Abstract
Proteolytic activation of the hemagglutinin (HA) protein is indispensable for influenza virus infectivity, and the tissue expression of the responsible cellular proteases impacts viral tropism and pathogenicity. The HA protein critically contributes to the exceptionally high pathogenicity of the 1918 influenza virus, but the mechanisms underlying cleavage activation of the 1918 HA have not been characterized. The neuraminidase (NA) protein of the 1918 influenza virus allows trypsin-independent growth in canine kidney cells (MDCK). However, it is at present unknown if the 1918 NA, like the NA of the closely related strain A/WSN/33, facilitates HA cleavage activation by recruiting the proprotease plasminogen. Moreover, it is not known which pulmonary proteases activate the 1918 HA. We provide evidence that NA-dependent, trypsin-independent cleavage activation of the 1918 HA is cell line dependent and most likely plasminogen independent since the 1918 NA failed to recruit plasminogen and neither exogenous plasminogen nor the presence of the A/WSN/33 NA promoted efficient cleavage of the 1918 HA. The transmembrane serine protease TMPRSS4 was found to be expressed in lung tissue and was shown to cleave the 1918 HA. Accordingly, coexpression of the 1918 HA with TMPRSS4 or the previously identified HA-processing protease TMPRSS2 allowed trypsin-independent infection by pseuodotypes bearing the 1918 HA, indicating that these proteases might support 1918 influenza virus spread in the lung. In summary, we show that the previously reported 1918 NA-dependent spread of the 1918 influenza virus is a cell line-dependent phenomenon and is not due to plasminogen recruitment by the 1918 NA. Moreover, we provide evidence that TMPRSS2 and TMPRSS4 activate the 1918 HA by cleavage and therefore may promote viral spread in lung tissue.
Influenza A viruses exhibit high genetic variability. The accumulation of relatively subtle changes in the surface proteins hemagglutinin (HA) and neuraminidase (NA) of currently circulating viruses, termed antigenic drift, is responsible for the annual influenza virus epidemics. However, the reassortment of genomic material between human and animal influenza A viruses can occasionally lead to emergence of viral variants with radically different antigenic properties, a phenomenon termed antigenic shift (9, 32). Due to the lack of preexisting immunity in the human population, these viruses can cause pandemics. Three influenza pandemics were recorded in the last century. The so-called Asian influenza in 1957 and the Hong Kong influenza in 1968 caused approximately >2 million and 1 million deaths (World Health Organization, Geneva, Switzerland; www.who.int/csr/disease/influenza), respectively, and the etiologic agents were reassortants between human and avian influenza A viruses (2). The third influenza pandemic, which occurred in 1918 and is commonly termed Spanish influenza, differed in several aspects from the previously mentioned pandemics (1, 30). First, the mortality associated with the 1918 pandemic was extraordinarily high, and it is estimated that about 20 to 50 million people died from the disease. Second, instead of infants and the elderly, who are usually the main populations affected in influenza virus epidemics, adults between the ages of 18 and 30 had to bear the brunt of the 1918 pandemic (1, 30). Third, evidence is accumulating that the 1918 virus has similarities with avian influenza viruses and was not the product of a reassortment between human and animal viruses (43, 44).
Reconstitution of the 1918 influenza virus by reverse genetics (45) showed that HA, NA, and PB1 critically contribute to high virulence (22, 31, 47). The HA protein mediates binding to the cellular receptor, alpha 2,6 sialylated glycans and, upon exposure to endosomal low pH, drives fusion of the virus and a cellular membrane, a prerequisite to infectious entry (39). Cleavage of the precursor protein HA0 into the covalently linked subunits HA1 and HA2 by a cellular protease is required for viral infectivity (20, 23) and is an important determinant of viral tropism (40). Usually a single arginine residue is present at the border between HA1 and HA2 and is part of the motif recognized by cellular proteases such as serine family proteases (17, 18), like the recently identified HA-processing proteases TMPRSS2 (transmembrane protease, serine 2) and HAT (human airway trypsin-like protease) (5). Since expression of these proteases is limited to the respiratory tract in mammalian hosts, virus replication is confined to this target site. However, an optimized, multibasic cleavage site is present in all highly pathogenic avian influenza viruses. The HA protein of these viruses is cleaved by ubiquitously expressed subtilisin-like proteases, and consequently the respective viruses can spread systemically in susceptible domestic poultry (17, 18). The requirement for addition of trypsin to support efficient virus replication in cell culture is determined by the nature of the cleavage site in the HA protein. Viruses harboring a multibasic cleavage site can spread in the absence of trypsin, while trypsin activation is essential for the replication of viruses that do not encode an HA protein with a multibasic cleavage site.
The 1918 influenza virus HA does not harbor a multibasic cleavage site, yet the virus replicates to high titers in MDCK cells irrespective of trypsin activation (45). Notably, NA is essential for trypsin-independent spread of the 1918 virus in MDCK cells (45). The NA of a related virus, A/WSN/33 (H1N1), also facilitates efficient viral replication in the absence of trypsin and is required for an expanded viral tropism (14, 15). The A/WSN/33 NA abrogates the need for trypsin by sequestering plasminogen which, upon conversion to plasmin, facilitates HA cleavage (14, 15). A comparable mechanism can be envisioned for the 1918 influenza virus NA. However, plasminogen binding by the A/WSN/33 NA critically depends on the absence of a N-linked glycosylation motif in NA that is otherwise conserved in all N1 subtype NA proteins of naturally occurring viruses, including the 1918 virus (34). The molecular mechanism behind NA-dependent, trypsin-independent replication of the 1918 influenza virus is therefore at present unclear.
We established a pseudotyping system to analyze the proteolytic activation of the 1918 HA. We report that the previously noted 1918 NA-dependent, trypsin-independent activation of the 1918 HA in MDCK cells does not occur in Huh-7 and 293T cells and is not due to plasminogen recruitment by the 1918 NA. In addition, we show that the transmembrane proteases TMPRSS2 and TMPRSS4 activate the 1918 HA by cleavage, suggesting that these proteases might facilitate replication of the 1918 influenza virus in lung cells.
MATERIALS AND METHODS
Cell culture and plasmids.
293T, Huh-7, and MDCK cells were maintained in Dulbecco's modified Eagle's medium (PAA) supplemented with 10% fetal bovine serum (FCS; Cambrex) and the antibiotics penicillin and streptomycin. Plasmids encoding the 1918 HA (South Carolina), NA, and M2 (Brevig Mission) proteins as well as the Zaire Ebola virus glycoproteins (ZEBOV-GP), A/WSN/33 HA, A/WSN/33 NA, vesicular stomatitis virus G protein (VSV-G), TMPRSS2, TMPRSS4, and mouse matriptase-3 have been described previously (11, 13, 19, 33, 34, 36, 38, 41, 46).
Production of virus-like particles (VLPs) and Western blot analysis.
293T cells were cotransfected with the human immunodeficiency virus type 1 (HIV-1) Gag (p55) encoding plasmid p96ZM651gag-opt (12) and the indicated combinations of the 1918 influenza virus HA, NA, and M2 expression plasmids or empty vector. The culture medium was changed 12 h after transfection, and VLPs in the supernatant were harvested 36 h later. VLPs were passed through 0.45-μm pore-size filters and concentrated by centrifugation through a 20% sucrose cushion for 2 h at 4°C and 14,000 rpm or by using Vivaspin centrifugal concentrators. VLPs were resuspended in medium and, if applicable, treated with trypsin and subsequently soybean trypsin inhibitor (Sigma, Germany). Sodium dodecyl sulfate (SDS) buffer was added, and the samples were incubated at 95°C for 20 min and loaded onto a 10% SDS-polyacrylamide gel electrophoresis gel. HA was detected using a mouse monoclonal antibody (13) at a 1:500 dilution.
Reporter viruses and infection assays.
For generating HIV-1 NL4-3-based luciferase reporter viruses, 293T cells were transiently cotransfected with pNL4-3 E−R− Luc (8) and combinations of 1918 influenza virus HA, NA, and M2 expression plasmids; A/WSN/33 HA and NA expression plasmids; or ZEBOV-GP and VSV-G expression plasmids (38) as controls. At 48 h posttransfection, cell culture supernatants were harvested, passed through 0.45-μm pore-size filters, aliquoted, and stored at −80°C. Subsequently, the p24 content of virus stocks was determined by enzyme-linked immunosorbent assay (Murex, Germany), or infectivity was assessed by inoculation of target cells with different dilutions of virus stocks, followed by quantification of luciferase activities in cellular lysates. For infection experiments, target cells were seeded in 96-well plates at a density of 1 × 104 cells/well 24 h prior to infection. The cellular monolayer was inoculated with 50 μl of virus stocks normalized for p24 content or infectivity. Medium was replaced 12 h after virus infection, and luciferase activities in cell lysates were determined 72 h postinfection employing a commercially available kit (Promega).
Influence of trypsin pretreatment and spinoculation on infectivity of HA pseudotypes.
Viruses pseudotyped with 1918 influenza virus proteins or A/WSN/33 HA and NA were pretreated with l-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (100 μg/ml final concentration) (Sigma, Germany) for 10 min at room temperature. Subsequently, trypsin was inactivated by adding soybean trypsin inhibitor (Sigma, Germany) to a final concentration of 100 μg/ml. Viruses were either added to the cells and the cells were directly placed in the incubator, or cells were infected by centrifugation (spinoculated) at 1,200 rpm and 25°C for 2 h. Medium was replaced 12 h after transduction, and luciferase activities in cell lysates were determined 72 h postinfection using a commercially available kit (Promega).
Protease inhibitor experiments.
Cells were pretreated with the indicated concentrations of E64d (Sigma, Germany) for 30 min at 37°C and infected with infectivity-normalized pseudotypes. In the case of HA NA pseudotypes, viruses were pretreated with trypsin as described above or treated with phosphate-buffered saline (PBS) and spinoculated onto cells. Medium was replaced 12 h postinfection, and luciferase activities in cell lysates were measured after 3 days using a commercially available kit (Promega).
Activation of pseudotype infectivity by cellular lysates.
MDCK and 293T cells were grown in 10-cm dishes, scraped off the culture dishes, and counted, and 5 million MDCK or 293T cells were resuspended in 1 ml of PBS containing 10 mM CaCl2 and 2% 1-deoxymannojirimycin hydrochloride (Sigma, Germany), as described previously (6). Cells were then sonicated on ice for 3 s. Pseudotyped viruses were pretreated with either the indicated amounts of cellular lysates for 1 h at 37°C or 100 μg/ml TPCK-treated trypsin for 10 min at room temperature. Virus particles were concentrated by centrifugation through a 20% sucrose cushion at 14,000 rpm for 2 h at 4°C. Virus pellets were resuspended in medium and used for infection of Huh-7 and 293T cells. Medium was replaced 12 h postinfection, and luciferase activities in cell lysates were determined 60 h later using a commercially available kit (Promega).
Plasminogen binding to HA and NA.
293T cells were seeded in T-25 flasks and transiently transfected with plasmids encoding the 1918 NA, 1918 HA, A/WSN/33 HA, A/WSN/33 NA, and ZEBOV-GP or were control transfected with empty vector. Culture medium was replaced at 12 h posttransfection, and at 2 h before analysis of plasminogen binding, the cells were washed with FCS-free medium and cultured in the absence of FCS. For assessment of plasminogen binding, cells were incubated with PBS or purified plasminogen (Sigma, Germany) at a final concentration of 10 μg/ml for 30 min at 4°C. Subsequently, unbound plasminogen was removed by washing, and bound plasminogen was detected employing a plasminogen-specific goat serum (Immunology Consultants Laboratory) and an Cy5-labeled anti-goat secondary antibody (Jackson ImmunoResearch). Cell staining was analyzed by fluorescence-activated cell sorting (FACS) using a Beckman Coulter Cytomics FC 500.
Replication of the 1918 influenza virus in MDCK and Huh-7 cells with or without trypsin activation.
Based on the published sequences of the 1918 influenza virus genes, the complete viral and complementary sequences were synthesized as 40-mer oligonucleotides, with a 20-nucleotide overlap between each forward and reverse primer. The ligase chain reaction was used for initial assembly of subgenomic fragments from the pooled primers for each gene, and the full-length genes were assembled by amplification from the pool of gene fragments by using PCR according to the protocol described by Rouillard and colleagues (35). Each gene was further amplified by PCR using the gene-specific universal primer pairs described by Hoffmann and colleagues (16) and subcloned into the pPolI vector for reverse genetics (16, 27). The 1918 influenza virus was rescued as previously described (22) and titers were determined by standard plaque assay on MDCK cells in the presence of TPCK-treated trypsin. MDCK or Huh-7 cells were seeded in six-well tissue culture plates and infected at a multiplicity of infection (MOI) of 0.001, and then culture medium was added with or without 1 μg/ml of TPCK-treated trypsin. Supernatants of the virus-infected MDCK cultures were collected at 24, 48, and 72 h and at 48 h postinfection (p.i.) from infected Huh-7 cultures and stored at −80°C. Virus titers were determined by the 50% tissue culture infective dose on MDCK cells seeded in 96-well plates. Samples from cultures maintained without trypsin were treated with trypsin (at a final concentration of 1 μg/ml) prior to titration. Procedures for the production and propagation of the 1918 virus and all subsequent experiments involving infectious 1918 influenza virus were performed in a biosafety level 4 facility of the National Microbiology Laboratory of the Public Health Agency of Canada.
Detection of TMPRSS4 mRNA in lung cells.
mRNA from cells in bronchoalveolar lavage was extracted, DNase treated, and reverse transcribed employing commercially available kits (Qiagen, Germany, and Invitrogen, Germany). Subsequently, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and TMPRSS4 sequences were amplified by nested PCR. The following sets of primers were used for amplification of TMPRSS4: the pair 5′-GAGAGCTGGACTGTCCCTTG-3′ (p5 TMPRSS4 out) and 5′-TCGTACTGGATGCTGACCTG-3′ (p3 TMPRSS4 out) and the pair 5′-GACGAGGAGCACTGTGTCAA-3′ (p5 TMPRSS4 in) and 5′-CTTCCCACAGGCAAGACAGT-3′ (p3 TMPRSS4 in).
RESULTS
Trypsin treatment of 1918 influenza virus HA- NA-bearing pseudotypes is required for infectivity.
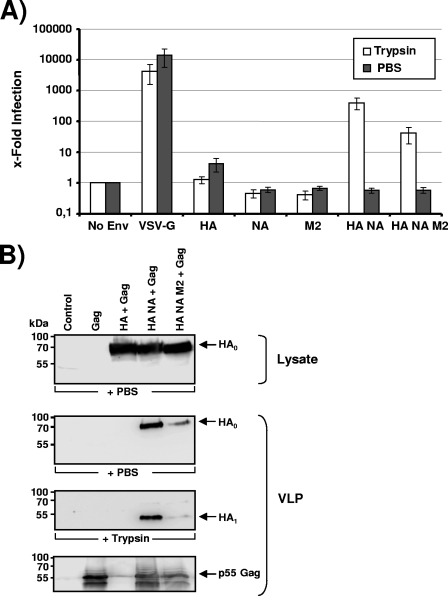
It has been shown previously that the genetically reconstituted 1918 H1N1 influenza virus replicates in MDCK cells to high titers irrespective of the presence of trypsin (45). The expression of the 1918 virus NA was essential for trypsin-independent growth (45), and it is possible that NA might recruit a cellular protease, which facilitates HA cleavage—a prerequisite to HA-dependent infectious cellular entry (39). In order to further characterize the role of NA in HA activation, we sought to establish a pseudotyping system, which allows convenient analysis of HA and NA in the absence of infectious 1918 influenza virus. Since the influenza virus M2 protein is present in the viral envelope and can prevent premature triggering of HA (3), we also included the 1918 influenza virus M2 protein in our analysis. We first investigated if Env-defective HIV-1 NL4-3-based reporter viruses (8) pseudotyped with the 1918 influenza virus HA, NA, and M2 or combinations thereof are infectious for 293T cells. To this end, virus preparations were normalized for equal content of p24 capsid protein, treated with TPCK-trypsin or PBS, and, after the addition of a trypsin inhibitor, used to infect 293T cells. No luciferase activities above background levels were observed in lysates of cells infected with Env-negative “bald” pseudotypes, while VSV-G-bearing pseudotypes allowed efficient virus entry independent of the presence of trypsin (Fig. 1A). Robust infection of 293T cells with pseudotypes bearing the membrane proteins of the 1918 influenza virus was observed but only when both HA and NA were expressed in the cells used for pseudotype production and when pseudotypes were treated with trypsin before addition to target cells (Fig. 1A). In contrast, pseudotypes produced in the absence of NA were not infectious. This defect was most likely due to a lack of virion incorporation of HA. Thus, lentiviruses are inefficiently released from influenza virus HA-transfected cells in the absence of NA (Fig. 1B, lane 3) (4), and the particles that are released under these conditions do not harbor detectable amounts of HA (4), consistent with the established key role of NA in facilitating virus release due to its receptor-destroying function (40). Finally, we observed that coexpression of HA and NA together with the M2 protein reduced HA incorporation into pseudotypes and thus diminished pseudotype infectivity (Fig. 1B, lane 5).
FIG. 1.
Trypsin activation of 1918 influenza virus HA pseudotypes is required for infectivity independent of the presence of NA. (A) 293T cells were infected with env-defective, p24-normalized HIV-1 NL4-3 reporter virus pseudotyped with the indicated glycoproteins. Prior to infection, the virions were treated with trypsin or PBS, and after 10 min soybean trypsin inhibitor was added. Three days after infection, luciferase activities in cellular lysates were determined. Infection observed with glycoprotein harboring pseudotypes is shown relative to infection detected with control particles bearing no glycoprotein. The average of four independent experiments with different virus stocks is shown. Error bars indicate standard errors of the means. (B) HIV-1-based VLPs with the indicated combinations of surface proteins were generated in 293T cells and concentrated by centrifugation through a 20% sucrose gradient. The VLP preparations were normalized for comparable content of HIV-1 Gag (p55), pretreated with trypsin or PBS, separated by SDS-polyacrylamide gel electrophoresis, and analyzed by Western blotting. The 1918 influenza virus HA was detected with a mouse monoclonal antibody. Similar results were obtained in two independent experiments with different VLP preparations.
We next sought to confirm whether trypsin treatment indeed resulted in cleavage of HA. Western blot analysis of V5 epitope-tagged versions of HA, NA, and M2 showed robust and comparable expression in 293T cell lysates (Fig. 1B and data not shown), and efficient incorporation of HA, NA, and M2 into HIV-1 Gag p55-based VLPs could be detected with V5-specific (for NA and M2) or HA-specific monoclonal antibodies (Fig. 1B and data not shown). Importantly, trypsin treatment of VLPs did not impact gel migration of NA or M2 (data not shown), while trypsin treatment reduced the size of HA from approximately 75 kDa to 50 kDa, consistent with cleavage at the border between HA1 and HA2 (Fig. 1B).
Augmentation of cellular attachment allows inefficient infectious entry of the 1918 HA NA pseudotypes in the absence of trypsin activation.
We next investigated if the infectivity of the 1918 influenza virus HA and NA pseudotypes for different cell lines is invariably trypsin dependent. Moreover, we asked if the requirement for trypsin can be overcome when virus attachment and, thus, infection efficiency are increased. For this, 293T, Huh-7, and MDCK cells were infected with pseudotypes harboring the 1918 influenza virus HA, NA, and M2 proteins or VSV-G in their envelopes. All pseudotypes were pretreated with trypsin or PBS and used for infection of two plates in parallel. One plate was instantly incubated at 37°C; the other was used for spinoculation (2 h at 25°C and 1,200 rpm) (28) and then kept at 37°C. All cell lines were readily susceptible to infection with VSV-G (independent of trypsin treatment) and trypsin-activated HA NA pseudotypes, while none of the cell lines was susceptible to HA NA pseudotypes in the absence of trypsin treatment and spinoculation (Fig. 2A). Spinoculation substantially augmented entry of HA- NA-bearing viruses in the absence of trypsin treatment (Fig. 2A), and analysis of p24-normalized, non-trypsin-treated virus preparations confirmed that the presence of HA and NA was required for appreciable infectious entry under these conditions (data not shown). However, even upon spinoculation, infectious entry of non-trypsin-treated virus was less efficient than entry of trypsin-activated virus (Fig. 2A). Finally, in agreement with our previous observations (25), spinoculation had little effect on infection by pseudotypes exhibiting high infectivity in the absence of experimentally optimized attachment, i.e., reporter viruses bearing trypsin-activated HA, NA, or VSV-G proteins (Fig. 2A). Thus, increased attachment efficiency allows some infectious entry of HA- NA-bearing viruses in the absence of trypsin but does not abrogate the requirement for trypsin activation for high infectivity.
FIG. 2.
Impact of spinoculation and protease inhibitors on 1918 influenza virus HA-driven viral entry. (A) 293T, Huh-7, and MDCK cells were seeded in 96-well plates and infected with p24-normalized pseudotypes bearing 1918 influenza virus HA, NA, and M2 or VSV-G. The virions were pretreated with trypsin or PBS and, subsequently, a trypsin inhibitor, before addition to target cells. Two plates were infected in parallel: one plate was incubated at 37°C immediately after addition of virus; the other plate was centrifuged for 2 h at 25°C and 1,200 rpm after infection and then incubated at 37°C. Three days after infection, luciferase activities in cellular lysates were determined. A representative experiment performed in triplicates is shown. Error bars indicate standard deviations. Similar results were obtained in two independent experiments. (B) Huh-7 cells were incubated with the indicated concentrations of the cysteine protease inhibitor E64d prior to infection with infectivity-normalized pseudotypes carrying the 1918 influenza virus HA, NA, M2, ZEBOV-GP, or VSV-G. Pseudotypes bearing influenza virus proteins were pretreated with PBS or trypsin for 10 min at room temperature, and soybean trypsin inhibitor was added. Three days after infection, luciferase activities in cellular lysates were determined. Infection of Huh-7 cells in the absence of protease inhibitors was set as 100%. The average of three independent experiments performed in triplicate is shown. Error bars indicate standard errors of the means. cps, counts per second.
The observation that HA NA pseudotypes can infect cell lines with low but detectable efficiency without prior trypsin activation (Fig. 2A) suggests that uncleaved HA might also be activated by proteases in target cells. The pH-dependent endosomal cysteine proteases cathepsin B and L activate the severe acute respiratory syndrome (SARS)-coronavirus and EBOV-GP (7, 37) and are required for infectious entry. In order to analyze if the activity of cysteine proteases also contributes to 1918 influenza virus HA- NA-mediated entry, Huh-7 cells were incubated with the cysteine protease inhibitor E64d prior to infection with pseudotypes in the absence of trypsin activation. Pseudotypes bearing ZEBOV-GP or VSV-G, as well as trypsin-activated HA NA pseudotypes served as controls. Huh-7 cells were chosen as targets because these cells were more susceptible to spinoculation with non-trypsin-activated HA NA pseudotypes than MDCK and 293T cells (Fig. 2A). As expected from previous reports, ZEBOV-GP-mediated entry was efficiently and dose-dependently reduced by E64d, while VSV-G-driven entry was unaffected (Fig. 2B). E64d had no appreciable impact on HA- NA-driven infection irrespective of trypsin treatment, indicating that activation by cysteine proteases in target cells is not required for trypsin-independent infectious entry of HA NA pseudotypes.
The A/WSN/33 NA but not 1918 NA sequesters plasminogen but fails to facilitate cleavage-activation of the 1918 HA.
The A/WSN/33 virus is related to the 1918 virus, and both viruses replicate in a trypsin-independent, NA-dependent fashion in cell culture. The NA of A/WSN/33 mediates trypsin-independent replication by capturing plasminogen, which is present in high concentrations in FCS, and, upon conversion to plasmin, facilitates HA cleavage activation. In order to assess a potential role of plasminogen in proteolytic activation of the 1918 HA, we compared trypsin dependence of pseudotypes bearing the 1918 HA and NA, A/WSN/33 HA and NA, or viruses in which the NA proteins were exchanged. In accordance with published data (14, 15) and the results described above (Fig. 1), we found that the A/WSN/33 HA and NA but not the 1918 HA and NA pseudotypes readily infected Huh-7 cells without prior trypsin activation (Fig. 3A). However, viruses bearing the 1918 HA and A/WSN/33 NA or A/WSN/33 HA and the 1918 NA were completely dependent on trypsin treatment for infectivity. In agreement with these observations, Western blot analysis of VLPs revealed that trypsin but not purified human plasminogen promoted 1918 HA cleavage (Fig. 3B), and incubation of 1918 HA-bearing pseudotypes with purified human plasminogen failed to render these viruses infectious (Fig. 3C), further indicating that plasminogen is unlikely to be involved in proteolytic activation of the 1918 HA. Finally, Western blot analysis of HA-transfected cells showed that plasminogen was able to cleave the A/WSN/33 HA but not the 1918 HA (data not shown), confirming that the plasminogen preparation employed for the studies described above was indeed active.
FIG. 3.
A/WSN/33 NA fails to promote cleavage activation of the 1918 HA despite efficient recruitment of plasminogen. (A) Pseudotypes bearing the indicated glycoproteins were PBS or trypsin treated and used for infection of Huh-7 cells. Three days after infection, luciferase activities in cellular lysates were determined. The results of a representative experiment are shown and were confirmed in two independent experiments. Error bars indicate standard deviations. (B) VLPs bearing the 1918 HA and NA were incubated with PBS, trypsin, or plasminogen, and the 1918 HA was visualized by Western blotting. (C) The experiment was carried out as described for panel A. However, pseudotypes were treated with PBS, trypsin, or plasminogen (100 μg/ml). The results of a representative experiment are presented and were confirmed within a separate experiment. (D) The indicated proteins were transiently expressed in 293T cells, the transfected cells were incubated with PBS or purified plasminogen (10 μg/ml), and plasminogen binding was detected by FACS. A representative experiment is shown. Similar results were obtained in two independent experiments. PMG, plasminogen; cps, counts per second.
To further characterize the contribution of plasminogen to 1918 HA cleavage, we performed FACS analysis to assess binding of purified plasminogen to cells transiently expressing HA or NA proteins. Plasminogen binding to control-transfected cells was low but detectable by FACS analysis (Fig. 3D), indicating that unmodified 293T cells express an inefficient plasminogen capture activity. Expression of the A/WSN/33 NA profoundly augmented plasminogen binding, as expected, with signals in the absence of purified protein being most likely due to incomplete removal of plasminogen contained in FCS. In contrast, plasminogen binding to 1918 NA-expressing cells was in the background range. To our surprise, expression of the 1918 HA also facilitated robust plasminogen sequestration (Fig. 3D), suggesting that plasminogen capture is not an exclusive function of NA expression. Finally, control cells expressing ZEBOV-GP did not exhibit specific binding to plasminogen, indicating that expression of an irrelevant viral glycoprotein does not alter endogenous plasminogen capture activity of 293T cells.
In summary, our analyses of pseudotype infectivity (Fig. 3A and C), proteolytic processing of HA (Fig. 3B), and plasminogen binding (Fig. 3D) suggest that under the conditions tested the A/WSN/33 NA but not the 1918 NA recruited plasminogen for HA cleavage and that A/WSN/33 NA-dependent plasminogen sequestration was insufficient to confer infectivity to pseudotypes bearing the 1918 HA and NA.
Lysates of MDCK but not 293T cells activate infectivity of pseudotypes bearing the 1918 HA and NA.
MDCK cells were found to allow for 1918 influenza virus replication in the absence of trypsin (45) and should thus express a protease, which is recruited by NA and mediates HA cleavage. We therefore asked if lysates from MDCK and, for comparison, 293T cells were capable of activating the 1918 influenza virus HA NA pseudotypes. Trypsin treatment of pseudotypes served as a positive control (Fig. 4). MDCK lysates had no appreciable impact on VSV-G-driven infection, albeit a slight decrease in infectivity was observed at the highest lysate concentrations. Similarly, lysates from 293T cells did not confer infectivity to the 1918 HA NA pseudotypes (Fig. 4). In contrast, MDCK cell lysates activated infectivity of 1918 HA- NA-bearing pseudotypes in a dose-dependent fashion, albeit clearly less efficiently than trypsin, suggesting a cell line-dependent expression of the protease(s) required for activation of HA of the 1918 virus.
FIG. 4.
Lysates from MDCK but not 293T cells activate 1918 HA-driven viral entry. Pseudotypes bearing the 1918 influenza virus HA, NA, and M2, or VSV-G were pretreated with trypsin and, subsequently, soybean trypsin inhibitor or lysates from MDCK or 293T cells. After 30 min the treated pseudotypes were used for infection of 293T and Huh-7 cells. Three days after infection, luciferase activities in cellular lysates were determined. The results of a representative experiment performed in triplicate are shown and were confirmed in two separate experiments. Error bars indicate standard deviations. Statistical significance was calculated employing a two-tailed t test for independent samples. cps, counts per second.
The 1918 influenza virus replicates efficiently in MDCK but not Huh-7 cells in the absence of trypsin.
Thus far, our results indicated that 1918 influenza virus HA-mediated infectivity requires protease activation before the addition of virions to target cells and that MDCK but not 293T cells expressed a protease capable of activating HA. Activation of HA by MDCK lysates was clearly less efficient than trypsin activation, raising the question of whether growth kinetics of replication-competent 1918 influenza virus in MDCK cells would be comparable in the presence and absence of trypsin. To address this question, MDCK cells were infected with 1918 H1N1 strain A/South Carolina/1/18 at an MOI of 0.001 and then cultured in medium with or without TPCK-treated trypsin (Fig. 5). Peak titers of virus, grown in the presence of trypsin, were seen at 24 h postinfection, coincident with complete lysis of cells by this time. A decrease in titer at later time points was observed in the absence of further replication and inactivation of virus at 37°C. Similarly high titers of virus were measured in cultures grown without trypsin at 24 h, and high titers persisted to 72 h p.i. Virus replication in the absence of trypsin was not significantly lytic, even at 48 h p.i., and unchanged titers of virus isolated between 24 and 72 h p.i. suggested continued replication of virus.
FIG. 5.
The 1918 influenza virus replicates efficiently in MDCK but not Huh-7 cells in the absence of trypsin. MDCK and Huh-7 cells were infected at an MOI of 0.001 and then cultivated in the absence or presence of trypsin. Supernatants were collected at 24, 48, and 72 h p.i. from MDCK cultures and at 48 h p.i. from Huh-7 cultures, and the viral titers were determined by a 50% tissue culture infective dose (TCID50) assay on fresh MDCK cells. A representative experiment performed in duplicate is shown; error bars indicate standard deviations.
Comparison of 1918 virus replication on Huh-7 cells under identical conditions showed efficient replication of virus in the presence of trypsin at 48 h p.i. although titers were slightly reduced compared to MDCK cultures (Fig. 5). However, in the absence of trypsin, replication was inefficient, and only low titers of virus were recovered, supporting the observation that efficient trypsin-independent growth of the 1918 virus is cell line dependent.
Proteolytic activation of the 1918 HA by TMPRSS2 and TMPRSS4.
Infection of MDCK, Huh-7, and 293T cells with the 1918 influenza virus or pseudotypes serves as a model for virus spread outside the lung. In order to investigate proteolytic activation of the 1918 HA in lung cells, we asked if transmembrane serine proteases expressed in lung tissue could activate 1918 HA by cleavage upon coexpression in 293T cells. We selected TMPRSS2 and TMPRSS4 for these studies since TMPRSS2 has already been shown to cleave and activate the HA of A/Hong Kong/1/68 (5) by cleavage, and TMPRSS4 shares 31% amino acid sequence identity with TMPRSS2. Murine matriptase-3, which is not expressed in the lung, was also tested. Transient transfection of the 1918 HA and NA alone did not result in HA cleavage, as expected, while cleavage was readily observed upon treatment of transfected cells with trypsin (Fig. 6A). Similarly, expression of matriptase-3 or the subtilisin-like protease furin did not promote proteolytic processing of the 1918 HA (Fig. 6B). However, coexpression of the 1918 HA with TMPRSS2 and TMPRSS4 resulted in efficient processing of HA independent of the presence of NA (Fig. 6A and B). In this context, it needs to be noted that total HA signals were generally reduced upon coexpression of TMPRSS4 and were often reduced upon coexpression of TMPRSS2, albeit to a lesser extent. The degree of signal reduction was dependent on the ratio of HA and protease plasmids used for transfection (3:1 for the experiment shown in Fig. 6A and 1:1 for the experiment shown in Fig. 6B) and might reflect “overdigestion” of HA under conditions of protease overexpression. In agreement with the specific cleavage of the 1918 HA by TMPRSS2 and TMPRSS4, the 1918 HA NA pseudotypes produced in the presence of TMPRSS2 or TMPRSS4 but not matriptase-3 were fully infectious for Huh-7 cells without prior trypsin activation (Fig. 6C). Finally, reverse transcription-PCR analysis of cells contained in bronchoalveolar lavage fluid (Fig. 6D) and PCR analysis of commercially available lung cDNA (Clontech) (data not shown) confirmed that TMPRSS4 is expressed in human lung tissue (Fig. 6D), suggesting that TMPRSS2 and TMPRSS4 might be able to support spread of the 1918 influenza virus in the human lung.
FIG. 6.
TMPRSS2 and TMPRSS4 activate 1918 HA by cleavage. (A) 293T cells were transiently cotransfected with the 1918 HA (jointly with pcDNA3 or the 1918 NA) and TMPRSS2, TMPRSS4, or mouse matriptase-3 (HA and protease expression plasmids were transfected at a 3:1 ratio). Subsequently, the transfected cells were treated with trypsin or PBS, and proteolytic processing of HA was analyzed by Western blotting. (B) 293T cells were transfected with the 1918 HA alone or in combination with the indicated proteases (HA and protease expression plasmids were transfected at a 1:1 ratio); the cells were treated with PBS or trypsin, and proteolytic processing of HA was analyzed by Western blotting. (C) Pseudotypes were generated in 293T cells expressing the empty vector or the indicated proteases (HA and protease expression plasmids were transfected at a 3:1 ratio), normalized to p24 (150 pg/well), and employed for infection of Huh-7 cells. Three days after infection, luciferase activities in cellular lysates were determined. A representative experiment is shown, and similar results were obtained in two independent experiments. Error bars indicate standard deviations. (D) RNA was obtained from cells present in bronchoalveolar lavage fluids and reverse transcribed within reaction mixtures containing reverse transcriptase (RT) enzyme or PBS, and GAPDH and TMPRSS4 were amplified by PCR. M, molecular weight marker. cps, counts per second.
DISCUSSION
The virus responsible for the devastating 1918 influenza virus pandemic has been “resurrected” by employing a reverse genetics approach (45). Exchange of specific genomic segments of the 1918 influenza virus against those of well-characterized influenza viruses revealed that HA and NA critically contribute to virus pathogenicity (22, 31, 47). Interestingly, the NA protein facilitates virus spread in MDCK cells in the absence of trypsin activation (45), most likely by recruiting a cellular protease which cleaves HA. Employing a lentiviral pseudotyping system, we show that the respective HA-activating protease is expressed in a cell-type-specific manner and is unlikely to be plasminogen. Moreover, we demonstrate that the transmembrane proteases TMPRSS2 and TMPRSS4, which are expressed in lung tissue, activate the 1918 HA by cleavage in an NA-independent fashion.
Analysis of the 1918 influenza virus will continue to provide important insights into the determinants of influenza virus transmission and pathogenicity but is hampered by the requirement for a high level of biocontainment. Taking advantage of the well-known ability of retroviruses to incorporate heterologous proteins in their envelopes during release from infected cells, we generated HIV-1-derived particles bearing the HA, NA, and M2 proteins of the 1918 influenza virus. The particles were infectious but replication defective and thus constitute a safe and convenient tool for analyzing cellular entry of the 1918 virus and its inhibition by, e.g., neutralizing antibodies. Small amounts of the M2 protein are incorporated into influenza virus particles, and the ion channel function of M2 plays an important role in uncoating and, for some viruses, in maturation (3). Increased infectivity of pseudotypes bearing the fowl plague virus HA has been described upon coexpression of M2 in virus-producing cells (26). We observed efficient M2 expression and incorporation into VLPs (data not shown). Nevertheless, the presence of M2 decreased incorporation but not cellular expression of HA in our system (Fig. 1B), resulting in reduced particle infectivity. The reasons for the discrepancy between the published observations (26) and the data presented here are at present unclear and might be due to differences in the influenza virus proteins and pseudotyping systems used or might be explained by differences in M2 expression levels.
Infectivity of pseudotypes bearing the surface proteins of the 1918 virus was dependent on the presence of both HA and NA. However, the requirement for NA was not due to NA facilitating HA cleavage, at least under the conditions tested. Instead, NA was necessary for efficient particle release (Fig. 1B), consistent with the well-established receptor-destroying function of this protein, and in the absence of NA most likely particles devoid of HA were released. Particle-associated HA exhibited a size of approximately 75 kDa, which is expected for uncleaved HA (10). Trypsin activation reduced the HA size to approximately 50 kDa and was indispensable for viral infectivity (Fig. 1), demonstrating that in the 293T cell-based system examined here particle-associated HA was uncleaved, most likely due to lack of expression of adequate proteases in this cell type (see below). Nevertheless, when viruses were concentrated onto target cells by centrifugation, a procedure termed spinoculation (28), some infectivity of HA NA pseudotypes was observed in the absence of trypsin activation. Thus, proteases in target cells might be able to process HA, albeit cleavage efficiency might be low. The endosomal/lysosomal cysteine proteases cathepsin B and cathepsin L activate the glycoproteins of EBOV (7), SARS-coronavirus (37), and the fusion protein of Hendra virus (29) by cleavage, and cathepsin B/L activity has been shown to be essential for virus infectivity. However, inhibitors of cysteine proteases, which were previously shown to block EBOV and SARS-coronavirus entry (7, 37), had no effect on infectious entry of HA NA pseudotypes, suggesting that a different class of proteases might be involved in HA cleavage in target cells (Fig. 2B).
In the absence of artificially enhanced entry, infectivity of the 1918 HA NA pseudotypes was strictly dependent on trypsin activation (Fig. 1A and 2). Thus, proteases that efficiently activate HA were not present in the FCS and were not secreted from the cell lines tested, at least not in sufficient concentrations to activate HA. Particularly the former finding is noteworthy since it has been shown previously that NA of A/WSN/33, a close relative of the 1918 influenza virus, recruits plasminogen present in FCS and facilitates generation of plasmin, which in turn activates HA (14, 15). This process is required for trypsin-independent growth and neurotropism of A/WSN/33 (14, 15). Our analysis confirmed efficient plasminogen binding by cells expressing the A/WSN/33 NA (Fig. 3D). In contrast, we failed to detect plasminogen capture by 1918 NA-positive cells, which is in agreement with the notion that the 1918 NA harbors a glycosylation site incompatible with plasminogen recruitment (42). Moreover, the A/WSN/33 NA failed to confer infectivity to 1918 HA-containing pseudotypes in the absence of trypsin treatment (Fig. 3A). In conjunction with the inability of purified human plasminogen or plasminogen-containing FCS to appreciably activate 1918 HA by cleavage and to render the 1918 HA NA pseudotypes infectious (Fig. 3B and C), these observations indicate that plasminogen is likely not involved in 1918 HA cleavage-activation. Nevertheless, 1918 HA-expressing cells displayed robust plasminogen binding activity (Fig. 3D). The significance of this finding for 1918 influenza virus biology is at present unclear. One can speculate, however, that influenza viruses might have evolved several independent strategies to recruit plasminogen either to ensure cleavage of HA or to facilitate a so far undiscovered proteolytic cleavage event involved in influenza virus replication. In fact, a recent study revealed that plasminogen enhances the replication of several influenza A viruses and that incorporation of annexin II into influenza virus particles promotes conversion of plasminogen to its proteolytically active form, plasmin (24). Collectively, our data suggest that the 1918 NA might allow trypsin-independent replication of the 1918 virus by recruiting a cellular protease other than plasminogen or, maybe less likely, that NA facilitates HA activation by an entirely different mechanism.
Lysates prepared from MDCK but not 293T cells activated pseudotype infectivity, albeit with low efficiency, suggesting that HA-activating proteases are expressed in the former but not the latter cell line. The observation that pseudotype infectivity for 293T and MDCK cells was not detectable in the absence of spinoculation and was comparable upon spinoculation (Fig. 2) indicates that the HA-activating protease detected in MDCK lysates (Fig. 4) might not be present (or active) in endocytic vesicles (and might thus be unable to activate HA on incoming viruses) but might be recruited by NA during transport in the secretory pathway or at the cell surface. Unfortunately, production of pseudotypes in MDCK cells was inefficient (data not shown), preventing us from investigating this hypothesis. The finding that MDCK lysates were clearly less efficient in activating pseudotype infectivity than trypsin suggests that insertion into an intact lipid membrane or the integrity of a specific subcellular compartment might be indispensable for efficient proteolytic processing of HA. In any case, infectious 1918 influenza virus replicated in MDCK cells with similar kinetics and, as reported previously (45), peak titers in the presence and absence of trypsin (Fig. 5), indicating efficient function of the activating protease in the context of intact MDCK cells. Furthermore, efficient replication of the 1918 virus in Huh-7 hepatoma cells in the presence but not in the absence of trypsin confirmed that the trypsin-independent growth of the virus in non-lung cells is a cell line-dependent phenomenon. This observation may have implications for understanding the tissue tropism of the virus which is primarily restricted to the respiratory tract in experimentally infected animals (21, 45). Further investigation as to the nature of the protease activity in MDCK cells is required, as is the proof that this protease activity is present and can facilitate virus spread in human cells.
It is believed that several proteases expressed in lung tissue can facilitate influenza virus spread. However, the nature of these HA-activating pulmonary proteases is incompletely understood. Böttcher and colleagues presented evidence that the transmembrane serine proteases TMPRSS2 and HAT activate the influenza viruses A/Memphis/14/96 (H1N1), A/Mallard/Alberta/205/98 (H2N9), and A/Texas/6/96 (H3N2). Coexpression of TMPRSS2 also facilitated cleavage of the 1918 HA independent of the presence of NA (Fig. 6A and B), and similar results were obtained with TMPRSS4, for which mRNA expression could be detected in human lung cells (Fig. 6A and B), suggesting that these proteases might support the spread of influenza viruses in human lung. In contrast, 293T, Huh-7, and MDCK cells were negative for TMPRSS4 transcripts when analyzed by direct reverse transcription-PCR (signals were observed for Huh-7 and MDCK cells upon nested PCR [data not shown]), indicating that these cell lines might express no TMPRSS4 protein or very small amounts thereof. These results are in agreement with the findings that infectivity of viruses produced in 293T and Huh-7 cells is trypsin dependent (Fig. 1 and 5) and that trypsin-independent virus spread in MDCK cells requires NA-dependent recruitment of a so far unidentified cellular protease (45) (note that TMPRSS2/ TMPRSS4 cleavage of the 1918 HA is NA independent) (Fig. 6A). Notably, cleavage of the 1918 HA by TMPRSS2 and TMPRSS4 proteases resulted in HA1 products of slightly different sizes (Fig. 6A and B), a finding that needs further investigation. One possible explanation might be that TMPRSS2 cleaves the 1918 HA at the border between HA1 and HA2 and, in addition, in HA1, thereby producing an HA1 fragment exhibiting slightly faster gel migration than the respective fragments produced by TMPRSS4 or trypsin cleavage. In any event, this study identifies TMPRSS4 as a novel influenza virus HA-processing protease expressed in lung tissue (Fig. 6C), and the impact of TMPRSS4, HAT, and TMPRSS2 on spread of the 1918 influenza virus and other influenza viruses in vivo deserves assessment.
Acknowledgments
We thank Adolfo García-Sastre for the kind gift of the 1918 influenza virus HA, NA, and M2 expression plasmids and Peter Palese for the kind gift of the A/WSN/33 HA and NA expression plasmids. In addition, we thank Yoshihiro Kawaoka for generously providing the pPolI and pCAGGS/MCS plasmids used for influenza virus reverse genetics. We also thank Thomas Moran and the members of the Center for Investigation of Viral Immunity and Antagonism for kindly providing us with anti-HA antibody, Heinz Feldmann for critically reading the manuscript, Franz Bange for bronchoalveolar lavage samples, Klaus Korn for the p24 enzyme-linked immunosorbent assay, and Bernhard Fleckenstein, Klaus von der Mark, and Thomas F. Schulz for support.
This study was supported by the NIH Intramural Program (T.H.B.), Korean Ministry of Health and Welfare grant BGC0800824 (to Y.P.), Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea (grant A080264 to S.K.), grants SFB466/SFB587 (A.M. and S.P.) and GK1071 (C.C.) from the Deutsche Forschungsgemeinschaft, the M.D./Ph.D. program in Molecular Medicine and Ph.D. program in Infection Biology at Hannover Medical School (T.S.T. and I.S.), and the Bundesministerium für Bildung und Forschung (I.G., S.B., and S.P.).
Footnotes
Published ahead of print on 21 January 2009.
REFERENCES
- 1.Ahmed, R., M. B. Oldstone, and P. Palese. 2007. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat. Immunol. 81188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belshe, R. B. 2005. The origins of pandemic influenza—lessons from the 1918 virus. N. Engl. J. Med. 3532209-2211. [DOI] [PubMed] [Google Scholar]
- 3.Betakova, T. 2007. M2 protein—a proton channel of influenza A virus. Curr. Pharm. Des. 133231-3235. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, V., B. Kramer, T. Pfeiffer, L. Starck, and D. A. Steinhauer. 2001. Inhibition of release of lentivirus particles with incorporated human influenza virus haemagglutinin by binding to sialic acid-containing cellular receptors. J. Gen. Virol. 822485-2494. [DOI] [PubMed] [Google Scholar]
- 5.Böttcher, E., T. Matrosovich, M. Beyerle, H. D. Klenk, W. Garten, and M. Matrosovich. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 809896-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boycott, R., H. D. Klenk, and M. Ohuchi. 1994. Cell tropism of influenza virus mediated by hemagglutinin activation at the stage of virus entry. Virology 203313-319. [DOI] [PubMed] [Google Scholar]
- 7.Chandran, K., N. J. Sullivan, U. Felbor, S. P. Whelan, and J. M. Cunningham. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 3081643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206935-944. [DOI] [PubMed] [Google Scholar]
- 9.Cox, N. J., and K. Subbarao. 2000. Global epidemiology of influenza: past and present. Annu. Rev. Med. 51407-421. [DOI] [PubMed] [Google Scholar]
- 10.Elliot, A. J., D. A. Steinhauer, R. S. Daniels, and J. S. Oxford. 2006. Functional and antigenic analyses of the 1918 influenza virus haemagglutinin using a recombinant vaccinia virus expression system. Virus Res. 12211-19. [DOI] [PubMed] [Google Scholar]
- 11.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 739679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, F., Y. Li, J. M. Decker, F. W. Peyerl, F. Bibollet-Ruche, C. M. Rodenburg, Y. Chen, D. R. Shaw, S. Allen, R. Musonda, G. M. Shaw, A. J. Zajac, N. Letvin, and B. H. Hahn. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retrovir. 19817-823. [DOI] [PubMed] [Google Scholar]
- 13.Glaser, L., J. Stevens, D. Zamarin, I. A. Wilson, A. Garcia-Sastre, T. M. Tumpey, C. F. Basler, J. K. Taubenberger, and P. Palese. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 7911533-11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goto, H., and Y. Kawaoka. 1998. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc. Natl. Acad. Sci. USA 9510224-10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto, H., K. Wells, A. Takada, and Y. Kawaoka. 2001. Plasminogen-binding activity of neuraminidase determines the pathogenicity of influenza A virus. J. Virol. 759297-9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 1462275-2289. [DOI] [PubMed] [Google Scholar]
- 17.Horimoto, T., and Y. Kawaoka. 2001. Pandemic threat posed by avian influenza A viruses. Clin. Microbiol. Rev. 14129-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horimoto, T., and Y. Kawaoka. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3591-600. [DOI] [PubMed] [Google Scholar]
- 19.Jung, H., K. P. Lee, S. J. Park, J. H. Park, Y. S. Jang, S. Y. Choi, J. G. Jung, K. Jo, D. Y. Park, J. H. Yoon, J. H. Park, D. S. Lim, G. R. Hong, C. Choi, Y. K. Park, J. W. Lee, H. J. Hong, S. Kim, and Y. W. Park. 2008. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene 272635-2647. [DOI] [PubMed] [Google Scholar]
- 20.Klenk, H. D., R. Rott, M. Orlich, and J. Blodorn. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68426-439. [DOI] [PubMed] [Google Scholar]
- 21.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, F. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445319-323. [DOI] [PubMed] [Google Scholar]
- 22.Kobasa, D., A. Takada, K. Shinya, M. Hatta, P. Halfmann, S. Theriault, H. Suzuki, H. Nishimura, K. Mitamura, N. Sugaya, T. Usui, T. Murata, Y. Maeda, S. Watanabe, M. Suresh, T. Suzuki, Y. Suzuki, H. Feldmann, and Y. Kawaoka. 2004. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431703-707. [DOI] [PubMed] [Google Scholar]
- 23.Lazarowitz, S. G., and P. W. Choppin. 1975. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68440-454. [DOI] [PubMed] [Google Scholar]
- 24.LeBouder, F., E. Morello, G. F. Rimmelzwaan, F. Bosse, C. Pechoux, B. Delmas, and B. Riteau. 2008. Annexin II incorporated into influenza virus particles supports virus replication by converting plasminogen into plasmin. J. Virol. 826820-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marzi, A., P. Moller, S. L. Hanna, T. Harrer, J. Eisemann, A. Steinkasserer, S. Becker, F. Baribaud, and S. Pöhlmann. 2007. Analysis of the interaction of Ebola virus glycoprotein with DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) and its homologue DC-SIGNR. J. Infect. Dis. 196(Suppl. 2)S237-S246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKay, T., M. Patel, R. J. Pickles, L. G. Johnson, and J. C. Olsen. 2006. Influenza M2 envelope protein augments avian influenza hemagglutinin pseudotyping of lentiviral vectors. Gene Ther. 13715-724. [DOI] [PubMed] [Google Scholar]
- 27.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 969345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 7410074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pager, C. T., and R. E. Dutch. 2005. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 7912714-12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palese, P. 2004. Influenza: old and new threats. Nat. Med. 10S82-S87. [DOI] [PubMed] [Google Scholar]
- 31.Pappas, C., P. V. Aguilar, C. F. Basler, A. Solorzano, H. Zeng, L. A. Perrone, P. Palese, A. Garcia-Sastre, J. M. Katz, and T. M. Tumpey. 2008. Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc. Natl. Acad. Sci. USA 1053064-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parrish, C. R., and Y. Kawaoka. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59553-586. [DOI] [PubMed] [Google Scholar]
- 33.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 961651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid, A. H., T. G. Fanning, T. A. Janczewski, and J. K. Taubenberger. 2000. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc. Natl. Acad. Sci. USA 976785-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouillard, J. M., W. Lee, G. Truan, X. Gao, X. Zhou, and E. Gulari. 2004. Gene2Oligo: oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 32W176-W180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirogane, Y., M. Takeda, M. Iwasaki, N. Ishiguro, H. Takeuchi, Y. Nakatsu, M. Tahara, H. Kikuta, and Y. Yanagi. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 828942-8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 10211876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmons, G., J. D. Reeves, C. C. Grogan, L. H. Vandenberghe, F. Baribaud, J. C. Whitbeck, E. Burke, M. J. Buchmeier, E. J. Soilleux, J. L. Riley, R. W. Doms, P. Bates, and S. Pöhlmann. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305115-123. [DOI] [PubMed] [Google Scholar]
- 39.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69531-569. [DOI] [PubMed] [Google Scholar]
- 40.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 2581-20. [DOI] [PubMed] [Google Scholar]
- 41.Szabo, R., S. Netzel-Arnett, J. P. Hobson, T. M. Antalis, and T. H. Bugge. 2005. Matriptase-3 is a novel phylogenetically preserved membrane-anchored serine protease with broad serpin reactivity. Biochem. J. 390231-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taubenberger, J. K. 1998. Influenza virus hemagglutinin cleavage into HA1, HA2: no laughing matter. Proc. Natl. Acad. Sci. USA 959713-9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taubenberger, J. K. 2006. The origin and virulence of the 1918 “Spanish” influenza virus. Proc. Am. Philos. Soc. 15086-112. [PMC free article] [PubMed] [Google Scholar]
- 44.Taubenberger, J. K., and D. M. Morens. 2006. 1918 Influenza: the mother of all pandemics. Emerg. Infect. Dis. 1215-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 31077-80. [DOI] [PubMed] [Google Scholar]
- 46.Tumpey, T. M., A. Garcia-Sastre, A. Mikulasova, J. K. Taubenberger, D. E. Swayne, P. Palese, and C. F. Basler. 2002. Existing antivirals are effective against influenza viruses with genes from the 1918 pandemic virus. Proc. Natl. Acad. Sci. USA 9913849-13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tumpey, T. M., A. Garcia-Sastre, J. K. Taubenberger, P. Palese, D. E. Swayne, and C. F. Basler. 2004. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc. Natl. Acad. Sci. USA 1013166-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]