Abstract
Enfuvirtide (ENF) prevents the entry of human immunodeficiency virus type 1 (HIV-1) into cells by binding to the HR-1 region of the viral envelope (Env) protein gp41 subunit. Resistance to ENF arises via mutations in the drug binding site in HR-1. In addition, HR-2 mutations are commonly observed in ENF-resistant Env proteins, though their role remains unclear. We explored the mechanistic basis for clinical resistance to ENF and the role of HR-2 mutations. Using panels of ENF resistance-associated mutants for two patients, we found that mutations in HR-1 slowed the fusion kinetics and that mutations in HR-2 restored fusion rates. We assessed the differences in the rates of fusion of these mutants from a temperature-arrested state and observed similar trends, suggesting that the step of delay occurs after coreceptor engagement. Sensitivity to neutralizing antibodies was unchanged by the HR-1 and HR-2 mutants in each panel. Since this result was in contrast to those of a previous in vitro analysis where enhanced sensitivity to neutralization was demonstrated for heterologous Envs with ENF resistance-associated HR-1 changes, we examined the context dependence of HR-1 and HR-2 mutations by transferring the mutations seen in one patient into the Env context of another. These studies revealed that some, but not all, HR-1 mutations, when placed out of context (i.e., in a patient Env where they did not originally arise), enhance sensitivity to neutralizing antibodies. However, in most cases, HR-1 mutations in ENF-treated patients evolve in a manner that preserves pretreatment neutralization sensitivity so as to evade the pressures of the immune system.
Infection of cells by human immunodeficiency virus type 1 (HIV-1) can be prevented by the use of inhibitors that target specific steps in the viral entry pathway (24). Viral resistance to entry inhibitors can arise via mutations in the viral envelope protein (Env), which exists as a trimer on the surfaces of virions. Each Env subunit is composed of a surface gp120 and a transmembrane gp41 protein. The gp120 surface protein is responsible for the interactions of the virus with CD4 and subsequently with a chemokine coreceptor (CCR5 or CXCR4) on the surface of the target cell. Receptor binding induces conformational changes in the gp41 transmembrane domain subunit, which contains an N-terminal fusion peptide that is inserted into the target cell membrane after coreceptor engagement and two helical heptad repeat regions (HR-1 and HR-2). The heptad repeat regions undergo a conformational rearrangement resulting in a six-helix bundle structure composed of the HR-1 and HR-2 regions from each of the three Env subunits (33). This structural transition is thought to bring the virus and cell membranes into close proximity, leading to fusion pore formation and membrane fusion. Enfuvirtide (ENF) is a 36-amino-acid peptide based on the sequence of the HR-2 region of gp41 (34, 35). ENF prevents six-helix-bundle formation by binding to the HR-1 regions of Env, which become exposed after coreceptor binding (11, 13, 21). Thus, ENF targets a conformational intermediate of the membrane fusion process and prevents virus infection (5, 10). ENF can effectively reduce virus loads in HIV-infected individuals and is typically used in treatment-experienced patients.
Resistance to ENF, either in vivo or in vitro, is almost always associated with one or more mutations in the HR-1 region of gp41 (9, 12, 15, 18, 20, 22, 27, 29, 30, 32). Presumably, these mutations impact the binding of ENF and hence its potency. Resistance to ENF is not associated with altered sensitivity to other antiretroviral agents, including other classes of entry inhibitors (27, 28). However, when HR-1 mutations are introduced into Env proteins, they can reduce the rate of membrane fusion and render the virus more sensitive to neutralization by antibodies that bind to the membrane-proximal region of gp41 (28). If this were to occur in vivo, virus fitness could be affected. The impact of ENF resistance on Env function could be minimized by the selection of resistance mutations that in a given context minimize impacts on Env function and by the selection of compensatory mutations that aid in the restoration of full Env function. Consistent with this, mutations in the HR-2 region of Env are commonly observed in virus strains that are resistant to ENF (2, 19, 26, 31, 36), yet these mutations usually do not contribute to drug resistance (27). In this study, we have explored the mechanistic basis for clinical resistance to ENF and the role of HR-2 mutations. We found that while mutations in HR-1 slow the kinetics of fusion, those in HR-2 restore fusion rates by accelerating the conformational changes that occur after coreceptor binding, most likely involving the formation of the six-helix bundle. The impact of HR-1 mutations on antibody-mediated neutralization was context dependent, as evidenced by the fact that mutations that evolved in vivo had little effect on neutralization by antibodies that bind to the membrane-proximal region of gp41. However, when the same mutations were introduced into heterologous Env backgrounds, neutralization sensitivity was enhanced. We conclude that HR-2 mutations commonly associated with ENF resistance play a compensatory role by restoring fusion kinetics, while selective pressures in vivo appear to minimize the impact of HR-1 mutations on virus sensitivity to antibody-mediated neutralization.
MATERIALS AND METHODS
Cells.
QT6 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% l-glutamine. HeLa/CD4/CCR5 cells (JC53) were obtained from D. Kabat and cultured in DMEM supplemented with 10% FCS. U87.CD4.CCR5 cells were also cultured in DMEM supplemented with 10% FCS.
Plasmids and antibodies.
env genes from patients were cloned into the pcDNA3.1 vector using TOPO cloning as described previously (27). ENF resistance-associated mutations were engineered into a pretreatment Env clone from the same patient from whom env genes were cloned, as described previously (27). For pseudotype production, env genes were subcloned into a pCI expression vector modified to contain hepatitis B virus posttranscriptional regulatory element for enhanced rev-independent Env expression and higher pseudotyping efficiency (3). Monoclonal antibodies (MAbs) 4E10 and 2F5 (6, 25) were obtained from the International AIDS Vaccine Initiative Neutralizing Antibody Consortium Repository.
Mutagenesis.
Site-directed mutagenesis was performed using specific oligonucleotides and the QuikChange site-directed mutagenesis kit (Stratagene). The entire env gene was sequenced after each round of mutagenesis to ensure the presence of the desired mutations and the absence of any second-site mutations.
Env fusion kinetics.
The fusion kinetics of pre- and posttreatment Envs, as well as those of the various mutant Env proteins, were determined using a β-lactamase reporter cell-cell fusion assay as described previously (17, 28). Briefly, effector QT6 cells were cotransfected with Env and codon-optimized β-lactamase-expressing plasmids and were infected with a T7 polymerase-encoding vaccinia virus. At 4°C, effector cells were added to HeLa/CD4/CCR5 cells (JC53) loaded with CCF2-AM (an acetoxymethylester derivative of CCF2 that has a donor fluorophore [coumarin] linked to an acceptor [fluorescein] by a β-lactam ring), and then the temperature was shifted immediately to 37°C. Cell-cell fusion in this assay can be detected as a shift from green to blue fluorescence, indicating the cleavage of CCF2 by β-lactamase. Fluorescence was detected using a fluorometer (FLUOstar Optima; BMG Labtech), and results are expressed as the difference between the blue/green fluorescence obtained with Env-transfected cells and the background blue/green fluorescence obtained with empty-vector-transfected cells.
Statistical analyses.
Envelope differences for the fusion kinetics experiments were assessed using mixed-effect models (16), in which random effects were used to account for the correlation among the repeated measures on each replicate. These are similar to repeated-measures analysis-of-variance methods but are particularly useful for handling staggered time points and the presence of noninformative missing observations. Polynomial functions were used to model linear trajectories over time.
TAS.
To dissect the steps of Env fusion that are delayed by HR-1 mutations, we arrested fusion at an intermediate point termed the “temperature-arrested state” (TAS), defined previously (21). To arrest fusion at the TAS, we mixed effector and target cells (as described in “Env fusion kinetics” above) and incubated the cells at 23°C for 2.5 h. To assay fusion kinetics from the TAS, we then immediately shifted the assay plate to a fluorometer prewarmed to 37°C and commenced measurements of green and blue fluorescence.
Virus infection assays.
Luciferase reporter pseudotype viruses bearing patient-derived Envs and HR-1 and HR-2 mutant Envs were produced by cotransfection of 293T cells with the pNL-luc (Env− Vpr+) and gp160 expression plasmids, as previously described (4, 7). For neutralization assays, pseudotypes were normalized for p24 content, incubated with serial dilutions of the MAb, and spinoculated onto U87.CD4.CCR5 cells. Cells were then incubated at 37°C for 3 h, and the virus inoculum was replaced with fresh medium. The infection was analyzed by assaying for luciferase expression 3 days postinfection.
RESULTS
HR-2 mutations compensate for the HR-1-induced delay in fusion rates.
Resistance to ENF results mostly or entirely from mutations in the ENF binding site in the HR-1 region of gp41 (9, 12, 15, 18, 20, 22, 29, 30, 32). In a previous report, we characterized Env proteins derived from five treatment-experienced patients prior to ENF treatment and at a time on treatment after virologic failure (27). Envs from all five patients harbored mutations in HR-1, and in four of these patients, full ENF resistance could be conferred by HR-1 mutations alone. In addition, all Envs obtained from patients after virologic failure had one (usually) or two mutations in the HR-2 region of gp41. In one instance, full ENF resistance required a mutation in HR-2 (N126K) in addition to HR-1 changes, but for the other four patients, the HR-2 mutations had no obvious impact on ENF resistance. We therefore reasoned that HR-2 mutations, which are commonly observed in patient-derived ENF-resistant proteins (2, 19, 26, 31, 36), in some way impact Env protein function in a manner that usually does not affect ENF sensitivity.
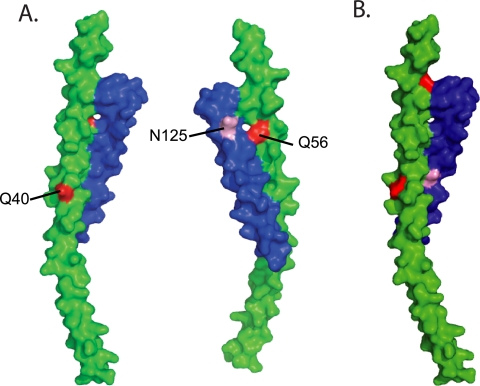
We chose to examine Env proteins derived from two patients, patients 3501 and 3518, both of whom initially responded to ENF therapy before virologic failure. For patient 3501, ENF resistance was associated with the acquisition of two mutations in HR-1 (Q40H and Q56R). In addition, a mutation in HR-2 (N125D) that did not affect ENF sensitivity was present in all Env clones (Fig. 1A) (27). For patient 3518, ENF resistance was accompanied by mutations N43D and Q66R, and a mutation in HR-2 (S138A) that did not obviously affect drug sensitivity was also present (Fig. 1B) (27). Since these HR-2 mutations did not affect sensitivity to ENF, we explored their impact on Env function and neutralization resistance in the presence and absence of their corresponding HR-1 mutations.
FIG. 1.
Structural mapping of the HR-1 and HR-2 mutations found in patients 3518 and 3501. The structure of the HIV-1 gp41 protein, showing the interaction between its two heptad repeat regions, is depicted. The HR-1 helix is shown in green, and HR-2 is shown in blue. The HR-1 and HR-2 mutations associated with ENF resistance in patients 3501 (A) and 3518 (B) are indicated in red and pink, respectively.
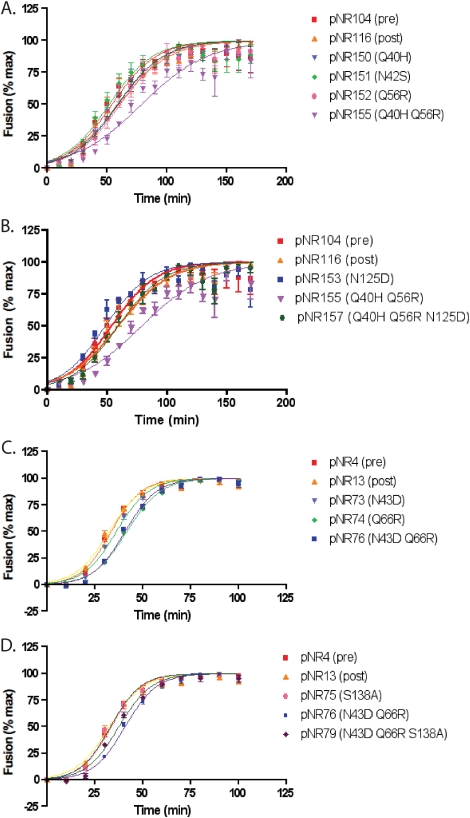
We have previously shown that HR-1 mutations commonly associated with ENF resistance can delay fusion kinetics in a cell-cell fusion assay when introduced into different env contexts (28). In addition, these mutations enhanced sensitivity to neutralization by MAbs that bind to epitopes in the membrane-proximal region of gp41. However, it is possible that ENF resistance mutations selected for in vivo impact Env function differently. To explore this, we examined the effects of our patient-derived HR-1 and HR-2 mutations in their native contexts on fusion rates by an assay in which cells expressing Env and β-lactamase are mixed with cells expressing CD4 and CCR5 and loaded with a fluorescent dye whose emission wavelength is changed upon the lactamase cleavage that follows cell-cell fusion. We found that the HR-1 mutations, when introduced into an Env obtained prior to ENF treatment, slowed fusion to a considerable extent (Fig. 2A to D; Table 1). For patient 3501, an Env protein (pNR155) bearing the HR-1 mutations Q40H and Q56R exhibited a delay of 19 min in the time to half-maximal fusion (t1/2max) relative to that for the pretreatment Env (pNR104), with high statistical significance (P = 0.0055) (Fig. 2A). The presence of the HR-2 mutation (N125D) in addition to these HR-1 mutations (pNR157) restored the kinetics of Env fusion to a rate comparable to those of the reference posttreatment Env (pNR116) and the pretreatment control (pNR104) (Fig. 2B).
FIG. 2.
Fusion kinetics of HR-1 and HR-2 mutant panels. The kinetics of fusion of two panels of patient-derived HR-1 (A and C) or HR-1 and HR-2 (B and D) mutant Envs were determined by a β-lactamase cell-cell fusion assay. The results for mutant panels derived from two patients, patient 3501 (A and B) and patient 3518 (C and D), are shown. Fusion is expressed as a percentage of the maximal fusion mediated by each Env. Results are averages ± standard errors of the means for at least three independent experiments. The kinetic parameters are described in Table 1.
TABLE 1.
Kinetic parameters of HR-1 and HR-2 mutant Env-mediated cell-cell fusiona
| Patient and envelope protein | t1/2max (min) | b |
|---|---|---|
| Patient 3501 | ||
| pNR104 (pre) | 47.7 ± 1.89 | 13.05 ± 1.64 |
| pNR116 (post) | 53.55 ± 1.83 | 15.69 ± 1.58 |
| pNR150 (Q40H) | 51.9 ± 1.87 | 13.66 ± 1.61 |
| pNR151 (N42S) | 44.83 ± 1.76 | 12.1 ± 1.53 |
| pNR152 (Q56R) | 56.18 ± 1.72 | 15.04 ± 1.48 |
| pNR153 (N125D) | 41.54 ± 1.68 | 11.56 ± 1.46 |
| pNR154 (Q40H N125D) | 39.5 ± 1.29 | 9.99 ± 1.12 |
| pNR155 (Q40H Q56R) | 66.48 ± 2.71 | 17.01 ± 2.29 |
| pNR156 (Q56R N125D) | 52.24 ± 2.31 | 14.09 ± 1.99 |
| pNR157 (Q40H Q56R N125D) | 54.6 ± 1.73 | 14.9 ± 1.49 |
| Patient 3518 | ||
| pNR4 (pre) | 32.72 ± 0.62 | 7.25 ± 0.53 |
| pNR13 (post) | 30.78 ± 0.6145 | 7.25 ± 0.53 |
| pNR73 (N43D) | 35.12 ± 0.5063 | 7.54 ± 0.44 |
| pNR74 (Q66R) | 41.01 ± 0.4936 | 8.28 ± 0.42 |
| pNR75 (S138A) | 32.06 ± 0.5541 | 7.03 ± 0.48 |
| pNR76 (N43D Q66R) | 39.85 ± 0.5678 | 7.59 ± 0.49 |
| pNR77 (N43D S138A) | 35.22 ± 1.309 | 7.09 ± 1.12 |
| pNR78 (Q66R S138A) | 34.99 ± 0.7469 | 6.15 ± 0.64 |
| pNR79 (N43D Q66R S138A) | 36.1 ± 0.7047 | 7.43 ± 0.60 |
The kinetic parameters of cell-cell fusion mediated by HR-1 and HR-2 mutant Env proteins for patients 3501 and 3518 were determined by a β-lactamase reporter assay (see Fig. 2). Fusion was assayed from 0 to 170 min (patient 3501) or from 0 to 100 min (patient 3518) in at least three independent experiments, and data were fitted to the equation Y = Ymax/{1 + exp[−(t − t1/2max)/b]}. pre, pretreatment; post, posttreatment; b, exponential rate constant.
The delay in fusion rates was more modest in the panel of Envs derived from patient 3518 (Fig. 2C and D). The HR-1 mutations (N43D and Q66R, in pNR76) slowed fusion by 7 min relative to that for the pretreatment Env (pNR4). However, this difference was very consistent and reached high statistical significance (P = 0.0005). Addition of the corresponding HR-2 mutation (S138A, in pNR79) observed in ENF-resistant Env proteins obtained from this patient restored fusion rates to levels that were not statistically distinguishable from that for the pretreatment pNR4 control (Fig. 2D). Thus, in two instances, we found that HR-2 mutations that were associated with ENF resistance functioned to restore fusion rates that would otherwise have been diminished as a result of HR-1 mutations that directly impact ENF binding.
Fusion kinetics of HR-1 and HR-2 mutants from a TAS.
Elegant studies aimed at understanding the detailed steps involved in HIV-1 membrane fusion have demonstrated that HIV-1 fusion can be arrested at a stage following CD4 and coreceptor binding but prior to membrane fusion (21, 23). This step in the fusion pathway, termed the TAS, can be achieved by incubating effector and target cells in a fusion assay at 23°C for 2.5 h. When the temperature is raised to 37°C from the TAS, fusion occurs within a few minutes, a much higher rate than that for cells that are bound and allowed to fuse at a constant 37°C. Fusion arrested at the TAS (after 2.5 h) is only marginally susceptible to coreceptor inhibitors, suggesting that the majority of Env proteins have already bound the coreceptor (23). In contrast, fusion following incubation at the TAS is fully susceptible to ENF (added at 23°C) with or without drug washout prior to the shift to 37°C, indicating that the HR-1 region of Env is exposed at the TAS and is not bound to HR-2 (21).
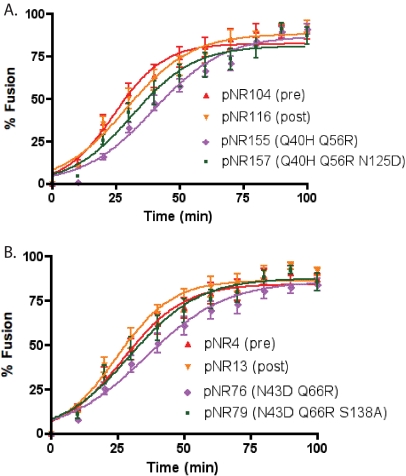
To determine the effects of HR-1 and HR-2 mutations on fusion kinetics from a step of fusion that is the conformational intermediate targeted by ENF, we performed our cell-cell fusion kinetics assay after arresting effector/target cells at the TAS. As expected, the lag phase seen prior to cell fusion from the TAS was significantly shorter than that under our normal conditions (compare Fig. 3 with Fig. 2). However, we saw the same relative trends in the kinetics of Env fusion from the TAS as we had observed under normal conditions. In both instances, the HR-1 mutations by themselves delayed fusion relative to that for the pre- and posttreatment Env controls upon release from the TAS by a shift to 37°C. Fusion rates were restored to near-control levels upon the addition of the HR-2 mutations (Fig. 3A and B; Table 2). These results indicate that HR-2 mutations can compensate for the delay in fusion kinetics that would otherwise be caused by HR-1 mutations alone and that this occurs after CD4 and coreceptor binding. In light of this, we speculate that HR-2 mutations accelerate the formation of the six-helix bundle that is required for the membrane fusion reaction.
FIG. 3.
Fusion kinetics of HR-1 and HR-2 mutants from a TAS. The fusion kinetics of panels of patient-derived HR-1 and HR-2 mutant Envs were determined from a TAS by a β-lactamase cell-cell fusion assay. The TAS was established by incubating effector and target cells at 23°C for 2.5 h and then raising the temperature to 37°C and commencing measurements of fusion. The results for mutant panels derived from two patients, patient 3501 (A) and patient 3518 (B), are shown. Fusion is expressed as a percentage of the maximal fusion mediated by each Env. Results are averages ± standard errors of the means for at least three independent experiments. The kinetic parameters are described in Table 2.
TABLE 2.
Kinetic parameters of HR-1 and HR-2 mutant Env-mediated cell-cell fusion from a TASa
| Patient and envelope protein | t1/2max (min) | b |
|---|---|---|
| Patient 3501 | ||
| pNR104 (pre) | 24.84 ± 2.21 | 9.64 ± 1.98 |
| pNR116 (post) | 29.38 ± 1.58 | 12.78 ± 1.43 |
| pNR155 (Q40H Q56R) | 40.18 ± 1.50 | 13.76 ± 1.33 |
| pNR157 (Q40H Q56R N125D) | 31.91 ± 2.03 | 11.99 ± 1.81 |
| Patient 3518 | ||
| pNR4 (pre) | 27.46 ± 1.67 | 11.33 ± 1.50 |
| pNR13 (post) | 25.12 ± 1.50 | 10.39 ± 1.35 |
| pNR76 (N43D Q66R) | 35.74 ± 1.66 | 15.11 ± 1.50 |
| pNR79 (N43D Q66R S138A) | 29.94 ± 1.68 | 12.99 ± 1.53 |
The kinetic parameters of cell-cell fusion mediated by HR-1 and HR-2 mutant Envs for patients 3501 and 3518, starting from a TAS, were determined by a β-lactamase reporter assay (Fig. 3). Fusion was assayed from 0 to 100 min in at least three independent experiments, and the data were fitted to the equation Y = Ymax/{1 + exp[−(t − t1/2max)/b]}. pre, pretreatment; post, posttreatment; b, exponential rate constant.
Mutations associated with clinical ENF resistance do not affect neutralization sensitivity.
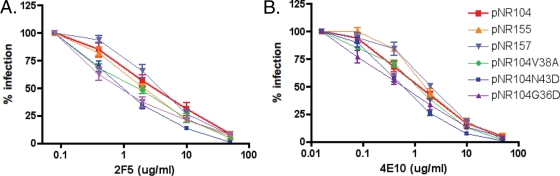
HR-1 mutations associated with ENF resistance can, when introduced into heterologous Envs, not only delay fusion kinetics but also lead to enhanced sensitivity to neutralization by broadly cross-reactive HIV antibodies that bind to the membrane-proximal region of gp41 (28). Whether this occurs in vivo as well, where drug resistance evolves in the face of the humoral immune response, is not known. To examine this, the effects of our patient-derived HR-1 and HR-2 mutations on neutralization sensitivity in their native Env contexts were studied using a virus pseudotype neutralization assay. Viral pseudotypes were produced bearing pretreatment or posttreatment Envs, as well as pretreatment Envs into which HR-1 mutations were introduced alone or in combination with their corresponding HR-2 mutations. Virus stocks were normalized for p24 content and were then used to infect U87 cells expressing CD4 and CCR5 in the presence or absence of the broadly neutralizing MAb 2F5 or 4E10. We found that the neutralization profiles of all viruses were similar: the HR-1 mutations had no discernible effect on the abilities of these antibodies to neutralize the virus (Fig. 4).
FIG. 4.
Susceptibilities of patient Envs and HR-1 and HR-2 mutant viruses to neutralizing antibodies. Shown is the neutralization of HR-1 and HR-2 mutant pseudotype virus infections by the neutralizing MAbs 2F5 (A, C, and E) and 4E10 (B, D, and F). Results are expressed as percentages of the level of Env-mediated infection in the absence of MAbs and are averages and standard errors of the means for at least three independent experiments.
Env context-dependent effects of HR-1 and HR-2 mutations.
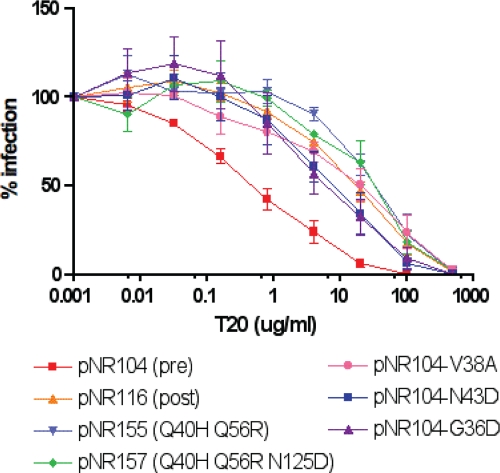
To study the context-dependent effects of HR-1 and HR-2 mutations, we constructed a panel of mutants in which we introduced into a pretreatment env clone from a given patient (pNR104 from patient 3501) either HR-1 mutations observed in another patient from our study (patient 3518) or other mutations commonly associated with ENF resistance. Specifically, we introduced the G36D, V38A, N43D, and Q66R mutations into the pNR104 Env background, produced virus pseudotypes, and performed infection assays in the presence of increasing concentrations of ENF. While all of these mutations increased resistance to ENF by 1 to 2 log units, the mutations that arose naturally (Q40H and Q56R) in this patient (patient 3501) had the greatest impact on ENF resistance (Fig. 5).
FIG. 5.
Susceptibilities of HR-1 swap mutants to ENF. The ENF sensitivities of HR-1 mutant Env proteins were determined in pseudotype virus infection inhibition assays. Results are expressed as percentages of the level of infection mediated by wild-type Env in the absence of ENF and are averages and standard errors of the means for at least three independent experiments.
We next performed pseudotype neutralization assays to assess the effects of these mutations on sensitivity to the broadly neutralizing antibodies 2F5 and 4E10 (Fig. 6A and B). As shown above, the naturally arising mutations of patient 3501, Q40H and Q56R, did not affect neutralization sensitivity. The V38A mutation also did not affect neutralization sensitivity. However, the G36D and N43D mutations rendered pNR104 reproducibly more susceptible to neutralization, albeit to a modest extent. These findings suggest that while HR-1 mutations generally impart resistance to ENF, their impacts on ENF fusion rates and on sensitivity to antibody-mediated neutralization are variable and context dependent.
FIG. 6.
Susceptibilities of HR-1 swap mutants to neutralizing antibodies. Shown is the neutralization of HR-1 and HR-2 swap mutant pseudotype virus infections by the neutralizing MAbs 2F5 (A) and 4E10 (B). Results are expressed as percentages of the level of Env-mediated infection in the absence of MAbs and are averages and standard errors of the means for at least three independent experiments.
DISCUSSION
The HIV-1 Env protein exists as a metastable structure that must be triggered to undergo the conformational changes needed to elicit membrane fusion. Sequential binding to CD4 and a coreceptor serves as the trigger needed for this process, ultimately resulting in interactions between the HR-1 and HR-2 regions of gp41 that lead to the formation of a six-helix bundle that is essential for membrane fusion induction (11, 21). ENF prevents this interaction and so provides a strong selective pressure for mutations in its binding site in the HR-1 domain (5, 10). However, mutations in HR-1 that prevent ENF binding are likely to impact the binding of the HR-2 domain as well, reducing the efficiency of membrane fusion (9, 12, 15, 18, 20, 22, 27, 29, 30, 32). Indeed, when mutations that confer ENF resistance (19) are introduced into HR-1, membrane fusion kinetics are delayed, there is enhanced sensitivity to neutralizing antibodies that bind to the membrane-proximal region in gp41 (28), and virus fitness is reduced as measured by in vitro assays (19). The fitness of ENF-resistant mutants in vivo is also reduced, since ENF-sensitive virus strains reemerge after the discontinuation of ENF therapy.
In vivo, the impact of ENF resistance-conferring mutations on fusion efficiency can be minimized in two obvious ways. First, compensatory mutations that mitigate the effects of mutations in HR-1 may arise. Second, a variety of mutations can confer ENF resistance on any given HIV-1 strain. Thus, there would likely be selection for mutations that in a given Env context have relatively modest effects on viral fitness while maximizing drug resistance (15). With regard to compensatory mutations, amino acid alterations in HR-2 are frequently observed in Envs cloned from individuals who have received ENF therapy only to have subsequent virologic failure (8, 31, 36). When introduced by themselves, such HR-2 mutations typically have either no impact or only a modest impact on ENF sensitivity (27, 36). In some instances, HR-2 mutations have been shown to enhance the stability of the six-helix bundle in either structural-modeling or peptide binding studies (1, 14). By using a variety of in vitro assays, we were able to probe the functional consequences of HR-2 mutations either in isolation or in conjunction with their corresponding HR-1 mutations that evolved in ENF-treated patients. In the panel of Env mutants studied here, uncompensated HR-1 mutations delayed fusion kinetics to variable extents. Fusion rates were restored to wild-type or near-wild-type levels when the corresponding HR-2 mutations were introduced. More specifically, the impact of HR-1 and HR-2 mutations on fusion rates occurred after Env had reached the TAS. Under these conditions, CD4 binding and coreceptor binding occur, but six-helix bundle formation is prevented. Thus, these findings are most consistent with HR-2 mutations having a direct role in mediating HR-1-HR-2 interactions and overcoming the deleterious effects that HR-1 mutations can have on membrane fusion rates.
The rate at which membrane fusion occurs could impact viral fitness in several ways. Once bound to the cell surface, the virus will likely be internalized and degraded or inactivated at some rate. Virus strains that fuse more slowly than other strains might be inactivated before undergoing the membrane fusion process. Since membrane fusion is likely to be a cooperative process involving not only all three subunits in an Env trimer, but perhaps several Env trimers as well, delayed fusion kinetics could make the entry process more asynchronous and less efficient. Finally, mutations that weaken HR-1-HR-2 interactions could increase the likelihood that Env will undergo conformational changes that cause inactivation rather than membrane fusion.
Delayed fusion kinetics have been associated with enhanced sensitivity to virus neutralization by some, but not all, well-characterized broadly neutralizing antibodies. We have interpreted this finding to mean that some neutralizing determinants, such as those in the membrane-proximal region of gp41, are more accessible to antibody binding in conformational intermediates of the fusion process. Thus, delayed fusion kinetics would result in prolonged, enhanced exposure of some neutralizing determinants. However, our results indicate that the impact of HR-1 mutations on neutralization sensitivity is context dependent. In contrast to the findings of our previous study, in which mutations linked to ENF resistance were placed in heterologous Env backgrounds (28), we found that HR-1 mutations selected for in vivo, and in their native Env contexts, had no discernible impact on the sensitivity of the virus to neutralization. When these mutations were introduced into a different Env background, some were associated with increased sensitivity to antibody-mediated neutralization. Thus, the evolution of ENF resistance is likely a complex balancing act: mutations must confer a sufficient degree of resistance to the drug while at the same time minimizing the consequences for membrane fusion kinetics and sensitivity to neutralization by antibodies.
Acknowledgments
We thank S. Ratcliffe for help with statistical analyses, C. Tilton for assistance with structural modeling, and G. Melikyan for informative discussions on the TAS experiments.
This work was supported by NIH grants F32-AI068442-01 to N.R. and AI-40880 to R.W.D.
Footnotes
Published ahead of print on 19 January 2009.
REFERENCES
- 1.Bai, X., K. L. Wilson, J. E. Seedorff, D. Ahrens, J. Green, D. K. Davison, L. Jin, S. A. Stanfield-Oakley, S. M. Mosier, T. E. Melby, N. Cammack, Z. Wang, M. L. Greenberg, and J. J. Dwyer. 2008. Impact of the enfuvirtide resistance mutation N43D and the associated baseline polymorphism E137K on peptide sensitivity and six-helix bundle structure. Biochemistry. doi: 10.1021/bi702509d. [DOI] [PubMed]
- 2.Baldwin, C. E., R. W. Sanders, Y. Deng, S. Jurriaans, J. M. Lange, M. Lu, and B. Berkhout. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 7812428-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, C. B., X. Shen, M. A. Egan, T. C. Pierson, C. M. Walker, and R. F. Siliciano. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, B. K., K. Saksela, R. Andino, and D. Baltimore. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 68654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C. H., T. J. Matthews, C. B. McDanal, D. P. Bolognesi, and M. L. Greenberg. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 693771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y. H., and M. P. Dierich. 1996. Identification of a second site in HIV-1 gp41 mediating binding to cells. Immunol. Lett. 52153-156. [DOI] [PubMed] [Google Scholar]
- 7.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206935-944. [DOI] [PubMed] [Google Scholar]
- 8.Eshleman, S. H., S. E. Hudelson, R. Bruce, T. Lee, M. R. Owens, J. Hackett, P. Swanson, S. G. Devare, and N. Marlowe. 2007. Analysis of HIV type 1 gp41 sequences in diverse HIV type 1 strains. AIDS Res. Hum. Retrovir. 231593-1598. [DOI] [PubMed] [Google Scholar]
- 9.Fikkert, V., P. Cherepanov, K. Van Laethem, A. Hantson, B. Van Remoortel, C. Pannecouque, E. De Clercq, Z. Debyser, A. M. Vandamme, and M. Witvrouw. 2002. env chimeric virus technology for evaluating human immunodeficiency virus susceptibility to entry inhibitors. Antimicrob. Agents Chemother. 463954-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5276-279. [DOI] [PubMed] [Google Scholar]
- 11.Gallo, S. A., A. Puri, and R. Blumenthal. 2001. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 4012231-12236. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg, M. L., and N. Cammack. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54333-340. [DOI] [PubMed] [Google Scholar]
- 13.He, Y., R. Vassell, M. Zaitseva, N. Nguyen, Z. Yang, Y. Weng, and C. D. Weiss. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 771666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenwitheesuk, E., and R. Samudrala. 2005. Heptad-repeat-2 mutations enhance the stability of the enfuvirtide-resistant HIV-1 gp41 hairpin structure. Antivir. Ther. 10893-900. [PubMed] [Google Scholar]
- 15.Labrosse, B., L. Morand-Joubert, A. Goubard, S. Rochas, J. L. Labernardiere, J. Pacanowski, J. L. Meynard, A. J. Hance, F. Clavel, and F. Mammano. 2006. Role of the envelope genetic context in the development of enfuvirtide resistance in human immunodeficiency virus type 1-infected patients. J. Virol. 808807-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laird, N. M., and J. H. Ware. 1982. Random-effects models for longitudinal data. Biometrics 38963-974. [PubMed] [Google Scholar]
- 17.Lineberger, J. E., R. Danzeisen, D. J. Hazuda, A. J. Simon, and M. D. Miller. 2002. Altering expression levels of human immunodeficiency virus type 1 gp120-gp41 affects efficiency but not kinetics of cell-cell fusion. J. Virol. 763522-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, J., S. G. Deeks, R. Hoh, G. Beatty, B. A. Kuritzkes, J. N. Martin, and D. R. Kuritzkes. 2006. Rapid emergence of enfuvirtide resistance in HIV-1-infected patients: results of a clonal analysis. J. Acquir. Immune Defic. Syndr. 4360-64. [DOI] [PubMed] [Google Scholar]
- 19.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. R. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 784628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews, T., M. Salgo, M. Greenberg, J. Chung, R. DeMasi, and D. Bolognesi. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3215-225. [DOI] [PubMed] [Google Scholar]
- 21.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mink, M., M. Greenberg, and S. Mosier. 2002. Impact of HIV-1 gp41 amino acid substitutions (position 36-45) on susceptibility to T20 (Enfuvirtide) in vitro; analysis of primary virus isolates recovered from patients during chronic Enfuvirtide treatment and site-directed mutants in NL4-3. Antivir. Ther. 7S17-S18. [Google Scholar]
- 23.Mkrtchyan, S. R., R. M. Markosyan, M. T. Eadon, J. P. Moore, G. B. Melikyan, and F. S. Cohen. 2005. Ternary complex formation of human immunodeficiency virus type 1 Env, CD4, and chemokine receptor captured as an intermediate of membrane fusion. J. Virol. 7911161-11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, J. P., and R. W. Doms. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. USA 10010598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 676642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Alvarez, L., R. Carmona, A. Ocampo, A. Asorey, C. Miralles, S. Perez de Castro, M. Pinilla, G. Contreras, J. A. Taboada, and R. Najera. 2006. Long-term monitoring of genotypic and phenotypic resistance to T20 in treated patients infected with HIV-1. J. Med. Virol. 78141-147. [DOI] [PubMed] [Google Scholar]
- 27.Ray, N., J. E. Harrison, L. A. Blackburn, J. N. Martin, S. G. Deeks, and R. W. Doms. 2007. Clinical resistance to enfuvirtide does not affect susceptibility of human immunodeficiency virus type 1 to other classes of entry inhibitors. J. Virol. 813240-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves, J. D., F.-H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 794991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sista, P. R., T. Melby, D. Davison, L. Jin, S. Mosier, M. Mink, E. L. Nelson, R. DeMasi, N. Cammack, M. P. Salgo, T. J. Matthews, and M. L. Greenberg. 2004. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS 181787-1794. [DOI] [PubMed] [Google Scholar]
- 31.Su, C., T. Melby, R. DeMasi, P. Ravindran, and G. Heilek-Snyder. 2006. Genotypic changes in human immunodeficiency virus type 1 envelope glycoproteins on treatment with the fusion inhibitor enfuvirtide and their influence on changes in drug susceptibility in vitro. J. Clin. Virol. 36249-257. [DOI] [PubMed] [Google Scholar]
- 32.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387426-430. [DOI] [PubMed] [Google Scholar]
- 34.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 8910537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 919770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, L., A. Pozniak, A. Wildfire, S. A. Stanfield-Oakley, S. M. Mosier, D. Ratcliffe, J. Workman, A. Joall, R. Myers, E. Smit, P. A. Cane, M. L. Greenberg, and D. Pillay. 2005. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob. Agents Chemother. 491113-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]