Abstract
After fusion of the envelope of herpesvirus particles with the host cell plasma membrane, incoming nucleocapsids are transported to nuclear pores. Inner tegument proteins pUL36, pUL37, and pUS3 remain attached to the nucleocapsid after entry and therefore might mediate interactions between the nucleocapsid and cellular microtubule-associated motor proteins during transport. To assay for the role of pUL37 in this process, we constructed a pUL37-deleted pseudorabies virus mutant, PrV-ΔUL37/UL35GFP, which expresses a fusion protein of green fluorescent protein (GFP) and the nonessential small capsid protein pUL35, resulting in the formation of fluorescently labeled capsids. Confocal laser-scanning microscopy of rabbit kidney cells infected with PrV-ΔUL37/UL35GFP revealed that, whereas penetration was not affected in the absence of pUL37, nuclear translocation of incoming particles was delayed by approximately 1 h compared to PrV-UL35GFP, but not abolished. In contrast, phenotypically complemented pUL37-containing virions of PrV-ΔUL37/UL35GFP exhibited wild type-like entry kinetics. Thus, the presence of pUL37 is required for rapid nuclear translocation of incoming nucleocapsids.
The herpesvirus replication cycle starts with attachment of extracellular virions to cellular receptors, which is followed by fusion of the virion envelope either with the cellular plasma membrane (20) or, after endocytosis, with the endosomal membrane (49), resulting in release of the nucleocapsid into the cytosol. During this process, most proteins of the tegument, which in the herpes virion resides between the envelope and nucleocapsid, are released from the incoming particle. Several tegument proteins prime the cell for virus infection by shutting off cellular protein synthesis (virion host shut-off factor, pUL41) (29) or, after translocation into the nucleus, boost viral transcription (α-transinducing factor, pUL48) (2). Other tegument proteins, however, remain bound to the incoming nucleocapsid during its transit to the nuclear pore, where the viral genome is released into the nucleus to start viral gene expression. Thus, efficient transport of nucleocapsids to the nuclear pore is crucial for herpesvirus infectivity.
For efficient nuclear targeting of incoming nucleocapsids, herpesviruses recruit cellular motor proteins to move along microtubules (MT) (9, 11, 17, 43, 45, 48). This process is particularly relevant to overcome even long distances, such as between neuronal axon termini and the nucleated cell bodies. MT are polar cytoskeletal filaments with a fast-growing plus end, extended toward the cell periphery, and a less dynamic minus end, attached to the MT organizing center (38). Minus end-directed transport catalyzed by the huge dynein-dynactin motor complex (44, 53) is used by a variety of viruses (9, 15, 30, 35, 40, 41, 45, 48, 50). Studies of epithelial and neuronal cells revealed that incoming herpes simplex virus type 1 (HSV-1) capsids colocalize with cytoplasmic dynein and dynactin and that capsid transport to the nucleus is inhibited by destabilization of the dynein motor complex, e.g., by overexpression of the dynactin subunit dynamitin (12, 28, 33, 48, 52). Dynein-mediated transport requires a physical interaction between the intracellular viral particle and the motor complex, and several herpesviral proteins, like pUL34, pUL35, pUL56, and pUS11, have been found to interact with cellular motor components (7, 8, 13, 26, 31, 32, 34, 55). However, the relevance of these interactions for intracellular transport of herpesvirus particles remains unclear. Proteins of the “inner,” capsid-proximal tegument (37) are prime candidates for mediating nuclear translocation by interacting with cellular motors. They encompass pUL36 and pUL37, which are conserved within the Herpesviridae and form a physical complex (16, 18, 21, 24), as well as the alphaherpesvirus-specific pUS3 protein kinase (19, 31). Although pUL36 and pUL37 of HSV-1 failed to interact with subunits of the dynein motor complex in a yeast two-hybrid assay (13), exposure of these proteins after removal of the outer tegument by chemical treatment increased the efficiency of MT-dependent transport of capsids in vitro (54). Proteins of the inner tegument are also involved in intracellular transport of nucleocapsids during egress. As shown for the porcine alphaherpesvirus pseudorabies virus (PrV), this plus end-directed motion of nucleocapsids is abolished in the absence of pUL36, whereas the presence of pUL37 was not essential but increased the efficiency of transport (32). Studies of the role of pUL37 during entry have not yet been performed. In contrast to pUL37-deleted HSV-1 (6), PrV-ΔUL37 is able to replicate productively on noncomplementing cells, though at significantly lower levels than the wild-type virus, demonstrating that pUL37 is not strictly essential for any crucial step during PrV replication (25). Thus, extracellular PrV virions lacking pUL37 can be obtained from noncomplementing cells and assayed for early steps in subsequent infection. Nevertheless, the presence of pUL37 is a prerequisite for efficient secondary nucleocapsid envelopment in the cytoplasm during morphogenesis of PrV and HSV-1 particles (6, 25).
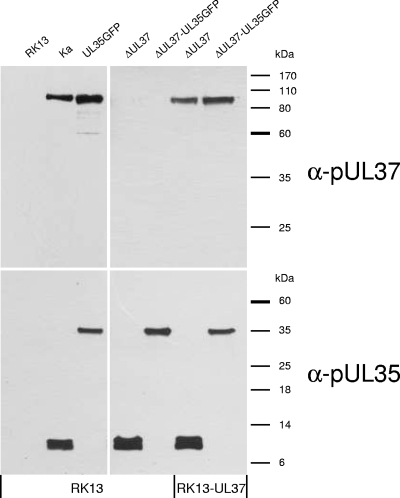
To analyze the role of pUL37 in early events of herpesvirus infection, we constructed and characterized a novel pUL37 deletion mutant of PrV based on wild-type PrV strain Kaplan (PrV-Ka) (22), which expresses a fusion protein of the nonessential small capsid protein pUL35 with enhanced green fluorescent protein (eGFP) (4, 5, 27). Although eGFP fusion can result in loss of pUL35 function (27), it has been shown for HSV-1 that dynein-mediated transport is not impaired, even by complete loss of pUL35 (1, 10). Genome analyses of the obtained recombinant PrV-ΔUL37/UL35GFP confirmed the expected deletion of UL37 codons 15 to 778 (of 920) (25), as well as in-frame fusion of the eGFP and pUL35 coding sequences (results not shown). For protein characterization, RK13 or complementing RK13-UL37 cells (25) were infected with PrV-ΔUL37/UL35GFP, PrV-UL35GFP (27), PrV-ΔUL37 (25), or PrV-Ka at a multiplicity of infection (MOI) of 1 for 30 h, and infected cell proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blot analyses were performed as described with monospecific anti-pUL35 (27) or anti-pUL37 rabbit sera (25) (Fig. 1). As expected, UL37 gene products were not detectable in RK13 cells infected with PrV-ΔUL37/UL35GFP or PrV-ΔUL37, whereas PrV-UL35GFP- and PrV-Ka-infected cells expressed the 100-kDa pUL37 (Fig. 1). Since a complete UL37 knockout would impair the promoter of the essential downstream UL36 gene, the 3′-terminal 142 codons were retained. However, the deletion brings these codons out of frame with the first 15 codons remaining from the 5′ end, which, theoretically, could still be expressed. pUL36 was incorporated into virions in the absence of pUL37. However, a more precise quantitation is problematic due to the extreme size (>300 kDa) of the protein. We did not observe an alteration in virion composition besides absence of pUL37 and presence of pUL35GFP compared to wild-type PrV (data not shown). After infection of RK13-UL37 cells with pUL37 deletion mutants, pUL37 was detected at levels similar to those of wild-type virions (Fig. 1). Independent of the cell line used, PrV-Ka- and PrV-ΔUL37-infected cells exhibited native pUL35 (ca. 12 kDa), and PrV-UL35GFP- or PrV-ΔUL37/UL35GFP-infected cells expressed the 38-kDa pUL35-eGFP fusion protein (27) (Fig. 1). Thus, the protein expression pattern of the mutant viruses was as expected.
FIG. 1.
Western blot analyses. Extracts of RK13 or RK13-UL37 cells infected with PrV-Ka, PrV-UL35GFP, PrV-ΔUL37, and PrV-ΔUL37/GFP were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins were transferred to nitrocellulose filters. Blots were incubated with monospecific rabbit antisera against pUL37 and pUL35 at dilutions of 1:100,000. Binding of peroxidase-conjugated anti-rabbit antibodies (Dianova) were detected by chemiluminescence (SuperSignal; Thermo Scientific). Locations of molecular mass marker proteins are indicated.
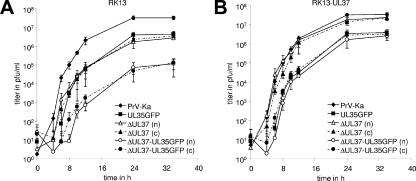
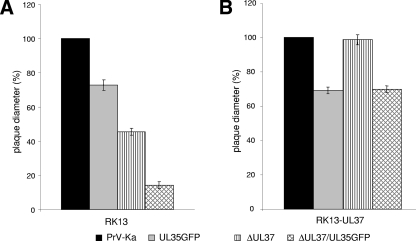
One-step replication kinetics and plaque sizes of PrV-Ka, PrV-UL35GFP, PrV-ΔUL37, and PrV-ΔUL37/UL35GFP were compared on RK13 and RK13-UL37 cells. In noncomplementing cells, the maximum titers of PrV-ΔUL37/UL35GFP were reduced ca. 100-fold compared to those of PrV-Ka, whereas the two single mutants exhibited ca. 10-fold titer reductions each (Fig. 2A). In cells expressing pUL37, the defect of PrV-ΔUL37 could be almost completely corrected, and PrV-ΔUL37/UL35GFP exhibited similar growth properties as PrV-UL35GFP (Fig. 2B). On either cell line, the growth kinetics after infection with noncomplemented or phenotypically complemented UL37 deletion mutants were similar (Fig. 2), indicating that pUL37 plays its main role after viral de novo protein synthesis during virion maturation and egress. In agreement with earlier results (25, 27), cell-to-cell spread of PrV in RK13 cells was also affected by deletion of UL37, as well as by eGFP tagging of pUL35. Compared to those of wild-type PrV-Ka, the plaque diameters of PrV-ΔUL37 and PrV-UL35GFP were reduced by ca. 50% and 25%, respectively, whereas plaques of PrV-ΔUL37/UL35GFP reached only 15% of the wild-type sizes (Fig. 3A). Analogous to the results of one-step growth studies, in RK13-UL37 cells, the plaque sizes of PrV-ΔUL37 were similar to those of PrV-Ka and the plaque sizes of PrV-ΔUL37/UL35GFP increased to those of PrV-UL35GFP (Fig. 3B). Thus, the defects caused by deletion of pUL37 and eGFP fusion of pUL35 were independent of each other, and the pUL37 defect could be complemented efficiently in cells that provided pUL37 in trans.
FIG. 2.
One-step growth kinetics. RK13 (A) or RK13-UL37 (B) cells were infected with noncomplemented (n) or phenotypically complemented (c) deletion mutants PrV-ΔUL37 and PrV-ΔUL37/UL35GFP as well as with PrV-Ka and PrV-UL35GFP at an MOI of 5. At the indicated times after infection (h p.i.), cells were harvested together with the supernatants, and progeny virus titers (PFU/ml) were determined by plaque assays on RK13-UL37 cells. Shown are mean titers and standard deviations of three independent experiments.
FIG. 3.
Plaque sizes. RK13 (A) or RK13-UL37 (B) cells were infected under plaque assay conditions with PrV-ΔUL37, PrV-ΔUL37/UL35GFP, PrV-UL35GFP, or wild-type PrV-Ka. After 2 days, 50 plaques per virus and cell line were measured microscopically. Mean diameters of three independent experiments are shown as percentages of wild-type PrV-Ka sizes, and standard deviations are also indicated.
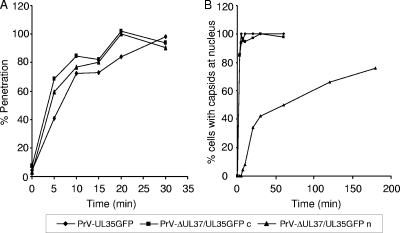
Analysis of penetration kinetics using citrate inactivation (36) showed similar entry phenotypes for all viruses tested (Fig. 4A), demonstrating that the presence or absence of pUL37 does not alter the ability of the virus to cross the plasma membrane.
FIG. 4.
Penetration kinetics and nuclear localization of capsids. (A) Penetration kinetics of PrV-UL35GFP, noncomplemented PrV-ΔUL37/UL35GFP (n), or phenotypically pUL37-complemented PrV-ΔUL37/UL35GFP (c) were assayed by citrate inactivation studies (36). Average results from three independent experiments are shown. (B) Assays shown in Fig. 5 were quantitated, indicating the percentage of cells exhibiting fluorescent capsids at the nucleus after infection with PrV-UL35GFP, noncomplemented PrV-ΔUL37/UL35GFP (n), or phenotypically pUL37-complemented PrV-ΔUL37/UL35GFP (c).
To assay for nuclear translocation of incoming nucleocapsids, subconfluent RK13 cells were precooled on ice for 15 min and subsequently infected with sucrose gradient-purified PrV-UL35GFP or either phenotypically complemented or noncomplemented PrV-ΔUL37/UL35GFP at an MOI of 200. After 1 h, inocula were replaced by prewarmed medium, and cells were further maintained at 37°C. Cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) at different times, from immediately after infection (0 h) to 8 h postinfection (p.i.), permeabilized with 0.3% Triton X-100, and incubated with a murine anti-α-tubulin antibody (diluted 1:1,000 in PBS; Sigma), which was detected with the fluorescence-tagged Alexa Fluor 647 goat-anti mouse-antibodies (diluted 1:1,000 in PBS; Molecular Probes). In addition, cellular F-actin was labeled with Alexa Fluor 546-tagged phalloidin (Molecular Probes). For scanning and photography, a confocal laser-scanning microscope (LSM510; Zeiss, Göttingen, Germany) was used. At least 10 randomly selected cells for each time point were analyzed in three independent experiments, and representative images are shown in Fig. 5 and 6.
FIG. 5.
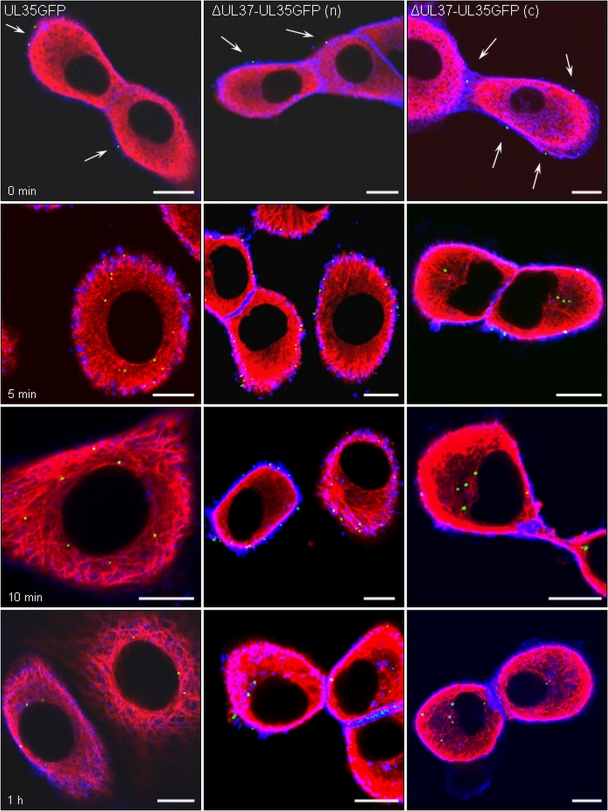
Nuclear translocation of incoming capsids. RK13 cells were infected at an MOI of 200 with PrV-UL35GFP (left panels), noncomplemented PrV-ΔUL37/UL35GFP (n) (middle panels), or phenotypically pUL37-complemented PrV-ΔUL37/UL35GFP (c) (right panels). Cells were fixed immediately after 1 h of virus adsorption on ice (0 min, top row) or 5 min (second row), 10 min (third row), and 1 h (bottom row) after the temperature shift to 37°C. Direct and indirect fluorescence reactions were analyzed by confocal laser-scanning microscopy. Green, pUL35-eGFP; red, MT; blue, actin. Viral capsids attached to the cell surface are marked by arrows. Bar, 10 μm.
FIG. 6.
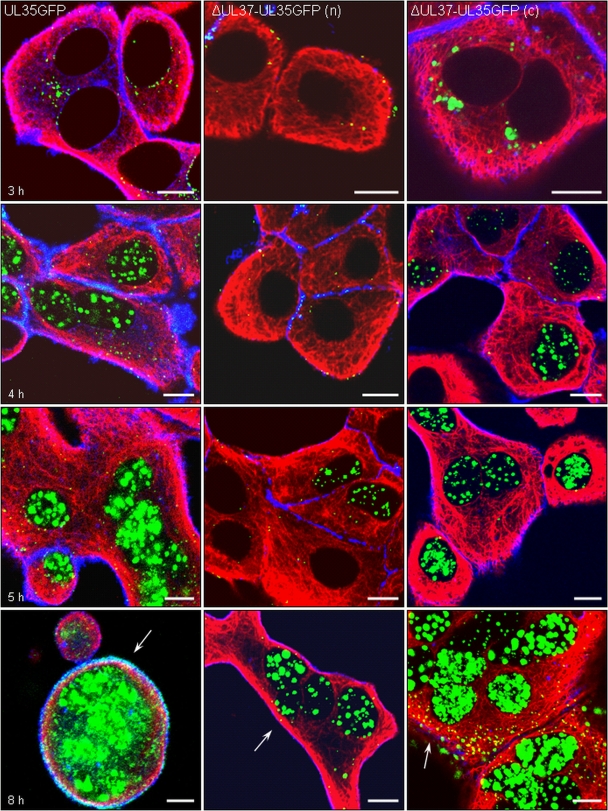
pUL35 expression, capsid formation, and viral egress. RK13 cells infected at an MOI of 200 with PrV-UL35GFP (left panels), noncomplemented PrV-ΔUL37/UL35GFP (n) (middle panels), or phenotypically complemented PrV-ΔUL37/UL35GFP (c) (right panels) were fixed 3 h (top row), 4 h (second row), 5 h (third row), and 8 h (bottom row) after infection. Direct and indirect fluorescence reactions were analyzed by confocal laser-scanning microscopy. Green, pUL35-eGFP; red, MT; blue, actin. Egressing viral capsids at the cell periphery are marked by arrows. Bar, 10 μm.
In the assays, blue indicates the actin-rich cortex underlying the plasma membrane, whereas MT (red) form a complex network within the cytosol. pUL35GFP-tagged capsids were detected as small green fluorescent spots. After 1 h of adsorption on ice, capsids of PrV-UL35GFP or PrV-ΔUL37/UL35GFP were found attached at the surface of the cells (Fig. 5, 0 min). As early as 5 min after the temperature shift to 37°C, numerous intracellular nucleocapsids could be detected, and many capsids of PrV-UL35GFP had moved to areas close to the nucleus (Fig. 5, 5 min), reflecting an efficient nuclear targeting. This parallels results from electron microscopic analyses demonstrating translocation of incoming PrV nucleocapsids to the nuclear pore by as early as 5 min after infection (19). After 20 min, almost all PrV-UL35GFP particles had accumulated at the nucleus and remained there at later times (Fig. 5, 1 h).
In contrast, nuclear targeting of capsids of PrV-ΔUL37/UL35GFP isolated from noncomplementing RK13 cells was significantly delayed (Fig. 5, middle panel). Intracellular capsids of PrV-ΔUL37/UL35GFP were detected inside the cell underneath the plasma membrane at 5 min p.i. (Fig. 5, 5 min), indicating that crossing of the plasma membrane and the actin cortex occurred unimpeded. However, capsids remained at the cell periphery for a considerable time (Fig. 5, 5 to 20 min) and were not detected at the nucleus until 1 to 2 h after the temperature shift (Fig. 5, 1 h).
In contrast, capsids of PrV-ΔUL37/UL35GFP passaged on RK13-UL37 cells traversed the cytoplasm with similar kinetics (Fig. 5, right panel) as PrV-UL35GFP (Fig. 5, left panel) and accumulated at the nuclei as early as 10 min p.i., demonstrating that the observed delay in nuclear translocation of phenotypically noncomplemented PrV-ΔUL37/UL35GFP capsids was due to the absence of pUL37. The percentage of cells with fluorescent capsids at the nucleus is given in Fig. 4B.
Beginning at 3 h p.i., the intensity of green fluorescence continuously increased in PrV-UL35GFP-infected cells, indicating de novo synthesis of the UL35-eGFP fusion protein (Fig. 6, left panel). The labeled protein was first restricted to the cytoplasm and accumulated in areas adjacent to the nucleus (Fig. 6, 3 h). From 4 h p.i., an extensive green fluorescence pattern was detected inside the nucleus, reflecting the targeting of newly synthesized pUL35GFP to intranuclear sites of capsid assembly (Fig. 6, 4 h). In addition, cytoplasmic accumulations of pUL35GFP decreased as intranuclear fluorescence intensity increased. However, although intranuclear fluorescence was predominant, intracytoplasmic fluorescent punctae, which could represent newly formed capsids during egress, were also observed (Fig. 6, 4 and 5 h). Late in infection, cells infected with PrV-UL35GFP showed typical morphological changes described for alphaherpesvirus infections. The cells assumed a more spherical shape and showed enlarged nuclei, which were filled with the UL35-eGFP fusion protein (Fig. 6, 5 and 8 h).
The course of infection by noncomplemented PrV-ΔUL37/UL35GFP was essentially the same, but with a ca. 1- to 2-h delay (Fig. 6, middle panel). This corresponds to the delay in capsid transport to the nucleus observed at earlier times after infection (Fig. 4B and 5). Interestingly, the delayed onset of replication of the pUL37 deletion mutant was corrected by phenotypic transcomplementation of PrV-ΔUL37/UL35GFP. After infection of RK13 cells with pUL37-containing virions of this mutant, first intracytoplasmic as well as intranuclear GFP fluorescence could be observed at 3 h and 4 h p.i., respectively (Fig. 6, right panel), similar to observations of PrV-UL35GFP-infected cells (Fig. 6, left panel).
Thus, our data show that the absence of pUL37 delays nuclear translocation of incoming capsids by about 1 h but does not abolish it. A delay between virus entry and immediate-early gene expression has also been observed in a human cytomegalovirus mutant lacking the pUL37 homolog UL47 (3), which might reflect a similar role for UL47 in human cytomegalovirus to that of pUL37 in PrV. The observed retention of capsids at the cellular periphery indicates that pUL37 may play an accessory role during formation of initial connections between viral capsids and MT-dependent transport proteins.
For active MT-mediated transport, herpesvirus particles have to interact directly or via adaptor proteins with cellular motor proteins. Interactions with dynein or kinesin subunits have been identified for several proteins of HSV-1 (13, 34, 37, 55), but up to now only the PrV homolog of tegument protein VP1/2 (pUL36) has been shown to be essential for intracellular capsid transport (32). Besides pUL36, its complex partner pUL37 also remains associated with incoming nucleocapsids during transport (19), and in this study we demonstrate that the conserved tegument protein pUL37 enhances nuclear trafficking of PrV capsids. However, since pUL37 is not essential for intracellular MT-mediated transport, a direct interaction with cellular motor proteins appears unlikely, although it cannot be completely excluded.
We consider it most likely that, in the absence of pUL37, binding of other viral protein(s), possibly pUL36, to the cellular MT motor system is impaired, resulting in the observed delay in nuclear translocation of incoming capsids. Since a similar defect has also been observed for egressing capsids (32), pUL37 may have similar roles during entry and egress. Although intracellular transport of herpesvirus capsids appears to be bidirectional, different directions are preferred during entry or egress (46, 47). At this time, it is not known by which molecular mechanism overall direction of capsid transport is controlled, but various compositions of the attached tegument proteins or their differential phosphorylation by cellular or viral kinases could be involved (31, 42, 47). Thus, pUL37 might also contribute to this regulatory process.
The similar virus titers and plaque sizes obtained with noncomplementing cells infected with phenotypically complemented or noncomplemented preparations of UL37-deleted PrV revealed that their different entry kinetics may not lead to drastic alterations in viral replication. However, a decrease in nucleocapsid velocity during egress could impair virion morphogenesis, e.g., the addition of other tegument proteins and final envelopment in different regions of the cytoplasm, which may not be reached “in time,” resulting in the observed accumulations of nonenveloped HSV-1 and PrV capsids in juxtanuclear regions (6, 25). Thus, it is conceivable that the observed defects associated with lack of pUL37, delayed transport and impaired virion morphogenesis, are indeed linked.
The entry delay of pUL37-deleted PrV that we observed in nonpolarized cultured cells may be significantly extended in sensory neurons because of the long distances between nerve endings embedded in viral replication centers in the periphery and the nucleated cell bodies in sensory ganglia (14, 51). A similar situation should then occur in anterograde capsid transport to the synapse for transneuronal spread (32). A combination of these defects could explain the complete block in neuroinvasion which was observed after deletion of PrV pUL37 in a murine infection model (23).
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (grant Me 854/9).
We thank C. Ehrlich and D. Werner for technical assistance.
Footnotes
Published ahead of print on 14 January 2009.
REFERENCES
- 1.Antinone, S. E., G. T. Shubeita, K. E. Coller, J. I. Lee, S. Haverlock-Moyns, S. P. Gross, and G. A. Smith. 2006. The herpesvirus capsid surface protein VP26, and the majority of tegument proteins are dispensable for capsid transport toward the nucleus. J. Virol. 805494-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J. Virol. 46371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtel, J., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 761043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai, P., A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247115-124. [DOI] [PubMed] [Google Scholar]
- 5.Desai, P., and S. Person. 1998. Incorporation of the green fluorescence protein into the herpes simplex virus type 1 capsid. J. Virol. 727563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, P., L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 7510259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diefenbach, R. J., E. Diefenbach, M. W. Douglas, and A. L. Cunningham. 2004. The ribosome receptor, p180, interacts with the kinesin heavy chain, KIF5B. Biochem. Biophys. Res. Commun. 319987-992. [DOI] [PubMed] [Google Scholar]
- 8.Diefenbach, R. J., M. Miranda-Saksena, E. Diefenbach, D. J. Holland, R. A. Boadle, P. J. Armati, and A. L. Cunningham. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 763282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Döhner, K., C. H. Nagel, and B. Sodeik. 2005. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 13320-327. [DOI] [PubMed] [Google Scholar]
- 10.Döhner, K., K. Radtke, S. Schmidt, and B. Sodeik. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated transport without the small capsid protein VP26. J. Virol. 808211-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Döhner, K., and B. Sodeik. 2005. The role of cytoskeleton during viral infection. Curr. Top. Microbiol. Immunol. 200567-108. [DOI] [PubMed] [Google Scholar]
- 12.Döhner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 132795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas, M. W., R. J. Diefenbach, F. L. Homa, M. Miranda-Saksena, F. J. Rixon, V. Vitton, K. Byth, and A. L. Cunningham. 2004. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 27928522-28530. [DOI] [PubMed] [Google Scholar]
- 14.Feierbach, B., M. Bisher, J. Goodhouse, and L. W. Enquist. 2007. In vitro analysis of transneuronal spread of an alphaherpesvirus infection on peripheral nervous system neurons. J. Virol. 816846-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florin, L., K. A. Becker, C. Lambert, T. Nowak, C. Sapp, D. Strand, R. E. Streeck, and M. Sapp. 2006. Identification of a dynein interaction domain in the papillomavirus minor capsid protein L2. J. Virol. 806691-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 7811879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garner, J. A. 2003. Herpes simplex virion entry into and intracellular transport within mammalian cells. 2003. Adv. Drug Deliv. Rev. 551497-1513. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39389-400. [DOI] [PubMed] [Google Scholar]
- 19.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 793200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication of pseudorabies virus in cell culture: a reassessment. J. Virol. 712072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmon, M. E., and W. Gibson. 1996. High molecular weight virion protein of human cytomegalovirus forms complex with product of adjacent open reading frame, abstr. W35-4, p.144. Proc. Am. Soc. Virol., London, Ontario, Canada.
- 22.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of the herpes simplex and pseudorabies viruses. Virology 7394-407. [DOI] [PubMed] [Google Scholar]
- 23.Klopfleisch, R., B. G. Klupp, W. Fuchs, M. Kopp, J. P. Teifke, and T. C. Mettenleiter. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J. Virol. 805571-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. The pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 763065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 758927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koshizuka, T., Y. Kawaguchi, and Y. Nishiyama. 2005. Herpes simplex virus type 2 membrane protein UL56 associates with the kinesin motor protein KIF1A. J. Gen. Virol. 86527-533. [DOI] [PubMed] [Google Scholar]
- 27.Krautwald, M., C. Maresch, B. G. Klupp, W. Fuchs, and T. C. Mettenleiter. 2008. Deletion or green fluorescent tagging of the pUL35 capsid component of pseudorabies virus impairs replication in cell culture and neuroinvasion in mice. J. Gen. Virol. 891346-1351. [DOI] [PubMed] [Google Scholar]
- 28.Kristensson, K., E. Lycke, M. Roytta, B. Svennerholm, and A. Vahlne. 1986. Neuritic transport of herpes simplex virus in rat sensory neurons in vitro. Effects of substances interacting with microtubular function and axonal flow [nocodazole, taxol and erythro-9-3-(2-hydroxynonyl)adenine)]. J. Gen. Virol. 672023-2028. [DOI] [PubMed] [Google Scholar]
- 29.Kwong, A. D., and N. Frenkel. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 634834-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leopold, P. L., G. Kreitzer, N. Miyazawa, S. Rempel, K. K. Pfister, E. Rodriguez-Boulan, and R. G. Crystal. 2000. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther. 11151-165. [DOI] [PubMed] [Google Scholar]
- 31.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 1025832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luxton, G. W., J. I. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mabit, H., M. Y. Nakano, U. Prank, B. Saam, K. Döhner, B. Sodeik, and U. F. Greber. 2002. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 769962-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Moreno, M., I. Navarro-Lerida, F. Roncal, J. P. Albar, C. Alonso, F. Gavilanes, and I. Rodriguez-Crespo. 2003. Recognition of novel viral sequences that associate with the dynein light chain LC8 identified through a pepscan technique. FEBS Lett. 544262-267. [DOI] [PubMed] [Google Scholar]
- 35.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of intracellular behavior of HIV in living cells. J. Cell Biol. 159441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171623-625. [DOI] [PubMed] [Google Scholar]
- 37.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113163-169. [DOI] [PubMed] [Google Scholar]
- 38.Nogales, E. 2001. Structural insight into microtubule function. Annu. Rev. Biophys. Biomol. Struct. 30397-420. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Ogawa-Goto, K., K. Tanaka, W. Gibson, E. Moriishi, Y. Miura, T. Kurata, S. Irie, and T. Sata. 2003. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J. Virol. 778541-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ploubidou, A., and M. Way. 2001. Viral transport and the cytoskeleton. Curr. Opin. Cell Biol. 1397-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quie, L., D. Macellino, and B. C. Herold. 1999. Herpes simplex virus entry is associated with tyrosine phosphorylation of cellular proteins. Virology 256220-227. [DOI] [PubMed] [Google Scholar]
- 43.Radtke, K., K. Döhner, and B. Sodeik. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell. Microbiol. 8387-400. [DOI] [PubMed] [Google Scholar]
- 44.Schroer, T. A. 2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20759-779. [DOI] [PubMed] [Google Scholar]
- 45.Smith, G. A., and L. W. Enquist. 2002. Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 18135-161. [DOI] [PubMed] [Google Scholar]
- 46.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. USA 983466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 10116034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sodeik, B., M. W. Ebershold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1361007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spear, P. G., S. Manoj, M. Yoon, C. R. Jogger, A. Zago, and D. Myscofski. 2006. Different receptor binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology 34417-24. [DOI] [PubMed] [Google Scholar]
- 50.Suikkanen, S., T. Aaltonen, M. Nevalainen, O. Valilehto, L. Lindholm, M. Vuento, and M. Vihinen-Ranta. 2003. Exploitation of microtubule cytoskel-eton and dynein during parvoviral traffic towards the nucleus. J. Virol. 7710270-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomishima, M. J., G. A. Smith, and L. W. Enquist. 2001. Sorting and transport of alpha herpesviruses in axons. Traffic 2429-436. [DOI] [PubMed] [Google Scholar]
- 52.Topp, K. S., K. Bisla, N. D. Saks, and J. H. Lavail. 1996. Centripetal transport of herpes simplex virus in human retinal pigment epithelial cells in vitro. Neuroscience 711133-1144. [DOI] [PubMed] [Google Scholar]
- 53.Vallee, R. B., J. C. Williams, D. Varma, and L. E. Barnhart. 2004. Dynein: an ancient motor protein involved in multiple modes of transport. J. Neurobiol. 58189-200. [DOI] [PubMed] [Google Scholar]
- 54.Wolfstein, A., C. H. Nagel, and K. Radtke. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7227-237. [DOI] [PubMed] [Google Scholar]
- 55.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 741355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]