory of Earl Brown contains Leu and Pro residues at positions 302 and 383, respectively (45). To determine whether sequence variability at μ2 positions 302 and 383 influences inclusion morphology, we generated T1L and T3D μ2-mutant viruses with substitutions at both sites (Table 1). Immunofluorescence analysis of L cells infected with rsT3D-T1Lμ2- and rsT3D-T1Lμ2F302L displayed filamentous inclusion morphology typical of T1L μ2 (Fig. 9A). However, infection with rsT3D-T1Lμ2L383P revealed only globular viral inclusions, whereas uniformly filamentous inclusions were observed in rsT3D-μ2S208P+L383P-infected cells (Fig. 9C). These findings show that an amino acid position in μ2 other than 208 modulates phenotypic effects of the Ser208/Pro208 polymorphism in a sequence context-dependent fashion. Furthermore, the Phe302-to-Leu substitution in rsT3D-T1Lμ2F302L serves as a specificity control for the effects of engineered changes on protein folding, which provides confidence that alterations in inclusion morphology associated with substitutions at positions 208 and 383 are unlikely to result from gross conformational changes in μ2. Thus, amino acid residues 208 and 383 are part of a functional, and possibly structural, domain involved in μ2-microtubule interactions.
FIG. 9.
Effect of altering μ2 amino acids other than at position 208 on viral inclusion morphology and growth. (A) Inclusion morphology of viruses with alterations in T1L μ2. L cells were infected at an MOI of 2 PFU/cell with wt viruses or viral mutants with substitutions at amino acid positions 302 and 383 in the T1L μ2 protein. At 24 h postinfection, cells were imaged using confocal immunofluorescence microscopy after staining with μNS-specific antiserum (green). Nuclei were stained with TO-PRO3 (blue). rsT3D, wt strain; rsT3D-T1Lμ2, rsT3D containing T1L-derived μ2; rsT3D-T1Lμ2F302L, rsT3D-T1Lμ2 with a Phe302-to-Leu substitution in μ2; rsT3D-T1Lμ2L383P, rsT3D-T1Lμ2 with a Leu383-to-Pro substitution in μ2. (B) Growth of virus with substitutions at amino acid position 383 of T1L μ2. L cells were infected with the indicated viruses at an MOI of 2 PFU/cell, and titers in culture lysates were determined by plaque assay after 0, 12, and 24 h of incubation. Results are mean viral yields (relative to time zero) from three independent experiments. Error bars denote standard deviations. (C) Inclusion morphology of viruses with alterations in T3D μ2. L cells were infected with the indicated viruses and imaged 24 h later as described for panel A. rsT3D-μ2S208P, rsT3D containing a Ser208-to-Pro substitution in μ2; rsT3D-μ2S208P+L383P, rsT3D with Ser208-to-Pro and Leu383-to-Pro substitutions in μ2.
Inclusion morphology and viral growth.
To ascertain the relevance of inclusion morphology to viral replication, L cells were infected with wt and μ2-mutant rs viruses at an MOI of 2 PFU/cell, and viral growth was assessed by plaque assay at various times postinfection (Fig. 8B and 9B). Kinetics of viral growth and absolute titers of infectious particles did not differ significantly among wt and mutant viruses, indicating that neither μ2 sequence variability at positions 208, 302, or 383 nor inclusion morphology is a critical modulator of reovirus replication efficiency in L cells infected with isogenic μ2 mutant viruses.
DISCUSSION
The μNS protein forms distinct structures resembling viral inclusions when expressed in the absence of other viral proteins (12). Minimum sequences required for autoassembly of μNS protein into inclusion-like structures are contained within the 161 to 251 C-terminal amino acids (9). The C-terminal two to eight amino acid residues and a putative metal-chelating motif (perhaps selective for Zn2+) involving His570 and Cys572 appear to play critical roles in this process (2, 9). These sequences may contribute to a dimerization domain perhaps involved in μNS homotypic or heterotypic interactions required to nucleate inclusions (9). We found that preservation of the μNS extreme C terminus and conservation of His570 and Cys572 are required for autoassembly of μNS into structures resembling viral inclusions. Furthermore, results of shRNA-based trans-complementation assays indicate that mutant μNS proteins failing to form inclusion-like structures are incapable of supporting viral growth in 293T cells, consistent with a recent study using a different trans-complementation methodology (2). Therefore, concordant results obtained in independent laboratories using different methods provide compelling functional evidence that establishment of cytoplasmic inclusions by μNS is a prerequisite to reovirus replication.
Although formation of inclusions is necessary for viral replication, results reported here indicate that inclusion formation is not sufficient. Our findings and those previously reported by Arnold et al. (2) show that μNSC, though capable of self-assembly into inclusion-like structures, cannot support viral replication. Such aspects of μNS structure-function relationships point to compulsory heterointeractions between μNS and μ2 and σNS proteins at one or more steps in the viral RNA life cycle, perhaps recruitment or retention of (+)-strand RNAs at sites of viral replication, dsRNA synthesis, or genome packaging. In support of this model, we previously found that selective RNAi-mediated elimination of μNS, μ2, or σNS from reovirus-infected cells inhibits viral dsRNA synthesis and production of infectious particles (28). We show here that removal of μNS N-terminal sequences specifically required for interactions with either σNS (1 to 13) or μ2 (14 to 40) (10, 39) also eliminates or drastically reduces the capacity of μNS to support viral growth, yet without compromising its capacity for inclusion formation.
Despite the failure of ectopically supplied μNSC to restore viral replication in μNS-depleted cells, μNSC might nevertheless contribute to viral inclusion formation or growth in infected cells. Thus, to unequivocally define the importance of μNSC in the viral infectious cycle, we characterized the replication of a μNSC-deficient mutant virus obtained using reverse genetics. In these experiments, we found that μNSC is neither required for normal viral inclusion development nor viral growth (Fig. 2). It is possible that μNSC promotes viral growth in certain cell or tissue types requisite to efficient spread within or between hosts. Alternatively, μNSC may modulate host defenses to reovirus infection at the intracellular or organismal level. We are currently investigating these possibilities.
RNAi-mediated reduction in μ2 expression is associated with marked retardation of viral inclusion development, inhibition of dsRNA synthesis, and virtual absence of progeny virion production (28). The exquisite responsiveness of reovirus replication to diminished μ2 levels permitted specific sequences in μ2 important for viral growth to be defined by complementation. We identified three short regions of sequence critical for viral replication: two predicted nucleotide-binding units with similarity to Walker A (A411VLPKGS FKS420) and B (D446EVG449) motifs of ATPases (26) and an N-terminal basic amino acid cluster conforming to a possible NLS (R100RLRKRLMLKKDLRK114) (15, 21). The Walker A- and B-like motifs are invariant among 14 reported μ2 sequences representing reovirus strains of all three serotypes, including the prototype strains (67). Single Ala substitutions at putative nucleotide-binding residues Lys415 and Lys419 in the A-like motif prevented viral replication, whereas mutation of the neutral position, Gly416, was replication compatible. These results strongly suggest that μ2 NTPase/RTPase activity is requisite to viral replication and agree with recent findings by Carvalho et al. (13), who reported that introduction of the double mutation, Lys415/Lys419-to-Ala415/Ala419, into μ2 caused an ∼10- to 100-fold viral titer reduction in CV-1 cells when they used an siRNA-based replication complementation system. Tolerance of a mutation at Gly449, but not Asp446, in the Walker B-like domain provides further evidence that viral replication is dependent on μ2 NTPase/RTPase functionality.
Our findings do not address specific steps in the viral infectious cycle disrupted by mutations in the Walker-like motifs. Considering that μ2 is a probable component of the core particle transcriptional machinery (8, 26, 66) as well as inclusion-associated protein (5, 10, 12, 37, 39, 40, 45) involved in genomic RNA synthesis and particle assembly (16), diminished NTPase/RTPase activities could interrupt progression of the replication program at multiple points during or following RNA (+)-strand synthesis. Introduction of Lys415/Lys419-to-Ala415/Ala419 mutations into T1L μ2 does not interfere with its capacity to form filamentous inclusion-like structures when coexpressed with μNS in CV-1 cells (26). Interestingly, the NTPase function of rotavirus viroplasm-forming protein NSP2 is essential for productive rotavirus replication, but not viroplasm formation, in trans-complemented MA104 cells (59). Taken together, these findings suggest that disruption of μ2 NTPase/RTPase activity blocks reovirus replication at a point subsequent to inclusion formation, for example, synthesis or packaging of dsRNA.
We found that nonconservative substitutions at charged, but not apolar, positions in the more N-terminal of two basic clusters in μ2 completely prevented transiently expressed μ2 from restoring viral growth in μ2-deficient cells. These changes produced a mild nuclear-to-cytoplasmic shift in the subcellular distribution of transiently expressed μ2 protein, suggesting that other sequences also regulate nuclear localization. One such possibility is the slightly more C-terminal NLS-like sequence element, K261RLR264, but additional nuclear-targeting activities may reside proximal to the N terminus of μ2, as very little mutant protein lacking the N-terminal 106 residues was present in the nucleus. Nevertheless, we found that the oligopeptide D99RRLRKRLMLKK110 is sufficient to direct a heterologous protein (GFP) to the nucleus. Thus, this region of μ2 possesses intrinsic capacity for nuclear localization. Classical (i.e., basic) NLSs are recognized by importin-α, and NLS-presenting proteins are translocated through the nuclear pore in a complex containing importin-α and importin-β (57). These findings are consistent with a physical interaction between μ2 and components of the nuclear import apparatus.
A functional role for μ2 nuclear entry is not apparent from our studies. However, it is noteworthy that μ2 protein has been genetically linked to viral strain-specific differences in the induction of alpha/beta interferon (IFN-α/β) expression in cardiac myocytes and reovirus sensitivity to the antiviral effects of IFN in these cells (55). A recent report by Zurney et al. (69) provided evidence that the μ2 protein of strain T1L, but not T3D, antagonizes signal transduction from the IFN-α/β receptor by favoring the nuclear accumulation of a key signaling molecule, IFN regulatory factor 9 (IRF9), which partners with signal transducer and activator of transcription 1 (STAT1) and STAT2 to form a heterotrimeric inducer of IFN-stimulated gene (ISG) expression (49). The nuclear phase of μ2 might benefit viral replication by directly or indirectly perturbing the normal cycle of IRF9 nucleocytoplasmic translocation, thereby suppressing the expression of ISGs. Activities of μ2 in the nucleus might also involve cellular processes unrelated to innate immunity, such as regulation of transcription or the cell cycle, congruous with diverse functional interactions between proteins encoded by cytoplasmically replicating RNA viruses and nuclear structures and proteins (23).
The most commonly used and best-characterized pathway for exporting nuclear cargo to the cytoplasm is dependent on Crm1, which recognizes hydrophobic, typically Leu-rich, NESs and translocates NES-containing proteins through the nuclear pore as part of a heteromeric complex containing GTP-bound Ran protein (57). A number of viral proteins are shuttled in a Crm1-dependent fashion (20, 46, 48, 63, 65), and like μ2, these proteins are frequently involved in viral RNA regulation or subversion of the IFN response. A specific inhibitor of Crm1 nuclear export activity, LMB (29, 30), did not significantly affect viral replication or the distribution of μ2 protein in T1L-infected L cells under treatment conditions adequate to induce nuclear retention of two different GFP-based reporter molecules containing the HIV-1 Rev NES. Nonetheless, our results do not exclude the possibility of bidirectional movement of μ2 between the cytoplasm and nucleus. It is possible that variation in the sequence of μ2 (or other proteins) among different viral strains (67) influences the extent or kinetics of μ2 nucleocytoplasmic shuttling and, therefore, sensitivity to LMB. Another possibility is that μ2 export from the nucleus occurs through a nonclassical pathway (43), reminiscent of Crm1-indpendent nuclear export of morbillivirus N proteins containing Leu-rich NESs (50). We note the presence of a sequence element in μ2, L328EMLGIEI335, which exhibits similarity to a Leu-rich NES (31) and is highly conserved among 14 published μ2 sequences except that belonging to strain T2J (67).
The μ2 protein binds and stabilizes microtubules and anchors inclusions of the majority of characterized reovirus strains to the cytoskeleton, resulting in filamentous inclusion morphology (12, 45, 67). However, a Pro208-to-Ser change present in some viral strains is associated with the covariant phenotypes of diminished microtubule binding by μ2 (45) and globular inclusion morphology (45, 67). We rescued isogenic viruses containing single amino acid substitutions in μ2 and found that the Pro208/Ser208 polymorphism is indeed an independent determinant of inclusion morphology. We further observed that a Leu/Pro polymorphism at position 383, identified in the cloned M1 cDNA sequence from a laboratory isolate of T1L (45), also determines inclusion morphology of rescued viruses expressing T1L μ2 and that the simultaneous presence of Pro and Leu at positions 208 and 383, respectively, is required to produce filamentous inclusions. Similar studies performed using viruses expressing T3D μ2 protein yielded the somewhat surprising result that the polymorphism at position 383 in T3D μ2 does not affect inclusion morphology dictated by amino acid 208. There are nine amino acid differences between the T1L- and T3D-derived μ2 proteins used for our studies (T1L/T3D amino acids: Val93/Ala93, Pro208/Ser208, Val300/Met300, Gln342/Arg342, Phe347/Leu347, Val434/Ile434, His458/Gln458, Met652/Ile652, and Asn726/Ser726), one or more of which must control the differential effects of residue 383 on inclusion morphology. Detailed structural models of the μ2 protein are needed to fully explain these strain-specific effects, but one logical hypothesis is that amino acid residues 208 and 383 are proximal in the μ2 structure and contribute to the microtubule-binding or microtubule-stabilizing region of μ2. Neighboring polymorphic positions may influence the interactions of amino acid residues 208 and 383 with microtubules or each other.
We were unable to correlate inclusion morphology with total viral yields or replication kinetics by using rescued μ2 mutant viruses, which grew equivalently to wt regardless of the inclusion morphotype or strain origin of μ2. These findings contrast with those reported by Carvalho et al. (13), which showed that viral growth in an RNAi-based trans-complementation system was significantly enhanced when complementation was performed with μ2 protein containing Pro rather than Ser at position 208. The mechanistic basis for these differential effects of Pro208 versus Ser208 on the capacity of transiently expressed μ2 to restore viral replication was not evident in those studies, but rescue of viral growth was sensitive to nocodazole, indicating dependence on intact microtubules. The requirements for microtubule binding by μ2 may be cell type dependent. In this regard, previous experiments were performed with primate kidney-derived (CV-1) cells, whereas we used murine fibroblast (L) cells. The μ2-encoding M1 gene is a determinant of differences between strains T1L and T3D with respect to viral growth in MDCK cells (47), murine cardiac cells (36), and bovine aortic endothelial cells (35); reovirus-induced myocarditis in immunocompetent mice (54); and organ-specific viral growth and injury in SCID mice (22). Thus, μ2-mediated recruitment of viral factories to microtubules might be necessary for efficient reovirus growth in some types of cells. Although a mechanistic explanation for such a requirement is not obvious, concentration of viral proteins and RNA on the cytoskeleton may facilitate genome replication and particle assembly or, perhaps, shield viral components from recognition by the intracellular pathogen surveillance system.
In this study, we combined RNAi-based and reverse genetics strategies to define the role of functional domains in the viral inclusion-forming proteins μNS and μ2 in reovirus replication. Both proteins display modular architectures built from what appear to be functionally and structurally unique sequence domains involved in discrete steps of inclusion development and viral replication. Recombinant reoviruses systematically altered in the μNS, μ2, and other viral proteins will foster new insights into the viral replication machinery and may reveal posttranscriptional determinants of viral pathogenesis.
Acknowledgments
We thank Sam Naik for his assistance with studies of μ2 subcellular localization and members of our laboratories for many helpful discussions about this project. We thank Ralph Kehlenbach, University of Gottingen, for kindly providing the GFP shuttling constructs.
This work was supported by Public Health Service awards K08 AI062862 (J.D.C.) and R01 AI32539 from the National Institute of Allergy and Infectious Diseases, the Naito Foundation (T.K.), the Elizabeth B. Lamb Center for Pediatric Research, and the Vanderbilt University Department of Pathology. Additional support was provided by Public Health Service awards CA68485 for the Vanderbilt-Ingram Cancer Center and DK20593 for the Vanderbilt Diabetes Research and Training Center. Acquisition and analysis of confocal imaging data were performed in part through the use of the Vanderbilt University Medical Center Imaging Shared Resource.
Footnotes
Published ahead of print on 28 January 2009.
REFERENCES
- 1.Antczak, J. B., and W. K. Joklik. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187760-776. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, M. M., K. E. Murray, and M. L. Nibert. 2008. Formation of the factory matrix is an important, though not a sufficient function of nonstructural protein μNS during reovirus infection. Virology 375412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, G. S., D. H. Ebert, C. J. Chung, A. H. Erickson, and T. S. Dermody. 1999. Mutant cells selected during persistent reovirus infection do not express mature cathepsin L and do not support reovirus disassembly. J. Virol. 739532-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, A. K., and A. J. Shatkin. 1970. Transcription in vitro by reovirus-associated ribonucleic acid-dependent polymerase. J. Virol. 61-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 751459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, M. M., T. R. Peters, and T. S. Dermody. 2003. Reovirus σNS and μNS proteins form cytoplasmic inclusion structures in the absence of viral infection. J. Virol. 775948-5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonner, W. M. 1978. Protein migration and accumulation in nuclei, p. 97-148. In H. Busch (ed.), The cell nucleus, vol. 6. Academic Press, New York, NY. [Google Scholar]
- 8.Brentano, L., D. L. Noah, E. G. Brown, and B. Sherry. 1998. The reovirus protein μ2, encoded by the M1 gene, is an RNA-binding protein. J. Virol. 728354-8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broering, T. J., M. M. Arnold, C. L. Miller, J. A. Hurt, P. L. Joyce, and M. L. Nibert. 2005. Carboxyl-proximal regions of reovirus nonstructural protein μNS necessary and sufficient for forming factory-like inclusions. J. Virol. 796194-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broering, T. J., J. Kim, C. L. Miller, C. D. Piggott, J. B. Dinoso, M. L. Nibert, and J. S. Parker. 2004. Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 781882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 745516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broering, T. J., J. S. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 768285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho, J., M. M. Arnold, and M. L. Nibert. 2007. Silencing and complementation of reovirus core protein μ2: functional correlations with μ2-microtubule association and differences between virus- and plasmid-derived μ2. Virology 364301-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, C. T., and H. J. Zweerink. 1971. Fate of parental reovirus in infected cell. Virology 46544-555. [DOI] [PubMed] [Google Scholar]
- 15.Chelsky, D., R. Ralph, and G. Jonak. 1989. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol. Cell. Biol. 92487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coombs, K. M. 1996. Identification and characterization of a double-stranded RNA-reovirus temperature-sensitive mutant defective in minor core protein μ2. J. Virol. 704237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coombs, K. M. 1998. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology. 243218-228. [DOI] [PubMed] [Google Scholar]
- 18.Ebert, D. H., J. Deussing, C. Peters, and T. S. Dermody. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 27724609-24617. [DOI] [PubMed] [Google Scholar]
- 19.Fields, B. N., C. S. Raine, and S. G. Baum. 1971. Temperature-sensitive mutants of reovirus type 3: defects in viral maturation as studied by immunofluorescence and electron microscopy. Virology 43569-578. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390308-311. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Bustos, J., J. Heitman, and M. Hall. 1991. Nuclear protein localization. Biochim. Biophys. Acta 107183-101. [DOI] [PubMed] [Google Scholar]
- 22.Haller, B. L., M. L. Barkon, G. P. Vogler, and H. W. Virgin, IV. 1995. Genetic mapping of reovirus virulence and organ tropism in severe combined immunodeficient mice: organ-specific virulence genes. J. Virol. 69357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiscox, J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 9513-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutten, S., S. Wälde, C. Spillner, J. Hauber, and R. H. Kehlenbach. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J. Cell. Sci., in press. [DOI] [PubMed]
- 25.Ishii, K., Y. Ueda, K. Matsuo, Y. Matsuura, T. Kitamura, K. Kato, Y. Izumi, K. Someya, T. Ohsu, M. Honda, and T. Miyamura. 2002. Structural analysis of vaccinia virus DIs strain: application as a new replication-deficient viral vector. Virology 302433-444. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J., J. S. Parker, K. E. Murray, and M. L. Nibert. 2004. Nucleoside and RNA triphosphatase activities of Orthoreovirus transcriptase cofactor μ2. J. Biol. Chem. 2794394-4403. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, T., A. A. R. Antar, K. W. Boehme, P. Danthi, E. A. Eby, K. M. Guglielmi, G. H. Holm, E. M. Johnson, M. S. Maginnis, S. Naik, W. B. Skelton, J. D. Wetzel, G. J. Wilson, J. D. Chappell, and T. S. Dermody. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi, T., J. D. Chappell, P. Danthi, and T. S. Dermody. 2006. Gene-specific inhibition of reovirus replication by RNA interference. J. Virol. 809053-9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 969112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell. Res. 242540-547. [DOI] [PubMed] [Google Scholar]
- 31.la Cour, T., L. Kiemer, A. Molgaard, R. Gupta, K. Skriver, and S. Brunak. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. 17527-536. [DOI] [PubMed] [Google Scholar]
- 32.Lee, P. W. K., E. C. Hayes, and W. K. Joklik. 1981. Characterization of anti-reovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology 108134-146. [DOI] [PubMed] [Google Scholar]
- 33.Li, J. K.-K., P. P. Scheible, J. D. Keene, and W. K. Joklik. 1980. The plus strand of reovirus gene S2 is identical with its in vitro transcript. Virology 105282-286. [DOI] [PubMed] [Google Scholar]
- 34.Love, D. C., T. D. Sweitzer, and J. A. Hanover. 1998. Reconstitution of HIV-1 Rev nuclear export: independent requirements for nuclear import and export. Proc. Natl. Acad. Sci. USA 9510608-10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matoba, Y., W. S. Colucci, B. N. Fields, and T. W. Smith. 1993. The reovirus M1 gene determines the relative capacity of growth of reovirus in cultured bovine aortic endothelial cells. J. Clin. Investig. 922883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matoba, Y., B. Sherry, B. N. Fields, and T. W. Smith. 1991. Identification of the viral genes responsible for growth of strains of reovirus in cultured mouse heart cells. J. Clin. Investig. 871628-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mbisa, J. L., M. M. Becker, S. Zou, T. S. Dermody, and E. G. Brown. 2000. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology 27216-26. [DOI] [PubMed] [Google Scholar]
- 38.McCrae, M. A., and W. K. Joklik. 1978. The nature of the polypeptide encoded by each of the ten double-stranded RNA segments of reovirus type 3. Virology 89578-593. [DOI] [PubMed] [Google Scholar]
- 39.Miller, C. L., T. J. Broering, J. S. Parker, M. M. Arnold, and M. L. Nibert. 2003. Reovirus σNS protein localizes to inclusions through an association requiring the μNS amino terminus. J. Virol. 774566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, C. L., J. S. Parker, J. B. Dinoso, C. D. Piggott, M. J. Perron, and M. L. Nibert. 2004. Increased ubiquitination and other covariant phenotypes attributed to a strain- and temperature-dependent defect of reovirus core protein μ2. J. Virol. 7810291-10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mustoe, T. A., R. F. Ramig, A. H. Sharpe, and B. N. Fields. 1978. Genetics of reovirus: identification of the dsRNA segments encoding the polypeptides of the μ and σ size classes. Virology 89594-604. [DOI] [PubMed] [Google Scholar]
- 42.Noble, S., and M. L. Nibert. 1997. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 717728-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ossareh-Nazari, B., C. Gwizdek, and C. Dargemont. 2001. Protein export from the nucleus. Traffic 2684-689. [DOI] [PubMed] [Google Scholar]
- 44.Paine, P. L., L. C. Moore, and S. B. Horowitz. 1975. Nuclear envelope permeability. Nature 254109-114. [DOI] [PubMed] [Google Scholar]
- 45.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 764483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasdeloup, D., N. Poisson, H. Raux, Y. Gaudin, R. W. H. Ruigrok, and D. Blondel. 2005. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334284-293. [DOI] [PubMed] [Google Scholar]
- 47.Rodgers, S. E., E. S. Barton, S. M. Oberhaus, B. Pike, C. A. Gibson, K. L. Tyler, and T. S. Dermody. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol. 712540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez, J. J., C. D. Cruz, and C. M. Horvath. 2004. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 785358-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadler, A. J., and B. R. G. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato, H., M. Masuda, R. Miura, M. Yoneda, and C. Kai. 2006. Morbillivirus nucleoprotein possesses a novel nuclear localization signal and a CRM1-independent nuclear export signal. Virology 352121-130. [DOI] [PubMed] [Google Scholar]
- 51.Schiff, L., M. Nibert, and K. L. Tyler. 2007. Orthoreoviruses and their replication, p. 1853-1915. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 52.Schonberg, M., S. C. Silverstein, D. H. Levin, and G. Acs. 1971. Asynchronous synthesis of the complementary strands of the reovirus genome. Proc. Natl. Acad. Sci. USA 68505-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shatkin, A. J., and M. Kozak. 1983. Biochemical aspects of reovirus transcription and translation, p. 79-106. In W. K. Joklik (ed.), The Reoviridae. Plenum Press, New York, NY.
- 54.Sherry, B., and M. A. Blum. 1994. Multiple viral core proteins are determinants of reovirus-induced acute myocarditis. J. Virol. 688461-8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherry, B., J. Torres, and M. A. Blum. 1998. Reovirus induction of and sensitivity to beta interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J. Virol. 721314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silverstein, S. C., and P. H. Schur. 1970. Immunofluorescent localization of double-stranded RNA in reovirus-infected cells. Virology 41564-566. [DOI] [PubMed] [Google Scholar]
- 57.Sorokin, A. V., E. R. Kim, and L. P. Ovchinnikov. 2007. Nucleocytoplasmic transport of proteins. Biochemistry (Moscow) 721439-1457. [DOI] [PubMed] [Google Scholar]
- 58.Sturzenbecker, L. J., M. L. Nibert, D. B. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 612351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taraporewala, Z. F., X. Jiang, R. Vasquez-Del Carpio, H. Jayaram, B. V. V. Prasad, and J. T. Patton. 2006. Structure-function analysis of rotavirus NSP2 octamer by using a novel complementation system. J. Virol. 807984-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Virgin, H. W., IV, R. Bassel-Duby, B. N. Fields, and K. L. Tyler. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 624594-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 656772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe, S., T. Watanabe, T. Noda, A. Takada, H. Feldmann, L. D. Jasenosky, and Y. Kawaoka. 2004. Production of novel Ebola virus-like particles from cDNAs: an alternative to Ebola virus generation by reverse genetics. J. Virol. 78999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wells, G., and A. Malur. 2008. Expression of human parainfluenza virus type 3 PD protein and intracellular localization in virus infected cells. Virus Genes. 37358-367. [DOI] [PubMed] [Google Scholar]
- 64.Wiener, J. R., J. A. Bartlett, and W. K. Joklik. 1989. The sequences of reovirus serotype 3 genome segments M1 and M3 encoding the minor protein μ2 and the major nonstructural protein μNS, respectively. Virology 169293-304. [DOI] [PubMed] [Google Scholar]
- 65.Wolff, B., J.-J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4139-147. [DOI] [PubMed] [Google Scholar]
- 66.Yin, P., M. Cheang, and K. M. Coombs. 1996. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J. Virol. 701223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin, P., N. D. Keirstead, T. J. Broering, M. M. Arnold, J. S. Parker, M. L. Nibert, and K. M. Coombs. 2004. Comparisons of the M1 genome segments and encoded μ2 proteins of different reovirus isolates. Virol. J. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zou, S., and E. G. Brown. 1996. Stable expression of the reovirus μ2 protein in mouse L cells complements the growth of a reovirus ts mutant with a defect in its M1 gene. Virology 21742-48. [DOI] [PubMed] [Google Scholar]
- 69.Zurney, J., T. Kobayashi, G. H. Holm, T. S. Dermody, and B. Sherry. 2008. The reovirus μ2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor-9. J. Virol. 822178-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zweerink, H. J., E. M. Morgan, and J. S. Skyler. 1976. Reovirus morphogenesis: characterization of subviral particles in infected cells. Virology 73442-453. [DOI] [PubMed] [Google Scholar]