Abstract
A limited number of human G6P[14] rotavirus strains that cause gastroenteritis in humans have been isolated in Europe and Australia. The complete genome sequences were determined for five of these human strains—B10925-97 (isolated in Belgium in 1997), 111/05-27 (Italy, 2005), PA169 (Italy, 1987), MG6 (Australia, 1993), and Hun5 (Hungary, 1997)—and their genetic relatedness to animal rotavirus strains was evaluated by sequencing the complete genome of the sheep rotavirus OVR762 (G8P[14]; Spain, 2002), the guanaco (Lama guanicoe) rotavirus strains Arg/Chubut/99 and Arg/Río Negro/98 (G8P[14] and G8P[1], respectively; Argentina, 1999 and 1998), the sable antelope strain RC-18/08 (G6P[14]; South Africa, 2008), and the bovine rotavirus strain Arg/B383/98 (G15P[11]; Argentina, 1998). These analyses revealed an overall consensus genomic constellation (G6/G8)-P[14]-I2-(R2/R5)-C2-M2-(A3/A11)-N2-T6-(E2/E12)-H3, together with a few gene reassortments, and the phylogenetic analyses confirmed that the P[14] human strains evaluated in this study were closely related to rotavirus strains isolated from sheep, cattle, goats, guanacos, and antelopes and to rabbits (albeit to a lesser extent), suggesting that one (or more) of these animal species might be the source of the human G6P[14] strains. The main feature of the genotype and phylogenetic analyses was the close overall genomic relatedness between the five human G6P[14] rotavirus strains and the ovine and antelope rotavirus strains. Taken together, these data strongly suggest a common origin for the human P[14] strains and those of the even-toed ungulates belonging to the mammalian order Artiodactyla, with sheep probably playing a key role in the interspecies transmission responsible for the introduction of P[14] rotavirus strains into the human population.
Group A rotaviruses are the major cause of diarrhea in young children and animals worldwide. Rotaviruses belong to the family Reoviridae and possess a genome of 11 segments of double-stranded RNA encoding six structural (VP) and six nonstructural (NSP) proteins (12). VP7 and VP4 have been widely used in a dual classification system, establishing 19 VP7 G (for glycoprotein) genotypes and 27 VP4 P (for protease-sensitive) genotypes (27). Recently, an extended group A rotavirus classification and nomenclature was established to encompass all 11 genome segments, defining genotypes for each genome segment (27). The nomenclature Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx represents the genotypes of, respectively, the VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5-encoding gene segments, with x indicating the numbers of the corresponding genotypes (27). The use of the new classification system, which agrees with the main group A rotavirus genogroups (Wa, DS-1, and AU-1), has enabled the identification of reassortment and interspecies transmission events, highlighting the role of animals as a source of rotavirus infection in humans (27). In fact, analyses of the complete rotavirus gene constellations have revealed a common origin for (i) human Wa-like rotavirus strains and porcine rotavirus strains and (ii) human DS-1-like rotavirus strains and bovine rotavirus strains (27). A Rotavirus Classification Working Group (RCWG) was created to preserve the classification system and to assign new genotypes that are identified for any of the 11 genome segments (28).
G1, G3, G4, and G9 (associated with P[8]) and G2 (associated with P[4]) are the epidemiologically most important human rotavirus genotypes worldwide (47). G12 might be the next important emerging strain, as it has been identified in sporadic cases and outbreaks in countries around the world (3, 6, 29, 43-46, 49, 51; G. Kulnis, M. Maliga, D. J. diStefano, M. Uvaydova, D. Lawley, and H. F. Clark, presented at the Int. Symp. Double-Stranded RNA Viruses, Cape Town, South Africa, 2006). Other rotavirus genotypes, such as P[6] in Africa or, more specifically, G8P[6] in Malawi and G5P[8] in Brazil, are regionally prevalent (1, 10, 29, 47). Additional G types (G6, G8, G10, and G11) and P types ([1], [3], [9], [10], [11], [14], [19], and [25]) have also been detected sporadically in humans (29, 47). Many of these genotypes are believed to have originated from animal rotaviruses that were introduced into the human populations through interspecies transmission and/or gene reassortments, one of the major ways in which genetic rotavirus diversity is generated (12, 27, 29, 43).
An increasing number of human P[14] rotavirus strains, mainly in combination with G6 (PA169, PA-5/89, MG6, ASG6.02, aG6.01, MG6.01, and Hun5) and, to a lesser extent, with G8 (HAL1166, HAL1271, EGY1850, DG8, and PR/1300/04), G10 (A64 and Mc35), G1 (GR55/87, GR475/87, GR506/87, GR951/87, and GR67/91), and G3 (PA710 and B4106), have been described across the globe (2, 4, 9, 11, 17-19, 21, 22, 30, 32, 33, 36, 37, 53). The P[14] genotype is commonly found in rabbits (7), but it has also been described in goats, antelope, cattle (15, 20), sheep (8), and guanacos (Lama guanicoe) (38). The G6 genotype is the most frequently detected G type in cattle, followed by G10, G8, and G15 (29). The G6 genotype has been described as infecting other ruminants, such as buffalo (40, 42) and sheep (13, 34). Other than humans, the G6P[14] combination has been found only in goats (GenBank accession number AY128708-9) and in a sable antelope (this report).
The first G6P[14] human rotavirus strains, PA169 and PA-5/89, were detected in Italy between 1987 and 1989 in young infants with gastroenteritis (2, 18). Several human G6P[14] strains (MG6, MG6.01, aG6.01, and ASG6.02) were isolated in south and central Australia between 1993 and 1997 (9, 11, 36), and one G6P[14] isolate (Hun5) was identified in Hungary in 1997 (4). The VP4 proteins of the human G6P[14] strains are more closely related to those of strains isolated from sheep (OVR762), goats (Cap455), cattle (Sun9, RUBV81), and guanacos (Arg/Chubut/99), all even-toed ungulates belonging to representative species of the order Artiodactyla, than to those from rabbits (ALA, C-11, BAP-2, and 30/96) (8, 15, 38).
To elucidate the roles of ungulate camelids and ruminants as sources of P[14] rotavirus infection in humans, the evolutionary origins and levels of genetic relatedness of the human G6P[14] rotavirus strains and P[14] animal rotavirus strains were investigated by determining the complete genome sequences of the human G6P[14] rotavirus strains PA169, MG6, Hun5, B10925-97, and 111/05-27, along with those of the ovine strain OVR762 (G8P[14]), the guanaco strains Arg/Chubut/99 (G8P[14]) and Arg/Río Negro/98 (G8P[1]), the sable antelope strain RC-18/08 (G6P[14]), and the bovine strain Arg/B383/98 (G15P[11]). The two non-P[14] strains Arg/Río Negro/98 and Arg/B383/98 were included in the study, as their overall genome constellation proved to be related to the genome constellation of the P[14] strains investigated.
MATERIALS AND METHODS
Rotavirus samples.
Rotavirus strain B10925-97 (G6P[14]) was identified in a stool sample from a hospitalized 2-year-old child with gastroenteritis in 1997 during a study conducted at the University hospital of Gasthuisberg in Leuven, Belgium (54). Rotavirus strain 111/27-05 (G6P[14]) was identified in a diarrheic sample from a 3-year-old child in March 2005 in Bari, Italy. Rotavirus strain RC-18/08 (G6P[14]) was identified in 2008 in a stool sample from an 8-month-old sable antelope with gastroenteritis living in captivity in a breeding program in Limpopo Province, South Africa. The human tissue culture-adapted G6P[14] rotavirus strains PA169, MG6, and Hun5 have been described previously (4, 18, 36). In addition, the Spanish ovine strain OVR762 (G8P[14]), the Argentinean guanaco strains Arg/Chubut/99 (G8P[14]) and Arg/Río Negro/98 (G8P[1]), and the Argentinean bovine strain Arg/B383/98 (G15P[11]) were also analyzed (8, 39).
RNA extraction.
One hundred and forty microliters of cell culture supernatant or fecal suspension was used to extract viral RNA using the QIAamp Viral RNA minikit (Qiagen/Westburg, Leusden, The Netherlands) according to the manufacturer's instructions.
RT-PCR.
The extracted RNA was denatured at 97°C for 5 min, and reverse transcription (RT)-PCR was carried out using the Qiagen OneStep RT-PCR kit (Qiagen/Westburg). The forward and reverse primers used for the amplification of different gene segments were developed based on alignments of known 5′ and 3′ sequences of the respective gene segments found in GenBank and are available upon request. The reaction was carried out with an initial RT step at 50°C for 30 min, followed by PCR activation at 95°C for 15 min, 40 cycles of amplification, and a final extension of 10 min at 72°C in a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems Group, Foster City, CA). The cycle conditions for the amplification of VP1, VP2, VP3, and VP4 were 30 s at 94°C, 30 s at 50°C, and 6 min at 70°C; for the other gene segments, the conditions were 30 s at 94°C, 30 s at 45°C, and 3 min at 72°C.
Nucleotide sequencing.
The PCR products were purified with the MSB Spin PCRapace kit (Invitek; Germany) and sequenced using the dideoxynucleotide chain termination method with the ABI Prism BigDye Terminator Cycle Sequencing Reaction kit (Applied Biosystems Group) on an ABI Prism 3100 automated sequencer (Applied Biosystems Group). The sequencing was performed with the forward and reverse primers used for the RT-PCR. Primer-walking sequencing was performed to cover the complete sequences of the respective fragments on both strands. The complete 5′- and 3′-terminal nucleotide sequences of the 11 gene segments were determined as previously described (31).
Amplification and sequencing of sable antelope strain RC-18/08.
The sable antelope strain genome was amplified in a single PCR by to the sequence-independent methods described previously, with significant improvements, by Potgieter and colleagues in South Africa (41). The genome was sequenced using Roche GS FLX technology. The genome was assembled de novo using Lasergene 7 software from DNAStar. Finally, the sequences were analyzed using BLAST analyses.
Nucleotide and protein sequence analyses.
The chromatogram sequencing files were analyzed using Chromas 2.23 (Technelysium, Queensland, Australia), and contigs were prepared using SeqMan II (DNAStar, Madison, WI). Multiple-sequence alignments were calculated using ClustalX 1.81 (50). The sequences were manually edited with the GeneDoc version 2.6.002 alignment editor (35).
Phylogenetic analyses.
Phylogenetic and molecular evolutionary analyses were conducted using the MEGA version 3.1 software (24). Genetic distances were calculated using the Kimura 2 correction parameter at the nucleotide level, and the phylogenetic trees were constructed using the neighbor-joining method with 500 bootstrap replicates.
SimPlot analyses.
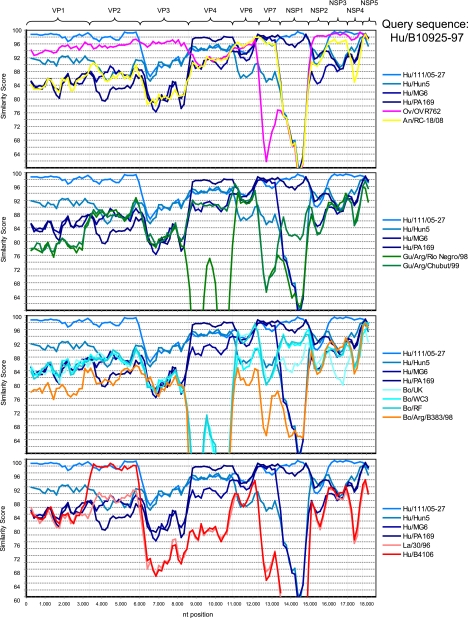
SimPlot analysis was carried out using the Kimura 2 parameter, with a window size of 500 bp and a step size of 150 bp. The concatenated genome sequences of the five human G6P[14] strains, B10925-97, 111/05-27, Hun5, MG6, and PA169, and the animal strains, OVR762 (ovine), RC-18/08 (antelope), Arg/Chubut/99 and Arg/Río Negro/98 (guanaco), Arg/B383/98, UK, WC3, RF (bovine), and B4106 (lapine-like) and 30/96 (lapine), were analyzed. The human strain B10925-97 was used as a query (see Fig. 5), and the other four human G6P[14] strains (111/05-27, Hun5, MG6, and PA169) were also used as separate query sequences (data available upon request).
FIG. 5.
SimPlot analyses comparing the human G6P[14] rotavirus strain B10925-97 with the four remaining human G6P[14] rotavirus strains (111/05-27, Hun5, MG6, and PA169). In the top panel, the animal strains OVR762 (ovine [Ov]) and RC-18/08 (antelope [An]) were added; in the upper-middle panel, the two guanaco (Gu) strains; in the lower- middle panel, the four bovine (Bo) strains; and in the bottom panel, the two lapine-like or lapine (La) rotavirus strains. Analyses were carried out using the Kimura 2 parameter, with a window size of 500 bp and a step size of 150 bp.
Assignment of newly identified genotypes.
The genotypes of each of the 11 genome segments for all 10 rotavirus strains that were completely sequenced in this study were determined according to the genotyping recommendations of the RCWG (28). Sequences of the VP1 and NSP4 gene segments of the three Argentinean strains, Arg/B383/98, Arg/Chubut/99, and Arg/Río Negro/98, did not belong to any of the established VP1 (R) and NSP4 (E) genotypes and were submitted to the RCWG for genotype assignment.
Nucleotide sequence accession numbers.
The nucleotide sequence data for the complete genomes of the rotavirus strains reported in this paper were submitted to GenBank under the following accession numbers: MG6, EF554093 to EF554103; Hun5, EF554104 to EF554114; B10925-97, EF554015 to EF554025; PA169, EF554026 to EF554036; 111/05-27, EF554037 to EF554047; OVR762, EF554148 to EF554158; Arg/Chubut/99, FJ347100 to FJ347110; Arg/Río Negro/98, FJ347122 to FJ347132; Arg/B383/98, FJ347111 to FJ347121; and RC-18/08, FJ495126 to FJ495136.
RESULTS
Genotype determination.
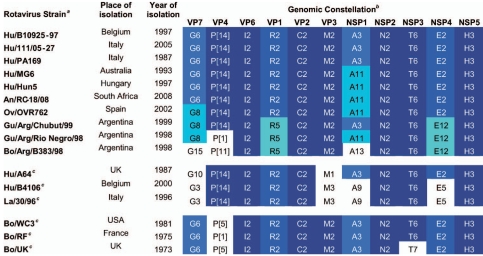
The entire genomes of five human (PA169, MG6, Hun5, 111/05-27, and B10925-97) and five animal (OVR762 [sheep], RC-18/08 [antelope], Arg/Chubut/99 and Arg/Río Negro/98 [guanaco], and Arg/B383/98 [bovine]) rotavirus strains were sequenced (approximately 185,000 bp in total). Table 1 shows the complete genotype assignment (Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx) of each of the rotavirus strains sequenced in this study, along with those of additional bovine and P[14] rotavirus strains whose complete genome sequences were previously described (21, 30). The genome segments of the majority of rotavirus strains sequenced in this study belonged to the already-established genotypes. However, the VP1 gene segments of the three Argentinean strains, Arg/B383/98, Arg/Chubut/99, and Arg/Río Negro/98, were highly conserved among each other but did not belong to any of the established R1 to R4 genotypes. They were confirmed by the RCWG to belong to a new VP1 genotype, R5. Similarly, the NSP4 gene segments of the same three Argentinean strains were highly conserved among each other but were only distantly related to the established E2 genotype. Since only a minimal number of pairwise identities among Arg/B383/98, Arg/Chubut/99, and Arg/Río Negro/98 and members of the E2 genotype were found above the 85% nucleotide cutoff, as established for NSP4 (27), the NSP4 sequences were referred to the RCWG. The few identities above the cutoff between strains belonging to different NSP4 genotypes (E2 to E12) were within the expected overlap between different NSP4 genotypes (27), and the NSP4 sequences of the three Argentinean strains were assigned to a new NSP4 genotype, E12.
TABLE 1.
Complete genomic constellations of the P[14] rotavirus strains sequenced in this study with those of selected human and animal rotavirus strains with known complete genomic constellations
Species of origin, followed by the name of the rotavirus strain. Bo, bovine; Gu, guanaco; Ov, ovine; Hu, human; La, lapine; An, antelope.
Dark blue indicates conserved genotypes, while lighter shades of blue indicate the less conserved genotypes or new genotypes (R5 and E12) identified in this study and validated by the RCWG as recommended (28). Uncolored genotypes reflect suspected reassortment events.
Analyses of full genome constellations.
Excluding the G and P types, the complete genotype determination of the 10 rotavirus strains analyzed in the study (Table 1) revealed an overall conserved consensus genotype constellation of I2-(R2/R5)-C2-M2-(A3/A11)-N2-T6-(E2/E12)-H3, consistent with the genetic constellation of bovine rotaviruses (27). Analyses of the patterns or constellations of genes revealed the following facts. (i) The five human G6P[14] strains possessed nearly identical genotypes for all genome segments, with the exception of the NSP1 genes, despite being isolated in different regions and at different times (Table 1 and Fig. 1). Strains MG6 and Hun5 belonged to the A11 NSP1 genotype, whereas strains B10925-97, 111/05-27, and PA169 belonged to the A3 NSP1 genotype. (ii) The ovine Spanish rotavirus strain OVR762 was very closely related to the five human strains; compared to strains MG6 (Australia) and Hun5 (Hungary), the OVR762 strain possessed only a different VP7 gene (G8 versus G6). (iii) The South African RC-18/08 antelope rotavirus strain showed an overall genotype constellation that was identical to those of the human MG6 and Hun5 strains. (iv) The G8P[14] strain Arg/Chubut/99, isolated from a guanaco, also possessed several genotypes in common with three of the G6P[14] human strains, B10925-97, 111/05-27, and PA169, with only the VP1, VP7, and NSP4 genotypes differing. Furthermore, an additional guanaco strain (Arg/Río Negro/98) possessed a genome constellation identical to that of the Arg/Chubut/99 strain, with the exception of the VP4 genotype, P[1], commonly found in cattle but, interestingly, not yet detected in cattle in Argentina (16). (v) The bovine rotavirus strain Arg/B383/98, detected in the Argentinean region of the Pampas, shared 8 out of 11 genotypes with the two guanaco strains detected in the same time period in the Patagonia region (a distant area, with a very low density of cattle), suggesting a relationship between rotaviruses from guanacos and cows, perhaps transmitted through a third species in contact with both animals, like goats or sheep. (vi) Other bovine strains also showed a large number of genotypes in common with Arg/B383/98 and the human and animal P[14] strains. Interestingly, when the genotypes of the human G6P[14] rotavirus strains were compared to those of additional P[14] strains, human A64 (G10), human lapine-like B4106 (G3), and lapine 30/96 (G3), whose complete genomes were previously determined (21, 27), five to nine genotypes were shared among the rotavirus strains (Table 1); any differences in genotypes may have arisen through gene reassortment. Taken together, these data strongly suggest a common origin for the human P[14] strains and those from the even-toed ungulates belonging to the mammalian order Artiodactyla.
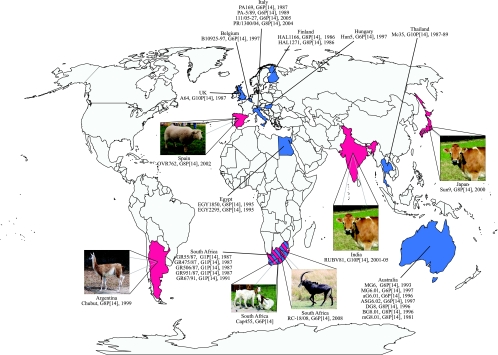
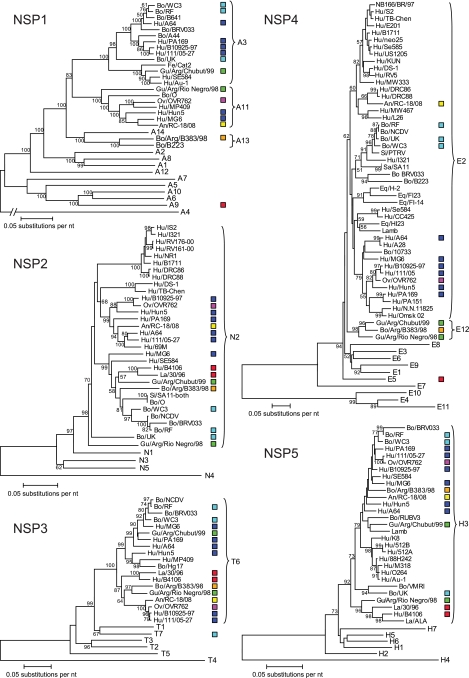
FIG. 1.
Geographic locations where human and nonlapine animal P[14] rotavirus strains have been reported. The countries in blue have reported human P[14] strains. The countries in red have reported nonlapine animal P[14] rotaviruses. For each country, the names of the P[14] strains isolated, together with their G/P genotypes and years of isolation, are shown.
Phylogenetic analyses.
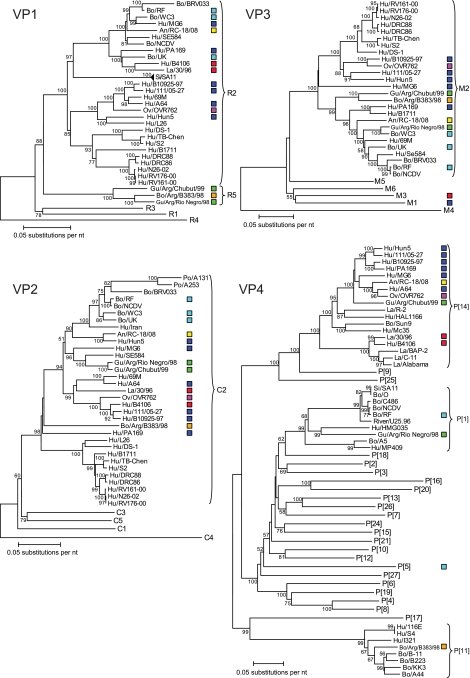
To further investigate the genetic relationships of the human and the different animal P[14] rotavirus strains in more detail, phylogenetic analyses, based on full-length nucleotide sequences of each of the 11 genome segments, were conducted (Fig. 2 to 4).
FIG. 2.
Phylogenetic trees based on the full-length nucleotide sequences of the rotavirus VP1, VP2, VP3, and VP4 genes. Bootstrap values (500 replicates) above 50 are shown. Only the genotypes in which the strains under investigation cluster are shown completely. The other genotypes are represented by their genotype numbers. For each strain, the following data are given: species of origin/strain name. The human (Hu) P[14] strains are represented by dark-blue boxes, the ovine (Ov) strain OVR762 by a purple box, the two guanaco (Gu) strains by green boxes, the bovine (Bo) strain Arg/B383/98 by an orange box, the lapine (La) strains by red boxes, and the bovine strains by light-blue boxes. An, antelope; Si, simiun.
FIG. 4.
Phylogenetic trees based on the full-length nucleotide sequences of the rotavirus NSP1, NSP2, NSP3, NSP4, and NSP5 genes. Bootstrap values (500 replicates) above 50 are shown. Only the genotypes in which the strains under investigation cluster are shown completely. The other genotypes are represented by their genotype numbers. For each strain, the following data are given: species of origin/strain name. The human (Hu) P[14] strains are represented by blue boxes, the ovine (Ov) strain OVR762 by a purple box, the two guanaco (Gu) strains by green boxes, the bovine (Bo) strain Arg/B383/98 by an orange box, the lapine (La) strains by red boxes, and the bovine strains by light-blue boxes. Fe, feline; An, antelope; Si, simian; Eq, equine.
The VP7, VP4, NSP4, and NSP5 gene segments of the five human G6P[14] strains clustered very closely together (Fig. 2 to 4), while for the VP1, VP3, VP6, NSP2, and NSP3 gene segments, most of the human G6P[14] strains clustered closely together, with one or two strains clustering more distantly within the same genotype (Fig. 2 to 4). The VP2 genes of the five human G6P[14] rotavirus strains were scattered across the C2 genotype (Fig. 2). The human G6P[14] strains clustered according to their respective NSP1 genotypes, A3 and A11 (Fig. 4). Of note, several gene segments (VP1, VP2, VP4, VP6, and NSP1 to NSP5) of the unusual human G10P[14] strain A64 clustered closely with those of all the human G6P[14] rotavirus strains (Fig. 2 to 4). The only exceptions were the VP3 and VP7 genes, since strain A64 belongs to genotypes M1 and G10, respectively, as opposed to genotypes M2 and G6 (Fig. 2 and 3, respectively). As the M1 genotype is typical of human Wa-like strains, a reassortment event with a human rotavirus strain most likely took place.
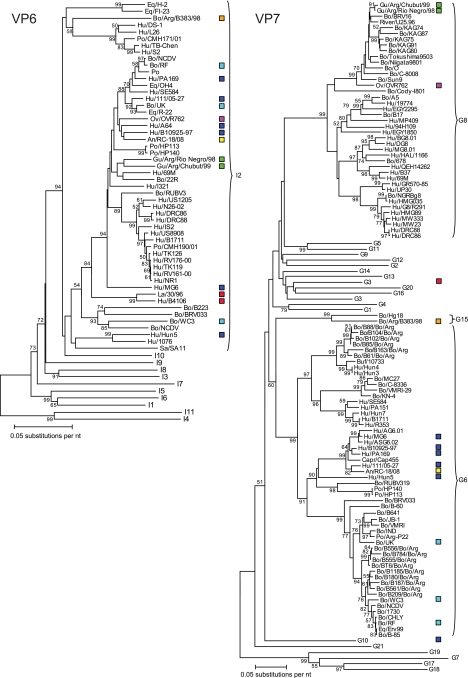
FIG. 3.
Phylogenetic trees based on the full-length nucleotide sequences of the rotavirus VP6 and VP7 genes. Bootstrap values (500 replicates) above 50 are shown. Only the genotypes in which the strains under investigation cluster are shown completely. The other genotypes are represented by their genotype numbers. For each strain, the following data are given: species of origin/strain name. The human (Hu) P[14] strains are represented by blue boxes, the ovine (Ov) strain OVR762 by a purple box, the two guanaco (Gu) strains by green boxes, the bovine (Bo) strain Arg/B383/98 by an orange box, the lapine (La) strains by red boxes, and the bovine strains by light-blue boxes. Eq, equine; Po, porcine; An, antelope; Buf, buffalo; Capr, caprine; Si, simian.
To evaluate the role of animals as a source of P[14] rotavirus infection in humans, we also evaluated the phylogenetic relatedness of animal P[14] strains of ovine, bovine, lapine, antelope, and South American camelid origin to human rotavirus strains across the entire genome. Remarkably, with the exception of the VP7 gene, the ovine rotavirus strain OVR762 clustered very closely with the human G6P[14] strains (Fig. 2 to 4), suggesting rather recent interspecies transmissions from sheep to humans (or vice versa). Also, the sable antelope strain, RC-18/08, showed moderately close (VP1, VP3, and NSP4) to very close (VP4, VP7, and NSP3) phylogenetic clustering with the human G6P[14] strains (Fig. 2 to 4). Similarly, the guanaco rotavirus strain Arg/Chubut/99 (G8P[14]) clustered closely with the G6P[14] human strains and the ovine and antelope strains when the gene segments VP2, VP4, VP6, NSP3, and NSP5 were analyzed (Fig. 2 to 4). In regard to the VP3, NSP1, and NSP2 gene segments, a more distant clustering within the same corresponding genotype was observed, while the VP7 gene segment of Arg/Chubut/99 was closely related to G8 of OVR762 (Fig. 2 to 4). Phylogenetic analyses of the VP1 and NSP4 genome segments of Arg/Chubut/99 confirmed that they belonged to new genotypes, R5 and E12, respectively (Fig. 2 and 4, respectively). The G8P[1] guanaco rotavirus strain, Arg/Río Negro/98, showed very close clustering with Arg/Chubut/99 for the VP1, VP2, VP6, VP7, and NSP4 gene segments, but for the VP3, NSP2, NSP3, and NSP5 gene segments, more distant clustering was observed within the corresponding genotype (Fig. 2 to 4). Phylogenetic analyses of the VP4 and NSP1 gene segments of Arg/Río Negro/98 confirmed their classification as P[1] and A11 genotypes, respectively (Fig. 2 and 4). The G15P[11] bovine rotavirus strain Arg/B383/98, isolated in Argentina from a dairy calf with diarrhea, was not intended to be included in this study; however, during our analyses, it was noted that many genotypes of this bovine strain were the same as those of the guanaco strains, also isolated in Argentina. Gene segments VP1, VP3, NSP2, NSP3, NSP4, and NSP5 of strain Arg/B383/98 also clustered closely with the guanaco strains Arg/Chubut/99 and Arg/Río Negro/98 (Fig. 2 and 4). The VP2 and VP6 gene segments of Arg/B383/98 clustered more distantly within the corresponding genotype, while the VP4, VP7, and NSP1 gene segments clearly belong to different genotypes (P[11], G15, and A13) (Fig. 2 to 4). The evolutionary relationship between the P[14] strains under investigation in this study and other bovine rotavirus strains, such as WC3, RF, and UK (Table 1), was also confirmed, since many gene segments clustered closely (VP6, NSP3, and NSP5) or distantly (VP2, VP3, VP7, NSP2, and NSP4) within the same corresponding genotypes (Fig. 2 to 4). Finally, for a number of gene segments (VP1, VP2, VP4, VP6, NSP2, NSP3, and NSP5), relatively close clustering was found between the lapine and human lapine-like strains 30/96 and B4106, respectively, and the human and/or ungulate P[14] strains, whereas for the VP3, VP7, NSP1, and NSP4 gene segments, different genotypes were evident (Fig. 2 to 4). Although the lapine P[14] strains and the nonlapine strains formed separate clusters in the VP4 phylogenetic tree, it should be noted that the VP4 gene segment of one lapine rotavirus strain, R-2, clustered more closely with human P[14] strains (Fig. 2). In addition, remarkably close clustering was observed for the lapine VP2 gene segments of 30/96 and B4106 and the human P[14] strains B10925-97, 111/05-27, OVR762, and A64 (Fig. 2).
SimPlot analyses.
To further study the overall genomic relationships among the human G6P[14] strains and the animal strains studied in this paper, Simplot analyses were conducted. The human G6P[14] strain B10925 was compared to the remaining human G6P[14] strains (Fig. 5), and the strains OVR762 (ovine) and RC-18/08 (antelope), the two guanaco strains, the four bovine strains, and the two lapine-like or lapine rotavirus strains were added. As shown in Fig. 5, various degrees of similarity exist between different strains and different gene segments. The close relationship between the human strain B10925-97 and the ovine strain OVR762 for gene segments VP1 to -3, VP6, and NSP2 to -5, as observed in the phylogenetic analyses, is precisely visualized in the top panel of Fig. 5. The various lower degrees of similarity for the remaining gene segments, VP4, VP7, and NSP1, might have been caused by intragenotype reassortments. Also, the close relationships of the VP1, VP2, VP4, VP7, NSP1, and NSP3 to -5 gene segments of the human B10925-97 and 111/05-27 strains were confirmed, and again, the slightly lower degrees of similarity for the VP3, VP6, and NSP2 genes might be attributed to intragenotype reassortments, albeit with rather closely related strains. Another striking observation in the bottom panel of Fig. 5 is the very high similarity between the VP2 gene segment of the human strain B10925-97 and the human lapine-like rotavirus strain B4106. As VP2 is the only gene segment for which the B4106 strain and the lapine strain 30/96 are not very closely related, this might indicate that the VP2 segment of B4106 might have a nonlapine origin or that the VP2 genes of B10925-97 and 111/27-05 might have a lapine origin (Fig. 5, bottom, and data not shown).
A few other remarkable observations are (i) the close relationship between the VP2 gene segments of the human G6P[14] strain Hun5 and the antelope strain RC-18/08, (ii) the close relationship between the NSP1 gene segments of the human G6P[14] strain MG6 and the antelope strain RC-18/08, (iii) the close relationships between the VP1 and NSP3 gene segments of the human G6P[14] strain MG6 and the bovine strains WC3 and RF, and (iv) the close relationships between the VP1 and VP6 gene segments of the human G6P[14] strain PA169 and bovine strains (data not shown).
DISCUSSION
The P[14] genotype was initially detected in humans (18, 19, 52, 53) and subsequently in rabbits (7). It was originally hypothesized that the lapine and human P[14] strains might have a common origin or that the P[14] human strains isolated in Italy (PA169 [G6]), Finland (HAL1166 [G8]), and Thailand (Mc35 [G10]) were naturally occurring reassortants between bovine, lapine, and human viruses, as all of these humans strains possessed a VP7 that is commonly found in cattle (7). Since then, the P[14] genotype has continued to be detected sporadically in humans (in association with G6, G8, G10, G1, or G3) and commonly in rabbits (always associated with G3), and it has now also been identified in several even-toed ungulate species belonging to the order Artiodactyla, such as cattle, goats, sheep, guanacos, and antelope (in association with G6, G8, or G10) (2, 4, 8, 9, 11, 14, 18-20, 22, 30, 32, 33, 36-38, 53) (Fig. 1). Interestingly, the VP4 proteins of the human P[14] strains are more closely related to those of the strains isolated from ungulates than to those of rabbits (8, 15, 38); therefore, large ruminants (cattle and antelope), small ruminants (goat and sheep), and other ungulate species, such as camelids, may play roles as sources of P[14] rotavirus infection in humans. Taken together with the demonstration that Wa-like human rotaviruses and porcine rotavirus have a common origin and that the DS-1-like human rotaviruses and bovine rotaviruses have a common origin (27), it was of interest to investigate whether the P[14] human strains have a common origin with rotaviruses isolated from members of the families Camelidae and/or Bovidae, both belonging to the order Artiodactyla.
The availability of several human G6P[14] strains isolated from different parts of the world at different times (Fig. 1) and several closely related animal strains allowed us to investigate the evolutionary origins and levels of genetic relatedness of the human G6P[14] rotavirus strains and the P[14] animal rotavirus strains by determining the complete genome sequences of five human G6P[14] rotavirus strains (PA169, MG6, Hun5, B10925-97, and 111/05-27), three animal P[14] rotavirus strains (associated with G6 or G8), and two animal rotavirus strains that, although not P[14], were closely related to P[14] animal strains when other genes besides VP4 were analyzed. Despite the widespread distribution (Belgium, Italy, Australia, and Hungary) of the five human G6P[14] rotavirus strains, genotype and phylogenetic analyses of all 11 genome segments of these strains, as well as those of the animal rotavirus strains sequenced in this and other studies, revealed a remarkable overall-conserved genotype constellation among the human G6P[14] strains: I2-R2-C2-M2-A3/A11-N2-T6-E2-H3.
Perhaps the most salient feature of the genotype and phylogenetic analyses is the close overall genomic relatedness between the five human G6P[14] strains and the ovine and antelope rotavirus strains OVR762 and RC-18/08. This could suggest that these human G6P[14] rotaviruses originated after interspecies transmissions from a common pool of ovine rotavirus strains, as antelopes are less likely to be the immediate donors because they live in rather limited geographical locations and do not live in close proximity to humans. Additional possible donors are goats, as a G6P[14] rotavirus strain, Cap455, was isolated in South Africa (unpublished data). Unfortunately, this strain could not be further analyzed, as no more sample was available. Another prominent feature is that the two Argentinean guanaco rotavirus strains, Arg/Chubut/99 and Arg/Río Negro/98, also have a relatively close relationship with each other and among the ovine, antelope, and G6P[14] human strains for several gene segments (Fig. 2 to 5). The human G10P[14] rotavirus strain A64, whose complete sequence was reported recently (21), was also found to be closely related (except the G and/or P types) to the P[14] human, ovine, and guanaco strains. Information on the genome constellation of the Argentinean bovine rotavirus strain Arg/B383/98 (G15P[11]) was obtained, since the G15 VP7 type is very rare in nature and the virus was suspected to harbor other unusual genome segments. Surprisingly, the gene constellation of the bovine strain Arg/B383/98, together with the other bovine strains WC3, UK, and RF (Table 1 and Fig. 2 to 5), proved to also have a number of gene segments closely related to the human and animal P[14] strains, suggestive of a distant common origin, although this cannot be directly inferred from its VP4 and VP7 genotypes, which might have been derived through gene reassortment between ruminants and/or camelids in South America in the case of strain Arg/B383/98. However, guanacos are normally not in close contact with cattle, which makes interspecies transmission unlikely. On the other hand, guanacos and cattle do both come in close contact with sheep and goats, which might be the intermediate carriers and might be the main hosts of the P[14] strains. The different G types (G6, G8, G10, and G15) identified in the strains analyzed in this study, bearing the conserved genotype constellation typical of bovine DS-1-like viruses, are all believed to be typical of cattle (29), although these G types are also found in a number of other members of the ruminant family Bovidae, such as buffalo, sheep, goats, and antelopes (13, 26, 38, 40, 42, 48), and in guanacos, members of the family Camelidae (39). The P[14] genotype is also prevalent in rabbits (7, 25), but the G3P[14] lapine rotavirus strain 30/96 and the human lapine-like G3P[14] rotavirus strain B4106 share only five to seven genotypes within the genomic constellation with the human and ungulate P[14] rotavirus strains investigated in this study, suggesting that lapine rotaviruses might have a distant evolutionary relationship with these nonlapine P[14] rotavirus strains. The VP2 gene segment of B4106 is a notable exception (Fig. 5). Further, no close relationship was observed between the P[14] rotavirus strains and any of the three human genogroup reference strains Wa, DS-1, and AU-1.
The phylogenetic trees and SimPlot analyses of all five human G6P[14] strains clearly show that the human G6P[14] strains are closely related; however, none of the strains shows similar levels of relatedness for all 11 gene segments, which does not support the hypothesis that one human strain was the direct progenitor of the other strains. For example, the strains B10925/97 and 111/05-27 are very closely related in a number of gene segments, such as VP1, VP2, NSP1, NSP3, and NSP4, but for other gene segments, such as VP3, VP4, VP6, VP7, NSP2, and NSP5, B10925/97 and 111/05-27 seem to be more closely related to other human P[14] strains (Fig. 2 to 5). This kind of observation of different levels of similarity between the gene segments of the different strains under investigation (Fig. 5) strongly suggests that reassortment events occur rather frequently among rotavirus strains possessing the conserved described genome constellation. This lack of a clear pattern, together with the fact that the strains were isolated in very distant geographical locations (Italy, Belgium, Hungary, and Australia) and over a period of nearly 2 decades (1987 to 2005), is an indication that the distinct human G6P[14] strains are more likely the result of individual interspecies transmissions from ruminants, as was suggested by Palombo and Bishop (36), rather than the result of human-to-human transmission after one interspecies transmission, as has also been suggested based only on VP4 and VP7 sequence data (4, 9). This questions the abilities of these human P[14] strains to successfully spread in the human population. However, a number of unusual G1P[14] strains isolated in South Africa were very closely related and isolated over a period of 4 years (1987 to 1991), which might indicate their capability for limited human-to-human transmission (33). Similarly, several closely related G6P[14] strains have been isolated in different locations and at different times in Australia, which also might be examples of limited human-to-human spread of these strains (9, 11, 36). However, P[14] rotavirus strains have not caused any large outbreaks, indicating the lack of high transmissibility among humans.
Altogether, these data point to an overall consensus genotype constellation, (G6/G8)-P[14]-I2-(R2/R5)-C2-M2-(A3/A11)-N2-T6-(E2-E12)-H3, that might circulate among sheep and other ruminants of certain families of the order Artiodactyla, such as cattle, goats, antelopes, and guanacos, which confirms the hypothesis of a close relationship between bovine and human P[14] rotaviruses derived from sequencing and hybridization assays (5, 15, 18, 20, 52). The hypothesis that a species other than humans or cattle might be the main host of certain G8 rotaviruses has been proposed by Browning and colleagues (5). However, as observed, the Artiodactyla-like genomic constellation is also very prone to intragenotype reassortment, but also to intergenotype reassortment, as is the case for VP3, VP4, VP7, and NSP1 (Table 1). This genomic constellation also seems to have been able to cross the host species barrier to humans on a number of occasions. Within the limits of the small number of P[14] human strains and those isolated from ungulates belonging to the families Bovidae (cattle, sheep, goats, and antelopes) and Camelidae (guanacos) of the order Artiodactyla, there was a tendency toward great similarity regardless of the geographic region or time interval. Although this could be a sampling artifact, it is suggestive of constant introduction of P[14]-like strains, with a genomic constellation typical of ungulates belonging to the order Artiodactyla, in humans.
Although additional surveillance in humans and, especially, in ruminants and camelids is warranted to confirm that these animals are the sources of human P[14] rotavirus strains, there is increasing evidence for the worldwide prevalence of P[14] rotavirus strains in humans and animals. Further complete genome analysis of additional G6- and non-G6-possessing P[14] rotavirus strains isolated from humans and from different ungulates, such as deer and giraffes, might shed more light on the history and genetic relatedness of all these P[14] rotavirus strains.
The data presented in this study suggest that human P[14] rotaviruses might have an ovine origin or an origin in another ruminant (cattle, goat, or antelope) or camelid (guanaco) artiodactyl, due to the conserved overall genetic constellation found in a variety of these animals. In this regard, it would be most likely that sheep (or goats or cattle) are the main reservoir of P[14] rotaviruses for humans across the world, because these animals live in close proximity to humans and are transported more frequently than antelopes or guanacos. Since rotavirus may be readily transmitted among members of the order Artiodactyla, the notion that human P[14] rotaviruses have a single common animal source may be too strict. The evolutionary relationships observed in this study suggest that rotaviruses from different members of the order Artiodactyla have intermingled on several occasions, and humans may be accidental hosts who are occasionally infected with the P[14]-like strains. This is supported by the lack of evidence for efficient human-to-human transmission of these viruses.
Acknowledgments
We are very grateful to Truuske Gerdes from the Virology Department at ARC-OVI for providing the sable antelope virus. In addition, we thank all our colleagues at the Laboratory of Clinical and Epidemiological Virology, Department of Microbiology and Immunology, Rega Institute for Medical Research, University of Leuven, Leuven, Belgium, for helpful comments and discussion.
J.M. was supported by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen).
Footnotes
Published ahead of print on 19 January 2009.
REFERENCES
- 1.Araujo, I. T., R. M. Assis, A. M. Fialho, J. D. Mascarenhas, M. B. Heinemann, and J. P. Leite. 2007. Brazilian P[8], G1, P[8], G5, P[8], G9, and P[4], G2 rotavirus strains: nucleotide sequence and phylogenetic analysis. J. Med. Virol. 79995-1001. [DOI] [PubMed] [Google Scholar]
- 2.Arista, S., E. Vizzi, C. Alaimo, D. Palermo, and A. Cascio. 1999. Identification of human rotavirus strains with the P[14] genotype by PCR. J. Clin. Microbiol. 372706-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bányai, K., A. Bogdan, P. Kisfali, P. Molnar, I. Mihaly, B. Melegh, V. Martella, J. R. Gentsch, and G. Szucs. 2007. Emergence of serotype G12 rotaviruses, Hungary. Emerg Infect. Dis. 13916-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bányai, K., J. R. Gentsch, D. D. Griffin, J. L. Holmes, R. I. Glass, and G. Szucs. 2003. Genetic variability among serotype G6 human rotaviruses: identification of a novel lineage isolated in Hungary. J. Med. Virol. 71124-134. [DOI] [PubMed] [Google Scholar]
- 5.Browning, G. F., D. R. Snodgrass, O. Nakagomi, E. Kaga, A. Sarasini, and G. Gerna. 1992. Human and bovine serotype G8 rotaviruses may be derived by reassortment. Arch. Virol. 125121-128. [DOI] [PubMed] [Google Scholar]
- 6.Castello, A. A., M. H. Arguelles, R. P. Rota, A. Olthoff, B. Jiang, R. I. Glass, J. R. Gentsch, and G. Glikmann. 2006. Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J. Clin. Microbiol. 442046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciarlet, M., M. K. Estes, and M. E. Conner. 1997. Comparative amino acid sequence analysis of the outer capsid protein VP4 from four lapine rotavirus strains reveals identity with genotype P[14] human rotaviruses. Arch. Virol. 1421059-1069. [DOI] [PubMed] [Google Scholar]
- 8.Ciarlet, M., C. Hoffmann, E. Lorusso, R. Baselga, M. A. Cafiero, K. Banyai, J. Matthijnssens, V. Parreño, S. de Grazia, C. Buonavoglia, and V. Martella. 2008. Genomic characterization of a novel group A lamb rotavirus isolated in Zaragoza, Spain. Virus Genes 37250-265. [DOI] [PubMed] [Google Scholar]
- 9.Cooney, M. A., R. J. Gorrell, and E. A. Palombo. 2001. Characterisation and phylogenetic analysis of the VP7 proteins of serotype G6 and G8 human rotaviruses. J. Med. Microbiol. 50462-467. [DOI] [PubMed] [Google Scholar]
- 10.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diwakarla, S., R. Clark, and E. A. Palombo. 2002. Expanding distribution of human serotype G6 rotaviruses in Australia. Microbiol. Immunol. 46499-502. [DOI] [PubMed] [Google Scholar]
- 12.Estes, M., and A. Kapikian. 2007. Rotaviruses, p. 1917-1974. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott, Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Fitzgerald, T. A., M. Muñoz, A. R. Wood, and D. R. Snodgrass. 1995. Serological and genomic characterisation of group A rotaviruses from lambs. Arch. Virol. 1401541-1548. [DOI] [PubMed] [Google Scholar]
- 14.Fukai, K., H. Onoda, T. Itou, M. Sato, Y. Miura, and T. Sakai. 2004. Genetic and serological characterization of novel serotype G8 bovine group A rotavirus strains isolated in Japan. J. Vet. Med. Sci. 661413-1416. [DOI] [PubMed] [Google Scholar]
- 15.Fukai, K., T. Saito, K. Inoue, and M. Sato. 2004. Molecular characterization of novel P[14], G8 bovine group A rotavirus, Sun9, isolated in Japan. Virus Res. 105101-106. [DOI] [PubMed] [Google Scholar]
- 16.Garaicoechea, L., K. Bok, L. R. Jones, G. Combessies, A. Odeon, F. Fernandez, and V. Parreño. 2006. Molecular characterization of bovine rotavirus circulating in beef and dairy herds in Argentina during a 10-year period (1994-2003). Vet. Microbiol. 1181-11. [DOI] [PubMed] [Google Scholar]
- 17.Gerna, G., A. Sarasini, A. Di Matteo, L. Zentilin, P. Miranda, M. Parea, F. Baldanti, S. Arista, G. Milanesi, and M. Battaglia. 1990. Serotype 3 human rotavirus strains with subgroup I specificity. J. Clin. Microbiol. 281342-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerna, G., A. Sarasini, M. Parea, S. Arista, P. Miranda, H. Brüssow, Y. Hoshino, and J. Flores. 1992. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J. Clin. Microbiol. 309-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerna, G., J. Sears, Y. Hoshino, A. D. Steele, O. Nakagomi, A. Sarasini, and J. Flores. 1994. Identification of a new VP4 serotype of human rotaviruses. Virology 20066-71. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, S., V. Varghese, S. Samajdar, M. Sinha, T. N. Naik, and N. Kobayashi. 2007. Evidence for bovine origin of VP4 and VP7 genes of human group A rotavirus G6P[14] and G10P[14] strains. J. Clin. Microbiol. 452751-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heiman, E. M., S. M. McDonald, M. Barro, Z. F. Taraporewala, T. Bar-Magen, and J. T. Patton. 2008. Group A human rotavirus genomics: evidence that gene constellations are influenced by viral protein interactions. J. Virol. 8211106-11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, J. L., C. D. Kirkwood, G. Gerna, J. D. Clemens, M. R. Rao, A. B. Naficy, R. Abu-Elyazeed, S. J. Savarino, R. I. Glass, and J. R. Gentsch. 1999. Characterization of unusual G8 rotavirus strains isolated from Egyptian children. Arch. Virol. 1441381-1396. [DOI] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 171244-1245. [DOI] [PubMed] [Google Scholar]
- 25.Martella, V., M. Ciarlet, A. Lavazza, A. Camarda, E. Lorusso, V. Terio, D. Ricci, F. Cariola, M. Gentile, A. Cavalli, M. Camero, N. Decaro, and C. Buonavoglia. 2005. Lapine rotaviruses of the genotype P[22] are widespread in Italian rabbitries. Vet. Microbiol. 111117-124. [DOI] [PubMed] [Google Scholar]
- 26.Martella, V., A. Pratelli, O. Pinto, G. Ferrara, M. Tempesta, and D. Buonavoglia. 1999. Typing by polymerase chain reaction of buffalo rotaviruses isolated in Italy. Zentralbl. Vetmed. B 46499-502. [DOI] [PubMed] [Google Scholar]
- 27.Matthijnssens, J., M. Ciarlet, E. Heiman, I. Arijs, T. Delbeke, S. M. McDonald, E. A. Palombo, M. Iturriza-Gómara, P. Maes, J. T. Patton, M. Rahman, and M. Van Ranst. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 823204-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthijnssens, J., M. Ciarlet, M. Rahman, H. Attoui, K. Banyai, M. K. Estes, J. R. Gentsch, M. Iturriza-Gómara, C. D. Kirkwood, V. Martella, P. P. Mertens, O. Nakagomi, J. T. Patton, F. M. Ruggeri, L. J. Saif, N. Santos, A. Steyer, K. Taniguchi, U. Desselberger, and M. Van Ranst. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 1531621-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthijnssens, J., M. Rahman, M. Ciarlet, and M. Van Ranst. 2008. Emerging human rotavirus genotypes, p. 171-219. In E. A. Palombo and C. D. Kirkwood (ed.), Viruses in the environment. Research Signpost, Trivandrum, India.
- 30.Matthijnssens, J., M. Rahman, V. Martella, Y. Xuelei, S. De Vos, K. De Leener, M. Ciarlet, C. Buonavoglia, and M. Van Ranst. 2006. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J. Virol. 803801-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthijnssens, J., M. Rahman, X. Yang, T. Delbeke, I. Arijs, J. P. Kabue, J. J. Muyembe, and M. Van Ranst. 2006. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1-like genogroup. J. Clin. Microbiol. 441801-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medici, M. C., L. A. Abelli, M. Martinelli, G. Dettori, and C. Chezzi. 2008. Molecular characterization of VP4, VP6 and VP7 genes of a rare G8P[14] rotavirus strain detected in an infant with gastroenteritis in Italy. Virus Res. 137163-167. [DOI] [PubMed] [Google Scholar]
- 33.Mphahlele, M. J., I. Peenze, and A. D. Steele. 1999. Rotavirus strains bearing the VP4P[14] genotype recovered from South African children with diarrhoea. Arch. Virol. 1441027-1034. [DOI] [PubMed] [Google Scholar]
- 34.Muñoz, M., I. Lanza, M. Alvarez, and P. Carmenes. 1995. Prevalence of neutralizing antibodies to 9 rotavirus strains representing 7 G-serotypes in sheep sera. Vet. Microbiol. 45351-361. [DOI] [PubMed] [Google Scholar]
- 35.Nicholas, K. B., H. B. Nicholas, and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. Embnet News. 414. [Google Scholar]
- 36.Palombo, E. A., and R. F. Bishop. 1995. Genetic and antigenic characterization of a serotype G6 human rotavirus isolated in Melbourne, Australia. J. Med. Virol. 47348-354. [DOI] [PubMed] [Google Scholar]
- 37.Palombo, E. A., R. Clark, and R. F. Bishop. 2000. Characterisation of a “European-like” serotype G8 human rotavirus isolated in Australia. J. Med. Virol. 6056-62. [PubMed] [Google Scholar]
- 38.Parreño, V., K. Bok, F. Fernandez, and J. Gomez. 2004. Molecular characterization of the first isolation of rotavirus in guanacos (Lama guanicoe). Arch. Virol. 1492465-2471. [DOI] [PubMed] [Google Scholar]
- 39.Parreño, V., V. Constantini, S. Cheetham, J. Blanco Viera, L. J. Saif, F. Fernández, L. Leoni, and A. Schudel. 2001. First isolation of rotavirus associated with neonatal diarrhoea in guanacos (Lama guanicoe) in the Argentinean Patagonia region. J. Vet. Med. B 48713-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pisanelli, G., V. Martella, U. Pagnini, L. De Martino, E. Lorusso, G. Iovane, and C. Buonavoglia. 2005. Distribution of G (VP7) and P (VP4) genotypes in buffalo group A rotaviruses isolated in Southern Italy. Vet. Microbiol. 1101-6. [DOI] [PubMed] [Google Scholar]
- 41.Potgieter, A. C., A. D. Steele, and A. A. van Dijk. 2002. Cloning of complete genome sets of six dsRNA viruses using an improved cloning method for large dsRNA genes. J. Gen. Virol. 832215-2223. [DOI] [PubMed] [Google Scholar]
- 42.Pratelli, A., V. Martella, M. Tempesta, and C. Buonavoglia. 1999. Characterization by polymerase chain reaction of ruminant rotaviruses isolated in Italy. New Microbiol. 22105-109. [PubMed] [Google Scholar]
- 43.Rahman, M., J. Matthijnssens, X. Yang, T. Delbeke, I. Arijs, K. Taniguchi, M. Iturriza-Gómara, N. Iftekharuddin, T. Azim, and M. Van Ranst. 2007. Evolutionary history and global spread of the emerging G12 human rotaviruses. J. Virol. 812382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramani, S., I. Banerjee, B. P. Gladstone, R. Sarkar, D. Selvapandian, A. M. Le Fevre, S. Jaffar, M. Iturriza-Gómara, J. J. Gray, M. K. Estes, D. W. Brown, and G. Kang. 2007. Geographic information systems and genotyping in identification of rotavirus G12 infections in residents of an urban slum with subsequent detection in hospitalized children: emergence of G12 genotype in South India. J. Clin. Microbiol. 45432-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray, P., S. Sharma, R. K. Agarwal, K. Longmei, J. R. Gentsch, V. K. Paul, R. I. Glass, and M. K. Bhan. 2007. First detection of G12 rotaviruses in newborns with neonatal rotavirus infection at all India Institute of Medical Sciences, New Delhi, India. J. Clin. Microbiol. 453824-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samajdar, S., V. Varghese, P. Barman, S. Ghosh, U. Mitra, P. Dutta, S. K. Bhattacharya, M. V. Narasimham, P. Panda, T. Krishnan, N. Kobayashi, and T. N. Naik. 2006. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. J. Clin. Virol. 36183-188. [DOI] [PubMed] [Google Scholar]
- 47.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 1529-56. [DOI] [PubMed] [Google Scholar]
- 48.Shen, S., B. Burke, and U. Desselberger. 1993. Nucleotide sequences of the VP4 and VP7 genes of a Chinese lamb rotavirus: evidence for a new P type in a G10 type virus. Virology 197497-500. [DOI] [PubMed] [Google Scholar]
- 49.Steyer, A., M. Poljsak-Prijatelj, T. L. Bufon, N. Marcun-Varda, and J. Marin. 2007. Rotavirus genotypes in Slovenia: unexpected detection of G8P[8] and G12P[8] genotypes. J. Med. Virol. 79626-632. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchida, R., B. D. Pandey, J. B. Sherchand, K. Ahmed, M. Yokoo, T. Nakagomi, L. E. Cuevas, N. A. Cunliffe, C. A. Hart, and O. Nakagomi. 2006. Molecular epidemiology of rotavirus diarrhea among children and adults in Nepal: detection of G12 strains with P[6] or P[8] and a G11P[25] strain. J. Clin. Microbiol. 443499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urasawa, S., A. Hasegawa, T. Urasawa, K. Taniguchi, F. Wakasugi, H. Suzuki, S. Inouye, B. Pongprot, J. Supawadee, S. Suprasert, et al. 1992. Antigenic and genetic analyses of human rotaviruses in Chiang Mai, Thailand: evidence for a close relationship between human and animal rotaviruses. J. Infect. Dis. 166227-234. [DOI] [PubMed] [Google Scholar]
- 53.Urasawa, T., K. Taniguchi, N. Kobayashi, K. Mise, A. Hasegawa, Y. Yamazi, and S. Urasawa. 1993. Nucleotide sequence of VP4 and VP7 genes of a unique human rotavirus strain Mc35 with subgroup I and serotype 10 specificity. Virology 195766-771. [DOI] [PubMed] [Google Scholar]
- 54.van der Donck, I., L. van Hoovels, K. de Leener, T. Goegebuer, L. Vanderwegen, J. Frans, M. Rahman, and M. van Ranst. 2003. Severe diarrhea due to rotavirus infection in a Belgian hospital 1981-2002. Acta Clin. Belg. 5812-18. [DOI] [PubMed] [Google Scholar]