Abstract
Entry of ebolavirus (EBOV) into cells is mediated by its glycoprotein (GP1,2), a class I fusion protein whose structure was recently determined (J. E. Lee et al., Nature 454:177-182, 2008). Here we confirmed two major predictions of the structural analysis, namely, the residues in GP1 and GP2 that remain after GP1,2 is proteolytically primed by endosomal cathepsins for fusion and residues in GP1 that are critical for binding to host cells. Mass spectroscopic analysis indicated that primed GP1,2 contains residues 33 to 190 of GP1 and all residues of GP2. The location of the receptor binding site was determined by a two-pronged approach. We identified a small receptor binding region (RBR), residues 90 to 149 of GP1, by comparing the cell binding abilities of four RBR proteins produced in high yield. We characterized the binding properties of the optimal RBR (containing GP1 residues 57 to 149) and then conducted a mutational analysis to identify critical binding residues. Substitutions at four lysines (K95, K114, K115, and K140) decreased binding and the ability of RBR proteins to inhibit GP1,2-mediated infection. K114, K115, and K140 lie in a small region modeled to be located on the top surface of the chalice following proteolytic priming; K95 lies deeper in the chalice bowl. Combined with those of Lee et al., our findings provide structural insight into how GP1,2 is primed for fusion and define the core of the EBOV RBR (residues 90 to 149 of GP1) as a highly conserved region containing a two-stranded β-sheet, the two intra-GP1 disulfide bonds, and four critical Lys residues.
Ebolaviruses (EBOVs) are filamentous, enveloped, negative-strand RNA viruses of the family Filoviridae; there are five reported species, namely, Zaire (ZEBOV), Sudan, Reston, Côte d'Ivoire, and Bundibugyo EBOVs (7, 28, 31). EBOV infections cause severe hemorrhagic fever in humans and nonhuman primates, with mortality rates reaching 88% (12, 20).
Entry of EBOV into host cells is initiated through its surface glycoprotein (GP1,2), a heavily glycosylated class I viral fusion protein that is cleaved during transit to the cell surface by a furin-like protease into a receptor binding subunit (GP1) and a fusion subunit (GP2). GP1 and GP2 remain associated through a disulfide bond and, like all class I fusion proteins, assemble as trimers on the virion surface (6, 8, 9, 26, 29, 30, 32). Unlike most class I fusion proteins, however, cleavage at the furin site is not essential for infection (22, 33). EBOV GP1,2-mediated entry into host cells involves binding to one or more unknown host receptors followed by delivery to an endosomal compartment (4, 19, 21, 26), where cathepsins B and L cleave GP1 from its initial 130-kDa form to a 19-kDa intermediate which is primed for fusion (19-kDa GP1) (21). Many aspects of the EBOV entry process remain obscure. For example, although several cell surface proteins have been shown to enhance infection (5), no definitive host cell surface receptor(s) has been identified. In addition, neither the sequence of 19-kDa GP1, the residues within 19-kDa GP1 that are critical for binding to host cells, nor the mechanism by which the primed EBOV glycoprotein (19-kDa GP1,2 [19-kDa GP1 disulfide bonded to GP2]) is triggered for fusion (4, 21) has yet been elucidated.
The focus of this study is the 19-kDa form of EBOV GP1 and its role in binding to host cells. Although the mucin-like domain at the C-terminal end of GP1 can facilitate initial virus attachment to cells (1, 16, 18, 23, 25), this domain is fully dispensable for entry. Moreover, 19-kDa GP1 clearly contains receptor binding activity, since virus binding and infection are actually enhanced after the mucin-like domain is removed (GP1,2Δ) (11, 21). Additionally, 19-kDa GP1 most likely includes residues from the amino-terminal portion of GP1, since mutations within the first 150 residues (residues 33 to 183) impair virus infection (3, 13, 15, 17) and since a recombinant protein including residues 54 to 201 binds specifically to permissive cells and inhibits EBOV GP1,2-mediated infection (13). Cys 53 should also be present, since 19-kDa GP1 remains attached to GP2 by a disulfide bond (4; data not shown).
In this study, we first purified the primed ZEBOV 19-kDa GP1,2 and identified its sequence by mass spectroscopy. We then generated several recombinant GP1 receptor binding regions (RBRs), including one encompassing the residues that we found to be present in 19-kDa GP1. Using a high-yield recombinant RBR, we characterized properties of RBR binding to host cells, identified an RBR that could inhibit ZEBOV GP1,2-mediated pseudoviral infection, and importantly, identified residues within the RBR that are critical for host cell binding. Our findings are fully consistent with and are discussed in terms of the recently solved prefusion structure of the EBOV glycoprotein trimer (14).
MATERIALS AND METHODS
Cells and cell culture conditions.
293T cells, a derivative of 293 cells (ATCC CRL1573), were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS), 1× penicillin-streptomycin (Gibco), and 500 μg/ml active G418 (Gibco). Jurkat cells (ATCC TIB-152) were maintained in RPMI 1640 medium with 10% FBS, 1× penicillin-streptomycin, and 1 mM sodium pyruvate (Gibco). Vero E6 cells (ATCC CRL-1586) were maintained in DMEM supplemented with 10% FBS and 1× penicillin-streptomycin.
Production and purification of ZEBOV pseudovirions.
Green fluorescent protein (GFP)-carrying vesicular stomatitis virus (VSVgfp) bearing ZEBOV GP1,2Δ or the VSV glycoprotein (G) was produced in BHK-21 cells essentially as described previously (21, 26, 27). The mucin-deleted ZEBOV GP1,2 was utilized throughout this study, since deletion of the mucin-like domain has no effect on entry or cell tropism (10, 24, 34), the viral titer of pseudovirions with GP1,2Δ is increased ∼10-fold compared to that of the wild type (WT), and 19-kDa GP1 can be produced equally well from pseudotypes with either full-length GP1,2 or GP1,2Δ (11; data not shown). For purification of primed 19-kDa GP1,2, a virus-containing supernatant was cleared by centrifugation at 2,500 rpm for 5 min at 4°C, concentrated by centrifugation through a Vivaspin-20 300-kDa-molecular-size-cutoff column (Sartorius Biolabs), and then banded on a step gradient of 25% and 60% sucrose (wt/vol) in HM buffer (20 mM HEPES, 20 mM morpholineethanesulfonic acid, 130 mM NaCl, pH 7.5) by centrifugation in an SW41 rotor at 21,000 rpm for ∼16 h at 4°C. The virus-containing band was then isolated, diluted to ∼20% sucrose in HM buffer, pelleted by centrifugation at 32,000 rpm in an SW55 rotor for 2 h at 4°C, and resuspended in approximately 500 μl of 10% sucrose in HM buffer.
Beta-lactamase-carrying human immunodeficiency virus (HIVblam) bearing ZEBOV GP1,2Δ or VSV G was produced in 293T cells as previously described (35). Briefly, a plasmid encoding mucin-deleted ZEBOV GP1,2 or VSV G was transfected along with plasmids pMM310 and pΔ8.2 into 293T cells with Fugene (Roche). After 24 h, cells were treated with 2 mM sodium butyrate and further incubated for 24 h prior to harvest of the virus-containing supernatant. The supernatant was cleared by centrifugation at 2,500 rpm for 5 min at 4°C, and the pseudovirions were pelleted by centrifugation through 20% sucrose (wt/vol) in HM buffer in an SW41 rotor at 21,000 rpm for 2 h at 4°C and resuspended in 200 μl of 10% sucrose in HM buffer.
Generation and purification of 19-kDa GP1,2.
To generate 19-kDa GP1,2, a sample of step-gradient-purified VSVgfp-GP1,2Δ pseudovirions (0.25 mg/ml) was incubated with thermolysin (0.25 mg/ml) in HM buffer containing 0.1 mM CaCl2 for 30 min at 37°C. The reaction was stopped by adding EDTA to a final concentration of 10 mM, and the mixture was centrifuged in an SW55 rotor at 33,000 rpm for 2 h at 4°C. The viral pellet was resuspended in 200 μl of HM buffer and disrupted by incubation in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, pH 7.5) at 37°C for 15 min. The detergent-disrupted particles were centrifuged in a TLA 100 rotor for 1 h at 32,000 rpm at 4°C to remove VSV cores (VSV L, N, and P proteins) and a fraction of the VSV M protein. The resulting supernatant, containing 19-kDa GP1-S-S-GP2, was then depleted of remaining VSV M protein by incubation with NHS-Fast Flow Sepharose beads (Pharmacia Biotech) to which the anti-VSV M monoclonal antibody 23H2 (a gift of Michael Whitt) had been covalently coupled following the manufacturer's instructions. The resulting sample was then concentrated by chloroform-methanol precipitation, resuspended in 80 μl of peptide n-glycosidase F digestion buffer, treated with 500 units of PNGase F (New England Biolabs) for 1 h at 37°C, and run in two lanes (∼20 μl per lane) of a 12.5% reducing sodium dodecyl sulfate (SDS) gel. The gel was stained with silver, and the region containing 19-kDa GP1 (17 kDa after PNGase F treatment) was excised and digested with either trypsin or ArgC. The resulting peptide fragments were analyzed by mass spectrometry (W. M. Keck Biomedical MS Core Facility, University of Virginia).
Construction of recombinant ZEBOV GP1 RBRs.
The protein sequence for ZEBOV GP1,2 (PDB accession no. U31033) was subjected to secondary structure predictions, using each of the secondary structure prediction programs available at http://www.expasy.org/tools/#secondary. The outputs were analyzed for regions corresponding or not corresponding to secondary structure elements. Regions outside predicted helices and strands were considered possible loops. We chose residues in the predicted loop regions, including three N-terminal residues (residues 57, 72, and 90) and three C-terminal residues (residues 149, 172, and 198), as the termini for recombinant RBRs, reasoning that truncations within loop segments would be least disruptive to GP1 structure and function. We also generated an RBR encompassing residues 54 to 201, essentially as described previously (11) (see Fig. 2A). Primers were designed for initiation or termination of the GP1 fragments at the chosen sites with an upstream or downstream BglII site, respectively. Primers for additional fragments starting at the N terminus of mature GP1 (residue 33, the first residue after signal sequence removal) and terminating at residue 149, 193 (a predicted C terminus of 19-kDa GP1 [see Results]), or 198 were also designed (see Fig. 2A). PCR products were then prepared using plasmid VRC6001 (a gift of Gary Nabel, NIH) as a template, purified, and inserted into the BglII site of pFUSE-rFc2 (Invivogen). pFUSE contains an interleukin-2 signal sequence, a multiple cloning site, and a rabbit immunoglobulin G Fc domain, allowing for the expression of secreted fusion proteins with a C-terminal rabbit Fc tag. GP1 fragments inserted into pFUSE-rFc2 in the correct orientation were selected by restriction digest analysis, and correct products were further confirmed by DNA sequence analysis.
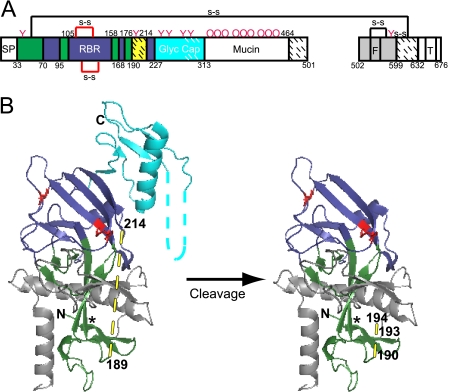
FIG. 2.
Model of primed 19-kDa ZEBOV GP1,2. (A) Domain architecture of EBOV GP1,2 (based on nomenclature and color coding in reference 14). The GP1 signal peptide (SP) (white; residues 1 to 32); base (green; residues 33 to 70, 96 to 105, 159 to 168, and 177 to 189), head (dark blue; residues 71 to 95, 106 to 158, 169 to 176, and 215 to 227), linker region (site of cathepsin B/L and thermolysin cleavage) (hatched yellow; residues 190 to 213), glycan cap (cyan; residues 228 to 313), mucin-like domain (white; residues 314 to 464), and C-terminal domain (hatched white; residues 465 to 501) are shown. The GP2 fusion peptide (F) and transmembrane domain (T) are labeled. Domains not present in the protein used for crystallography (14) are shown in white, and regions present but unresolved in the crystal structure are shown with hatches. Pink Ys and pink Os represent sites of N- and O-glycosylation. (B) Native structure of GP1,2Δ (left) and model of primed 19-kDa GP1,2 (right), based on Fig. 2 of reference 14 and colored as in panel A. Cysteine residues involved in disulfide bonds are in red, and predicted loop regions are shown with dashed lines. The asterisk (*) represents a contact point between the β1 and β13 strands (see the text for more details). Potential cathepsin B and cathepsin L cleavage sites (residues 190, 193, and 194) are labeled in the model (right). The graphic representations are based on PDB file 3CSY (14) and were produced with Pymol. Note that the depiction of primed GP1,2 in panel B (right) and in Fig. 7A, C, and D is strictly a model that assumes no conformational changes following proteolytic priming.
Expression of recombinant ZEBOV GP1 RBR-Fc proteins.
293T cells (1 to 15 dishes; 150 × 25 mm) were transfected with plasmids encoding ZEBOV GP1 RBR-Fc proteins by use of polyethylenimine as described previously (2). Briefly, cells (∼75% confluent) were washed in PBS++ (phosphate-buffered saline [PBS] containing Ca2+ and Mg2+) and then incubated in serum-free DMEM (18 ml/plate) with 1× penicillin-streptomycin and G418 (500 μg/ml). A mixture containing plasmid DNA (35 μg), polyethylenimine (105 μg), and 2 ml Optimem (Gibco) was added to each dish. Cell supernatants were harvested after 36 h, and cell debris was removed by centrifugation (2,500 rpm for 10 min at 4°C) and filtration through a 0.22-μm-pore-size filter (Millipore). Cleared supernatants were applied to a column (0.5 ml) of protein A-agarose beads (Roche) at 4°C by slow drip (∼18 h), using a peristaltic pump. The beads were washed with 50 bed volumes of PBS and eluted with 10 bed volumes of 50 mM sodium citrate-50 mM glycine at pH 2.5, neutralized with 2 M Tris, and concentrated in a Vivaspin 500 microcentrifuge tube (Argos). Purified proteins were analyzed for Fc concentration by enzyme-linked immunosorbent assay (Bethyl Laboratories) and for total protein concentration by either bicinchoninic acid assay (Pierce) or by measuring the absorbance at 280 nm (Nanodrop spectrophotometer; Thermo Scientific). Protein purity and size were further assessed by reducing SDS-polyacrylamide gel electrophoresis followed by Western blotting with peroxidase-conjugated protein A (Cappel) diluted 1:1,000 or by silver staining.
Cell binding assays.
Adherent cells (293T and Vero E6) were detached from their dishes with PEEG buffer (5 mM EDTA, 5 mM EGTA, 10 mM glucose in PBS) and diluted with an equal volume of PBS++. Jurkat cells were harvested by pipetting. Samples were kept at 4°C throughout all binding and washing steps. The cells (5 × 105 per sample) were then pelleted and blocked in PBS++-3% bovine serum albumin (BSA) (100 μl per sample) for 15 min. RBR-Fc proteins or control rabbit Fc was added to each sample at a concentration of 200 nM (unless otherwise stated) in PBS++-3% BSA and allowed to bind while rocking for 1.5 h. Cells were washed in PBS++-3% BSA and incubated for 45 min with protein A-Alexa Fluor 488 (Invitrogen) at a dilution of 1:250. Cells were washed twice with PBS++-3% BSA and fixed in 4% paraformaldehyde. Cell surface binding was quantified by flow cytometry. For experiments involving protease pretreatment of target cells, the cells were lifted with 0.5% trypsin-EDTA (Gibco) for 15 min at 4°C, and the assay continued as described above. Experiments examining the pH stability of binding were done as described above, with the inclusion of a 10-min incubation in medium at 4°C at the indicated pH values following the RBR-Fc binding step.
Mutagenesis of RBR-1-Fc and RBR-12-Fc.
Ala substitutions were made in RBR-1-Fc and RBR-12-Fc by use of a QuikChange mutagenesis kit (Stratagene) according to the manufacturer's protocol. Insertion of the mutations (and the lack of spurious mutations) was confirmed by sequencing the resulting DNAs in their entirety.
Inhibition of VSVgfp pseudovirion infection.
Vero E6 cells plated in a 96-well dish were washed in PBS++, and the medium was replaced with serum-free DMEM. VSVgfp-GP1,2Δ or control VSVgfp-G (with or without the indicated WT or mutant RBR-Fc at the indicated concentrations) was added to the cells at a low multiplicity of infection (between 0.1 and 1.0). Rabbit Fc was used as a negative control. The cells were incubated for 3 h at 37°C, washed twice with PBS++, and then further incubated in serum-containing growth medium. After 24 h, the cells were fixed with 4% paraformaldehyde and analyzed by flow cytometry.
Inhibition of HIVblam pseudovirion infection.
Vero E6 cells plated in a 48-well dish were washed in PBS++, and the medium was replaced with phenol red-free Optimem (Gibco). HIVblam-GP1,2Δ or control HIVblam-G was added to the cells (with or without RBR-12-Fc or RBR-12-4mer-Fc at 800 nM). The cells were incubated for 5 h at 37°C, washed twice with phenol red-free, serum-free DMEM, and loaded with the beta-lactamase substrate CCF2/AM. Cells loaded only with CCF2/AM served as a negative control. After 1 h at room temperature, the cells were washed and then incubated for 16 h at room temperature in the dark. Cells were fixed with 4% paraformaldehyde, and the extent of CCF2/AM cleavage by the virus-introduced cytoplasmic beta-lactamase, detected by the change in dye emission from green to blue, was evaluated by flow cytometry.
RESULTS AND DISCUSSION
Primed ZEBOV GP1,2 includes residues 33 to 190 of GP1 and 502 to 648 of GP2.
EBOV GP1,2 is primed for fusion in a low-pH endosomal compartment by cathepsins B and L. These proteases cleave GP1 (130 kDa) to a 20-kDa species and a key, 19-kDa species (4, 11, 21) that confer enhanced virus binding to and infection of the host cell (11, 21). In addition to cathepsins B and L, which function at pH ∼5, ZEBOV GP1,2 can be primed more quantitatively to 19-kDa GP1 with thermolysin, a protease that functions at neutral pH (21). Viral particles primed with either cathepsins or thermolysin show similarly enhanced infection and sensitivity to endosomal inhibitors (21).
The first goal of this study was to determine the sequences of 19-kDa GP1 and GP2 in the primed form of GP1,2. We therefore treated purified VSVgfp-GP1,2Δ particles with thermolysin, purified the resultant 19-kDa GP1,2 complex, and determined its sequence by mass spectrometry (see Fig. S1 in the supplemental material). As seen in Fig. 1, primed ZEBOV GP1,2 contains residues 33 to 190 of GP1 and residues 502 to 648 of GP2. Hence, the amino termini of both GP1 and GP2 are intact in 19-kDa GP1,2, and GP2 (the fusion subunit) is fully protected during proteolytic priming. Furthermore, based on cleavage site preferences (http://merops.sanger.ac.uk/) and calculated molecular masses, 20-kDa GP1 likely includes residues 33 to 200, while 19-kDa GP1 may terminate at residue 190, the last residue we found in the sequence analysis (Fig. 1), or extend to residue 193 or 194. These alternate possibilities likely correspond to the closely clustered multiple species of GP1 that are often seen on SDS gels following in vitro priming.
FIG. 1.
Amino acid sequence of primed 19-kDa ZEBOV GP1,2. The 19-kDa GP1,2 was purified from thermolysin-treated VSVgfp-GP1,2Δ and run on an SDS gel. A silver-stained band containing 19-kDa GP1 and GP2 was excised and treated with trypsin or ArgC. The sequences of the resulting proteolytic fragments, identified by mass spectrometry, are underlined (black, tryptic fragments; gray, ArgC fragments) beneath the complete amino acid sequences of GP1 and GP2. The sequences encompassed by the identified fragments are boxed in gray. The signal sequence cleavage site is denoted by an inverted black triangle. The potential C termini of cathepsin B- and L-cleaved 19-kDa GP1 (K190/F193/F194) are marked by gray triangles. The raw mass spectrometry data are shown in Fig. S1 in the supplemental material.
Our findings on 19-kDa GP1 are completely consistent with the recently solved structure of the EBOV GP1,2Δ trimer (14), in which three GP1 subunits form the bowl of a chalice supported by a trimeric GP2 stem. Based on the structure, Lee and coworkers predicted that cathepsins cleave within a disordered loop encompassing GP1 residues 190 to 213 (Fig. 2). Our sequence analysis (Fig. 1) confirmed this prediction. Cleavage within residues 190 to 194 (Fig. 1) would remove the mucin-like domain, the glycan cap, and the outer beta strand of the head domain, thereby exposing the RBR (Fig. 2; also see Fig. 7).
FIG. 7.
Model of 19-kDa GP1 and locations of key receptor binding residues. (A) Ribbon diagram of 19-kDa GP1 showing key recombinant RBRs used in this study, including RBR-7 (residues 90 to 149; dark blue), RBR-1 (residues 57 to 149; dark blue and green), and RBR-12 (residues 33 to 193; dark blue, green, and pink). The side chains of K95, K114, K115, and K140 are shown. Cysteines involved in disulfide bonds are shown in red. An asterisk (*) denotes a contact point between the β1 and β13 strands. (B) Surface rendering (top view) of the GP1,2Δ trimer structure. One monomer is colored dark gray for clarity. (C and D) Model of GP1,2Δ trimer after cleavage by cathepsins B and L, viewed from the top (C) and side (80° rotation) (D). Residues that decreased RBR-1-Fc binding when replaced with Ala are shown in all figures (K95, orange; K114, K115, and K140, cyan). The graphic representations were based on PDB file 3CSY (14) and rendered with Pymol. Note that the depictions in panels A, C, and D are only models that assume no conformational changes in GP1 following proteolytic priming.
Recombinant ZEBOV GP1,2 RBR-Fc proteins ending at residue 149 are produced in high yield.
A second goal of this study was to generate a ZEBOV RBR that can be produced in high yield, as a drawback of the previously identified truncation variant containing EBOV GP1 residues 54 to 201 tagged with human Fc is that it is expressed only at concentrations of <0.1 μg/ml (13; data not shown). We therefore designed a nested set of RBRs initiating and terminating within predicted loop regions of GP1. The first set of RBRs initiated at residue 57, 72, or 90, terminated at residue 149, 172, or 198, and were tagged with rabbit Fc at the C-terminal end. An RBR including residues 54 to 201, similar to that previously described (13) but tagged with rabbit Fc, was also created (Fig. 3A, set 1). These 10 RBR-Fc proteins were all expressed in 293T cells. Those terminating at residue 149 (RBR-Fc 1, 4, and 7) were efficiently secreted (∼10 μg/ml of culture medium, as determined by Fc enzyme-linked immunosorbent assay), approximately 100- to 1,000-fold better than those terminating at residue 172 or 198, RBR-10-Fc (residues 54 to 201), or the RBR containing residues 54 to 201 fused to human Fc (Fig. 3B) (13; data not shown).
FIG. 3.
Production of ZEBOV GP1,2 RBR-Fc proteins. (A) Domain structure of full-length EBOV GP1,2 and the regions of GP1 included in RBR-Fc proteins. Designations are as described in the legend to Fig. 2A. Thirteen RBR proteins conjugated to rabbit Fc were generated. Set 1, RBRs initiating at residue 57, 72, or 90 and terminating at residue 149, 172, or 198. An RBR containing residues 54 to 201 (13) conjugated to rabbit Fc was also created. Set 2, RBRs initiating at residue 33 and terminating at residue 149, 193, or 198. RBR-12-Fc represents a “19-kDa GP1-like” protein. Cys 53 (normally disulfide bonded to Cys 609) was mutated to Ser in all set 2 RBRs to avoid potential misfolding. (B) 293T cells were transfected with plasmids encoding each RBR-Fc. At 24 h posttransfection, lysates of cell pellets (P) and supernatants (S) (equal volumes for each RBR-Fc) were collected, immunoprecipitated with protein A-agarose beads, and analyzed by Western blotting for rabbit Fc. Western blots from one representative experiment of three or more are shown.
After deducing the sequence of 19-kDa GP1 (Fig. 1), we designed a second set of recombinant RBRs (Fig. 3A, set 2). These initiated at residue 33 (the N-terminal residue of 19-kDa GP1) and terminated at either residue 149 (the C-terminal residue that gave the best secretion), 193 (a predicted 19-kDa GP1 C terminus), or 198 (per recombinants 3, 6, and 9). Cys 53, which is normally linked to GP2 via a disulfide bond, was mutated to Ser in each of the set 2 RBRs to eliminate potential folding problems. As shown in Fig. 3B, all of the RBR-Fc proteins beginning at residue 33 were expressed in 293T cells. Again, the RBR-Fc that terminated at residue 149 (RBR-11-Fc) was the most efficiently secreted protein. Surprisingly, RBR-Fcs 12 and 13 were reasonably well secreted (∼1 μg/ml of culture medium for RBR-12-Fc), suggesting that constructs extending beyond residue 149 require an N-terminal extension (residues 33 to 56) to be secreted efficiently. The recently solved structure of the EBOV GP1,2Δ trimer (Fig. 2B) may support this hypothesis. A β-sheet (β1; residues 35 to 56) in the N-terminal extension interacts with the tip of a C-terminal β-sheet present in the longer constructs (β13; residues 175 to 186), perhaps improving the conformational stability of these proteins (Fig. 2B; see Fig. 7A [asterisks denote the point of interaction]).
Binding properties of RBR-Fc proteins.
We compared the abilities of selected RBR-Fc proteins to bind to permissive cells. We first tested the RBR-Fc proteins produced at the highest levels, i.e., constructs 1 (residues 57 to 149), 4 (residues 72 to 149), and 7 (residues 90 to 149). Equivalent concentrations of each RBR-Fc were incubated with permissive (293T and Vero E6) and nonpermissive (Jurkat) cells (32). Binding was quantified by flow cytometry. As shown in Fig. 4A, RBR-1-Fc bound efficiently to 293T and Vero cells but showed negligible binding to Jurkat cells. Binding was higher for Vero E6 cells than for 293T cells, which correlates well with the relative susceptibilities of these cells to infection with VSVgfp-GP1,2Δ pseudovirions (data not shown). Binding of RBR-1-Fc to 293T cells was comparable to that observed using the human Fc-tagged RBR 54-201 (13; data not shown). Furthermore, RBR-1-Fc bound to each of 12 other EBOV-permissive cell lines tested (6; data not shown). The shorter truncations, RBR-4-Fc (residues 72 to 149) and RBR-7-Fc (residues 90 to 149), also bound specifically to 293T and Vero cells (Fig. 4A), suggesting that residues critical for RBR binding lie between residues 90 and 149, in the head domain of GP1 (Fig. 2; also see Fig. 7). We next tested the binding of RBR-12-Fc (residues 33 to 193), the RBR that most closely corresponds to 19-kDa GP1 (Fig. 1; also see Fig. 7A). As shown in Fig. 4B, RBR-12-Fc bound specifically to permissive cells, but somewhat less efficiently than RBR-1-Fc.
FIG. 4.
Binding activities of selected ZEBOV GP1,2 RBR-Fc proteins. (A) RBR-Fcs 1, 4, and 7 (200 nM) were incubated with 293T or Vero E6 cells (permissive) or with Jurkat lymphocytes (nonpermissive). Cell surface binding was analyzed by flow cytometry, using protein A-Alexa Fluor 488 to detect the Fc portion of the RBR conjugate. (B) Binding of RBR-1-Fc and RBR-12-Fc to 293T, Vero E6, or Jurkat cells was determined and analyzed as in panel A. In all experiments, rabbit Fc served as a negative control. The averages of two or more experiments are shown. Bars represent the percentages of cells that bound the indicated RBR-Fc proteins. Error bars indicate standard deviations. Significance (relative to RBR-1-Fc) was determined by Student's t test. *, P < 0.05.
For reasons that are not clear, we observed that preparations of RBR-1-Fc were more stable during storage at 4°C than preparations of RBR-12-Fc. We therefore characterized additional binding properties of RBR-1-Fc, since it is reproducibly produced in high yield as a stable product that binds efficiently to EBOV-permissive cells. To test if binding was saturable, we incubated increasing concentrations of RBR-1-Fc with 293T cells. As shown in Fig. 5A, binding of RBR-1-Fc was saturated at a concentration between 2.0 and 2.5 μM, in terms of both the percentage of cells bound (Fig. 5A, panel i) and the mean fluorescence intensity (Fig. 5A, panel ii). Although only a few experiments were conducted (due to lower yields), binding of RBR-12-Fc, the “19-kDa GP1-like” RBR-Fc, was also saturated at ∼2.5 μM on 293T cells (data not shown), further supporting RBR-1-Fc as an appropriate binding model.
FIG. 5.
Binding properties of RBR-1-Fc. (A) RBR-1-Fc was incubated with 293T cells at the indicated concentration, and binding was determined as described in the legend to Fig. 4. (i) Percentage of cells that bound RBR-1-Fc (black) or Fc (gray). (ii) Mean fluorescence intensity (MFI) of cells incubated with RBR-1-Fc, with background values for control Fc subtracted. Data for one representative experiment of three are shown. (B) 293T cells were lifted as indicated with a solution containing EDTA (PEEG, as for all other binding experiments) or with 0.5% trypsin-EDTA for 15 min and then processed for RBR-1-Fc (200 nM) binding. The averages of three experiments are shown. Error bars indicate standard deviations. Significance (between RBR-1-Fc binding to trypsin- and EDTA-treated cells) was determined by Student's t test. *, P < 0.05. (C) 293T cells were incubated with RBR-1-Fc (200 nM), exposed to medium at the indicated pH for 10 min (at 4°C), returned to normal medium, and then processed for cell surface binding. Values were normalized to those for RBR-1-Fc at pH 7.0. The averages of two or more experiments are shown. Error bars indicate standard deviations. Significance (relative to RBR-1-Fc binding at pH 7) was determined by Student's t test. *, P < 0.05.
The susceptibility of cells to EBOV GP1,2-mediated infection was previously shown to be sensitive to pretreatment with proteases (26; data not shown). To investigate whether the binding sites for RBR-1-Fc are also protease sensitive, we pretreated cells with trypsin prior to incubation with RBR-1-Fc. As shown in Fig. 5B, pretreatment of both 293T and Vero E6 cells with trypsin strongly reduced binding.
Since EBOV must traffic to a low-pH compartment prior to virus-cell fusion, we examined the stability of RBR-1-Fc binding to 293T cells during a brief treatment across a range of pH values. As shown in Fig. 5C, binding in the cold was stable during a brief wash with buffers ranging in pH from 3 to 11, with small decreases at the extremes. This result suggests that during EBOV infection, the RBR-receptor interaction may be maintained as the virus moves along the endocytic pathway and is primed for fusion.
Residues K95, K114, K115, and K140 in the bowl of the GP1 chalice are critical for ZEBOV GP1,2-mediated binding.
Previous work has shown that many residues throughout GP1 are important for EBOV GP1,2-mediated entry into host cells (3, 15, 17), but to date there have been no direct binding studies with mutant GP1,2 proteins. Since the smallest RBR we analyzed (RBR-7; GP1 residues 90 to 149) bound specifically to permissive cells, we engineered Ala substitutions (into RBR-1-Fc) at each residue between residues 90 and 149 that had been suggested to be important for receptor binding (based on infectivity studies) and that did not compromise GP1,2 incorporation into pseudovirions: these were K95, K114/K115, K140, and G143 (see Fig. S2 in the supplemental material) (3, 15, 17). We also created three mutants with composite mutations, which we denoted 3mer (K114A/K115A/K140A), 4mer (K95A/K114A/K115A/K140A), and 5mer (K95A/K114A/K115A/K140A/G143A). All mutant RBR-Fc proteins were expressed and secreted from 293T cells at levels comparable to that of WT RBR-1-Fc (data not shown). The K95A, K114/K115A, and K140A mutations each decreased binding by a modest but significant degree (25 to 30%), but the substitution at G143 had no effect (Fig. 6). The 3mer mutant showed 60% decreased RBR-1-Fc binding, and the 4mer and 5mer mutant proteins both displayed ∼75% reduced RBR-1-Fc binding. Furthermore, in a preliminary experiment, introducing the 4mer mutations into RBR-12-Fc (RBR-12-4mer-Fc) also decreased binding to 293T cells by 78% (data not shown). These findings suggest that combined Ala substitutions at four Lys residues, K95, K114, K115, and K140, all of which are absolutely conserved throughout all five sequenced species of EBOV GP1,2, significantly impair RBR binding to host cells. However, even combined disruption of K95, K114, K115, and K140 did not completely eliminate RBR binding, suggesting that other residues within GP1 make important binding contacts.
FIG. 6.
Binding of WT and mutant RBR-1-Fc proteins. Ala substitutions were made individually and in combination within RBR-1-Fc at K95, K114 and K115, K140, and G143 (3, 15, 17). 3mer, K114A/K115A/K140A mutant; 4mer, K95A/K114A/K115A/K140A mutant; 5mer, K95A/K114A/K115A/K140A/G143A mutant. 293T cells were incubated with the indicated RBR-Fc or control Fc (200 nM) and analyzed for cell surface binding as described in the legend to Fig. 4. Binding values were normalized to those for WT RBR-1-Fc. The number of experiments performed is shown above each bar. Significance (relative to WT RBR-1-Fc) was determined by Student's t test. *, P < 0.04; **, P < 0.0004.
Lee and coworkers proposed that the receptor binding site occupies an area of ∼20 by 15 Å on the inner surface of the chalice bowl. Three of the residues that we found to be critical for binding—K114, K115, and K140—lie within this site, where they are clustered in a strip ∼20 Å in length (Fig. 7A and B). If the mucin-like domain and glycan cap move prior to proteolytic priming (to 19 kDa GP), these three lysines would lie near the top of the newly exposed surface of the chalice bowl (Fig. 2B). After priming (removal of the mucin-like domain, glycan cap, and outer beta strand of the head domain), K114, K115, and K140 would be found on the top edge of the chalice bowl (Fig. 7C and D). K95, which is also important for RBR-Fc binding (Fig. 6), lies outside the proposed receptor binding site, deeper in the chalice bowl (Fig. 7A). It may play a direct role in receptor binding or it may help provide the RBR-Fc with an optimal conformation for binding. In total, our mutational analysis using a physical binding assay is fully consistent with the prediction made by Lee and coworkers regarding the location of the receptor binding site of EBOV GP1.
RBR-12-Fc inhibits ZEBOV GP1,2Δ pseudovirus infection.
We next examined the ability of RBR-1-Fc and RBR-12-Fc to inhibit ZEBOV GP1,2-mediated infection. We first infected Vero E6 cells with VSVgfp-GP1,2Δ in the presence or absence of various concentrations of WT and 4mer mutant RBR-1-Fc, WT and 4mer mutant RBR-12-Fc, and rabbit Fc (as a negative control). At a concentration of 2.5 μM, RBR-1-Fc exerted a small degree of inhibition of infection (ranging from 3 to 15%). Although it was only a small effect, this inhibition was consistently greater than that seen with 4mer mutant RBR-1-Fc analyzed in parallel at the same concentration (Fig. 8A). RBR-12-Fc exerted a greater, but still limited, inhibition of infection (ranging from 16 to 41% at 2.5 μM). Again, although only modest, the inhibition by RBR-12-Fc was significantly greater than that by RBR-12-4mer-Fc analyzed in parallel at the same concentration. Similar inhibition profiles were seen for WT RBR-1-Fc and WT RBR-12-Fc when infections were performed with VSVgfp-19kDa GP1,2 or with VSV pseudovirions bearing marburgvirus GP (data not shown). There was no significant inhibition of VSVgfp-G infection with any of the RBR-Fc proteins tested at any concentration (data not shown).
FIG. 8.
Inhibition of EBOV GP1,2Δ-mediated infection by RBR-1-Fc and RBR-12-Fc. (A) Vero E6 cells were incubated for 3 h with VSVgfp-GP1,2Δ in the presence or absence of RBR-1-Fc, RBR-12-Fc, or control rabbit Fc at the indicated concentration. Unbound virus was removed by washing, the cells were incubated overnight, and GFP expression was quantified by flow cytometry. Samples were analyzed in triplicate. The average values from one representative experiment are shown. Error bars represent standard deviations. Significance (relative to the rabbit Fc control tested at the same concentration) was determined by Student's t test. *, P < 0.05. The exact same experiment was conducted two times with similar results. RBR-1-Fc and RBR-12-Fc were tested five additional times (with HIV gp120-Fc as a negative control), with very similar results. (B) Vero E6 cells were infected with HIVblam-GP1,2Δ or HIVblam-G in the presence or absence of WT or 4mer mutant RBR-12-Fc at 800 nM. Cells were loaded with the beta-lactamase substrate CCF2/AM. Cells loaded only with CCF2/AM served as a negative control (no virus). The extent of CCF2/AM cleavage by beta-lactamase introduced into the cytoplasm was evaluated by flow cytometry (detected by the change in dye emission from green to blue). Samples were analyzed in duplicate. The averages for duplicate samples from one representative experiment are shown. Error bars represent standard deviations.
The RBR containing residues 54 to 201 tagged with human Fc was previously shown to inhibit infection with EBOV GP1,2-bearing murine leukemia virus (MLV) pseudovirions (13). We therefore tested our RBR-1-Fc and RBR-12-Fc proteins for inhibition of infection by retrovirus pseudovirions bearing EBOV GP1,2. Vero E6 cells were infected with HIVblam-GP1,2Δ or HIVblam-G in the presence or absence of WT and 4mer mutant RBR-12-Fc at a concentration of 800 nM. As shown in Fig. 8B, in this system RBR-12-Fc showed strong inhibition (ranging from 52 to 67%), which was ∼3-fold greater than that seen with 1 μM RBR-12-Fc, using VSV pseudovirions. Importantly, the 4mer mutant of RBR-12-Fc did not inhibit infections by HIVblam-GP1,2Δ when tested in parallel at the same concentration. There was no significant inhibition of HIVblam-G with either WT or 4mer mutant RBR-12-Fc. Previous work has suggested that EBOVs and marburgvirus may utilize a common receptor, as a human Fc-tagged RBR containing EBOV GP1,2 residues 54 to 201 inhibited MLV pseudovirions bearing marburgvirus GP more strongly than they inhibited MLV pseudovirions bearing EBOV GP1,2 (13). In one preliminary experiment, RBR-12-Fc (800 nM) completely inhibited infection by MLV pseudovirions bearing marburgvirus GP, with stronger inhibition than that seen with the human Fc-fused RBR 54-201 tested in parallel (J. Kuhn, data not shown).
Since RBR-12-Fc inhibits ZEBOV GP1,2 pseudovirion infection more strongly than RBR-1-Fc does, residues 150 to 193 may be critical for inhibition of infection (Fig. 8), though not for receptor binding (Fig. 4B). Additionally, the location of the Fc tag, which neighbors important binding contacts, could limit RBR-1-Fc from competing with virus particles bearing multiple GP1,2 trimers. Another possibility is that EBOV GP1,2 can utilize multiple receptors to gain entrance into the cell and that blocking the receptor that interacts with RBR-1-Fc is not sufficient to block these additional interactions. Furthermore, the ability of RBR-12-Fc to more strongly inhibit retroviral pseudovirion infection than VSV pseudovirion infection could be due to differences in the copy number or orientation of the GP1,2 trimers on the respective viral particles. Further studies will help to clarify these differences.
Conclusions.
There are several conclusions of our study. First, ZEBOV GP1,2 that has been primed for fusion contains residues 33 to 190 of GP1 (possibly extending to residue 193 or 194) and all residues of GP2. Second, an Fc-tagged protein containing the residues present in primed GP1 can inhibit ZEBOV GP1,2-mediated infection. Third, four Lys residues within primed (19-kDa) GP1 are critical for binding to a host cell receptor. Three (K114, K115, and K140) are located in a small strip on the inner surface of the GP1 chalice and are predicted to lie on the top surface after cathepsin priming; the fourth (K95) lies deeper in the chalice bowl (14). Our findings should aid future investigations into how EBOVs enter and infect host cells.
Supplementary Material
Acknowledgments
We thank Erica Ollmann Saphire for sharing the structure of the ZEBOV GP1,2Δ trimer ectodomain shortly prior to publication, Dusan Turk for discussions regarding cathepsin cleavage sites, and Dennis Burton, Gary Nabel, Michael Whitt, and Paul Bates for gifts of reagents.
This work was supported by research grants from the NIH to J.W. and S.D. (AI22470 and U54 AI57168) and by training grants from the NIH in infectious diseases (5T32 AI055432) and biodefense (5T32 AI0704627) to K.S., M.B., and D.D.
Footnotes
Published ahead of print on 14 January 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alvarez, C. P., F. Lasala, J. Carrillo, O. Muniz, A. L. Corbi, and R. Delgado. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 766841-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boussif, O., F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix, and J. P. Behr. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA 927297-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brindley, M. A., L. Hughes, A. Ruiz, P. B. McCray, Jr., A. Sanchez, D. A. Sanders, and W. Maury. 2007. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J. Virol. 817702-7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandran, K., N. J. Sullivan, U. Felbor, S. P. Whelan, and J. M. Cunningham. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 3081643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolnik, O., L. Kolesnikova, and S. Becker. 2008. Filoviruses: interactions with the host cell. Cell. Mol. Life Sci. 65756-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dube, D., K. L. Schornberg, T. S. Stantchev, M. I. Bonaparte, S. E. Delos, A. H. Bouton, C. C. Broder, and J. M. White. 2008. Cell adhesion promotes Ebola virus envelope glycoprotein-mediated binding and infection. J. Virol. 827238-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldmann, H., T. Geisbert, B. Jahrling, H. Klenk, S. Netsov, C. Peters, A. Sanchez, R. Swanepoel, and V. Volchkov. 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, San Diego, CA.
- 8.Ito, H., S. Watanabe, A. Sanchez, M. A. Whitt, and Y. Kawaoka. 1999. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J. Virol. 738907-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito, H., S. Watanabe, A. Takada, and Y. Kawaoka. 2001. Ebola virus glycoprotein: proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J. Virol. 751576-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffers, S. A., D. A. Sanders, and A. Sanchez. 2002. Covalent modifications of the Ebola virus glycoprotein. J. Virol. 7612463-12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaletsky, R. L., G. Simmons, and P. Bates. 2007. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 8113378-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn, J. H. 2008. Filoviruses: a compendium of 40 years of epidemiological, clinical, and laboratory studies. Springer, Berlin, Germany. [PubMed]
- 13.Kuhn, J. H., S. R. Radoshitzky, A. C. Guth, K. L. Warfield, W. Li, M. J. Vincent, J. S. Towner, S. T. Nichol, S. Bavari, H. Choe, M. J. Aman, and M. Farzan. 2006. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 28115951-15958. [DOI] [PubMed] [Google Scholar]
- 14.Lee, J. E., M. L. Fusco, A. J. Hessell, W. B. Oswald, D. R. Burton, and E. O. Saphire. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manicassamy, B., J. Wang, H. Jiang, and L. Rong. 2005. Comprehensive analysis of Ebola virus GP1 in viral entry. J. Virol. 794793-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marzi, A., P. Moller, S. L. Hanna, T. Harrer, J. Eisemann, A. Steinkasserer, S. Becker, F. Baribaud, and S. Pohlmann. 2007. Analysis of the interaction of Ebola virus glycoprotein with DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) and its homologue DC-SIGNR. J. Infect. Dis. 196(Suppl. 2)S237-S246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mpanju, O. M., J. S. Towner, J. E. Dover, S. T. Nichol, and C. A. Wilson. 2006. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 121205-214. [DOI] [PubMed] [Google Scholar]
- 18.Powlesland, A. S., T. Fisch, M. E. Taylor, D. F. Smith, B. Tissot, A. Dell, S. Pohlmann, and K. Drickamer. 2008. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J. Biol. Chem. 283593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez, A. 2007. Analysis of filovirus entry into Vero E6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J. Infect. Dis. 196(Suppl. 2)S251-S258. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez, A., T. W. Geisbert, and H. Feldmann. 2007. Filoviridae: Marburg and Ebola viruses, p. 1409-1448. In D. M. Knipe et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 21.Schornberg, K., S. Matsuyama, K. Kabsch, S. Delos, A. Bouton, and J. White. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 804174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimojima, M., A. Takada, H. Ebihara, G. Neumann, K. Fujioka, T. Irimura, S. Jones, H. Feldmann, and Y. Kawaoka. 2006. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 8010109-10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmons, G., J. D. Reeves, C. C. Grogan, L. H. Vandenberghe, F. Baribaud, J. C. Whitbeck, E. Burke, M. J. Buchmeier, E. J. Soilleux, J. L. Riley, R. W. Doms, P. Bates, and S. Pohlmann. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305115-123. [DOI] [PubMed] [Google Scholar]
- 24.Simmons, G., R. J. Wool-Lewis, F. Baribaud, R. C. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 762518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takada, A., K. Fujioka, M. Tsuiji, A. Morikawa, N. Higashi, H. Ebihara, D. Kobasa, H. Feldmann, T. Irimura, and Y. Kawaoka. 2004. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J. Virol. 782943-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 9414764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takada, A., S. Watanabe, H. Ito, K. Okazaki, H. Kida, and Y. Kawaoka. 2000. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology 27820-26. [DOI] [PubMed] [Google Scholar]
- 28.Towner, J. S., T. K. Sealy, M. L. Khristova, C. G. Albarino, S. Conlan, S. A. Reeder, P. L. Quan, W. I. Lipkin, R. Downing, J. W. Tappero, S. Okware, J. Lutwama, B. Bakamutumaho, J. Kayiwa, J. A. Comer, P. E. Rollin, T. G. Ksiazek, and S. T. Nichol. 2008. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe, S., A. Takada, T. Watanabe, H. Ito, H. Kida, and Y. Kawaoka. 2000. Functional importance of the coiled coil of the Ebola virus glycoprotein. J. Virol. 7410194-10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.WHO. 2007. Ebola haemorrhagic fever in Uganda. World Health Organization, Geneva, Switzerland.
- 32.Wool-Lewis, R. J., and P. Bates. 1998. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol. 723155-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wool-Lewis, R. J., and P. Bates. 1999. Endoproteolytic processing of the Ebola virus envelope glycoprotein: cleavage is not required for function. J. Virol. 731419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Z. Y., H. J. Duckers, N. J. Sullivan, A. Sanchez, E. G. Nabel, and G. J. Nabel. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6886-889. [DOI] [PubMed] [Google Scholar]
- 35.Yonezawa, A., M. Cavrois, and W. C. Greene. 2005. Studies of Ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J. Virol. 79918-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.