Abstract
Phylogenetic studies of the emergence and spread of natural recombinants in herpesviruses infecting humans and animals have been reported recently. However, despite an ever-increasing amount of evidence of recombination in herpesvirus history, the recombination process and the consequences on the genetic diversity of the progeny remain poorly characterized. We addressed this issue by using multiple single-nucleotide polymorphisms (SNPs) differentiating the two subtypes of an alphaherpesvirus, bovine herpesvirus 1 (BoHV-1). Analysis of a large sample of progeny virions obtained in a single growth cycle of coinfected BoHV-1 strains provided a prospective investigation of the recombination dynamics by using SNPs as recombination markers. We found that the simultaneous infection with two closely related herpesviruses results in a highly diversified recombination mosaic. From the analysis of multiple recombinants arising in the progeny, we provide the first evidence of genetic interference influencing the recombination process in herpesviruses. In addition, we report striking differences in the levels of recombination frequency observed along the BoHV-1 genome. With particular emphasis on the genetic structure of a progeny virus population rising in vitro, our data show to which extent recombination participates to the genetic diversification of herpesviruses.
Genetic variation within a species arises through the process of mutation. If the mutation is nonlethal and if the new variant is not lost, genetic, demographic, and evolutionary processes determine its population frequency and its nonrandom association (linkage disequilibrium) with adjacent sites along the DNA segment on which it arose. Recombination is the primary genetic process that influences linkage disequilibrium over time, enabling the creation of new combinations of genetic material through the pairing and shuffling of related DNA sequences (2). In contrast with most RNA viruses, DNA replication in herpesviruses leads to rare spontaneous mutations because of an efficient proofreading activity of the DNA polymerase (6, 9, 10, 26). With regard to the low rate of nucleotide substitution, recombination can be seen as an evolutionary driving force increasing the probability of a rare nonsynonymous mutation spreading within a herpesvirus species (44, 45). In accordance with this hypothesis, recently reported phylogenetic evidence demonstrated both the high degree of gene conservation in natural herpesvirus populations and the emergence and spread of several natural recombinants in herpesvirus species that infect humans (1, 24, 25, 29, 33-35, 37) and animals (8, 18, 36).
Two types of recombination are described for herpesviruses: illegitimate and homologous recombinations (48). However, the precise mechanism of herpesvirus recombination is poorly understood. It is most likely coupled with viral DNA replication and may require both viral and cellular factors (4, 11, 43). Homologous recombination can occur only between closely related genomes (16, 27). Nonetheless, the creation of new genomes requires some sequence heterogeneity in the genomes involved in recombination. These sequence variations are used as recombination markers. As described above, an intrinsic characteristic of herpesviruses is their high level of intraspecies gene conservation. In the current study, we detected rare single-nucleotide polymorphisms (SNPs) differentiating two wild-type strains of an alphaherpesvirus, bovine herpesvirus 1 (BoHV-1). BoHV-1 is a major pathogen in cattle (31). Primary infection is associated with various clinical manifestations such as infectious bovine rhinotracheitis, infectious pustular vulvovaginitis, abortion, and systemic infection (21, 46). According to antigenic and genomic characteristics, BoHV-1 has been subdivided into two distinct but closely related subtypes: subtypes 1 (BoHV-1.1) and 2 (BoHV-1.2) (31). Although most BoHV-1.1 strains have been isolated from respiratory tract diseases or abortion cases and most BoHV-1.2 strains have been isolated from genital organ lesions, the only reliable distinctive criterion has emerged through the results of viral DNA analysis by restriction endonuclease fingerprinting (13, 42). To date, natural BoHV-1 recombinants generated from the two BoHV-1 subtypes have never been observed.
In this study, we aimed to investigate the impact of recombination on genetic diversity in the BoHV-1 progeny issued from coinfection with two wild-type BoHV-1 strains distinguished by SNP. SNP for virus populations was previously defined as the nucleotide changes shared by a minimum of two isolates in a virus population (14). TaqMan genotyping assays were set up, allowing the allelic discrimination of seven SNPs that spanned the BoHV-1 genome and differentiating two wild-type strains of each of the BoHV-1 subtypes. These SNPs were further used to characterize the progeny virus genotype of around 300 virions randomly sorted from an in vitro coinfection experiment involving BoHV-1.1 and BoHV-1.2. The tool developed in this study overcomes several biases induced by previously used recombination markers such as deletion markers (15, 27, 28, 40), marker rescue techniques (3, 20, 32), and restriction endonuclease (RE) sites (4, 47). The main limits of the latter markers are possible recognition failures of some of the RE sites and a star effect with some of the REs. Moreover, the number of RE sites that differentiates wild-type genomes is not exhaustive. When they are artificially introduced, RE sites do not allow the investigation of recombination between natural variants. In contrast, TaqMan genotyping assays developed in our study showed high sensitivity, specificity, and robustness to investigate recombination between natural BoHV-1 isolates.
This original recombination assay contributed to the discovery of unknown characteristics of recombination in herpesvirus. We found that simultaneous infection with two wild-type strains of BoHV-1 results in a highly diversified recombination mosaic. The high frequencies of multiple recombinants observed in the progeny provided the first evidence of a negative genetic interference in an animal DNA virus. In addition, our findings support the idea that the localization of a gene in the BoHV-1 genome influences its recombination rate.
MATERIALS AND METHODS
Viruses and cell culture.
Madin-Darby bovine kidney cells (MDBK; ATCC CCL-22) were grown as previously described (27). Viral stocks were produced by infection of MDBK cells at a multiplicity of infection (MOI) of 0.01, a condition known to minimize the rise of defective interfering particles. At 72 h after infection, the culture medium was removed and clarified by centrifugation. Aliquots of supernatants were frozen at −80°C and titrated by plaque assay on MDBK cells. Virus titers exceeded 108 PFU/ml for each parental virus used in coinfection experiments in order to minimize the potential effect of defective interfering particles. Four BoHV-1 strains were used in the present study. BoHV-1 strains Ciney (22) and Cooper (49) belong to BoHV-1.1, while strains K22 (49) and ST (23) belong to BoHV-1.2.
Coinfection experiment.
Monolayers of MDBK cells prepared in 25-cm2 flasks were coinfected with the parental viruses BoHV-1.1 Ciney and BoHV-1.2 K22 at a total MOI of 10 using a ratio of 50:50 of each the parental strains. Virus attachment was allowed for 1 h at 4°C. Cells were then further incubated at 37°C. Two hours after the temperature shift, the inoculum was removed, and the cells were washed with phosphate-buffered saline and treated for 1 min using low-pH inactivation of extracellular virions (with a solution at pH 3 containing 40 mM citric acid, 10 mM KCl, and 135 mM NaCl). Cells were then washed twice with Earle's minimal essential medium (MEM) and further incubated at 37°C in 6 ml of MEM supplemented with 5% heat-inactivated fetal calf serum. Twenty-four hours after the temperature shift (when the monolayers showed extensive cytopathic effects), the culture medium was removed, briefly sonicated, clarified twice by centrifugation (1,000 × g), aliquoted, and stored at −80°C.
Isolation and screening of progeny viruses resulting from coinfections.
An aliquot of the supernatant from the situation of coinfection was diluted serially in MEM in order to identify the appropriate dilution for individual plaque isolation. Each dilution was used to infect MDBK monolayers cultured in six-well plates (Multiwell; Becton Dickinson). After a 1-h incubation at 37°C, the supernatant was removed, and cell monolayers were overlaid with MEM containing penicillin-streptomycin, 1% (wt/vol) agarose (agar bacteriological; Oxoid), and 10% bovine serum containing BoHV-1-neutralizing antibodies. After 72 h of incubation, individual plaques were picked and propagated individually by the inoculation of MDBK cells grown in 24-well plates with MEM containing penicillin-streptomycin and 2% horse serum.
Virus growth analysis.
MDBK cells were infected with the respective viruses at an MOI of 10. After 1 h at 4°C, prewarmed medium was added, and cells were further incubated for 2 h at 37°C to allow virus penetration. The inoculum was then removed, and the cells were washed with phosphate-buffered saline and treated for 1 min using low-pH inactivation of extracellular virions. Cells were then washed twice with MEM and further incubated at 37°C in MEM supplemented with 5% heat-inactivated fetal calf serum. Immediately thereafter and after 5, 10, 15, 20, 25, 30, and 35 h of incubation at 37°C, an aliquot of the culture medium was removed and clarified twice by centrifugation (1,000 × g). Titrations of these supernatants determined by plaque assay on MDBK cells determine extracellular virus production. Infected cell monolayers were frozen with the rest of the medium after the same incubation times. Titrations of the unfrozen cell lysates gave the total virus production. Intracellular virus production was obtained by subtracting the extracellular production from the total production.
Viral DNA preparation.
Viral DNA of the parental strains was prepared from clarified supernatants of infected MDBK cell cultures as previously described (22). To characterize the genotype of the progeny viruses obtained in the coinfection experiment, DNA was extracted together with the cellular DNA from approximately 200,000 infected cells by using a small-scale extraction-purification procedure (Qiamp DNA minikit; Qiagen). DNA extracts were stored at −20°C until use.
PCR and sequencing.
Seven genes were selected along the BoHV-1 genome (Table 1): UL44 (gC), UL27 (gB), UL11 (tegument protein), LRORF2 (latency-related product), US2 (US2 protein), US6 (gD), and US8 (gE). In order to maximize the rate of detection of single point mutations differentiating BoHV-1.1 and BoHV-1.2, the PCR primers used were designed to target the portions of the genes that showed the greatest divergence between the BoHV-1 and BoHV-5 genomes, with BoHV-5 being the virus most closely related to BoHV-1 (GenBank accession numbers AJ004801 and AY261359, respectively) (Table 1) (7). PCRs were carried out using 5 ng of purified viral DNA as a template in 50 μl containing 15 pmol of both forward and reverse primers, 200 μM of each deoxynucleoside triphosphate, 3% dimethyl sulfoxide, 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris HCl, 2 mM MgSO4, 0.1% Triton X-100, and 1 U Taq DNA polymerase (New England Biolabs). PCR started with denaturation for 4 min at 95°C, followed by 35 cycles as follows: denaturation for 30 s at 95°C, annealing for 30 s, and elongation for 1 min at 72°C. Annealing temperatures were set up for each primer pair (Table 1). PCR products were purified by using Wizard PCR preps (Promega). Sequencing reactions for purified PCR products were performed with a BigDye Terminator v3.0 apparatus (Applied Biosystems). An ABI Prism 3730 DNA analyzer (Applied Biosystems) was used for analysis of the samples. All PCR product sequences were verified on both strands.
TABLE 1.
PCR amplification and sequencing primers used to detect mutations in seven BoHV-1 genes
| Target sequence | Primer sequence (5′-3′)b | Tm (°C)c | Product length (bp) (position in BoHV-1a) |
|---|---|---|---|
| US8/gE (N terminal) | ACGGCGCACGCGAGAGGGTTC (F) | 61 | 300 (121676-121976) |
| CGTCGAGGCAGACGGGCTCCG (R) | |||
| US6/gD (N terminal) | AACATGCAAGGGCCGACATTGG (F1) | 58 | 376 (118893-119269) |
| CGGTGTACTCCATGTAGTAC (R1) | |||
| AACATGCAAGGGCCGACATTGG (F2) | 55 | 552 (118893-119445) | |
| GACCGTGCCGTCGATGTACAG (R2) | |||
| US2/tegument protein (C terminal) | CGAGTTCAGCAAGTTGTAGCC (F) | 57 | 314 (115340-115654) |
| ATAGTCACGTGTGC[G/A]GATAG (R) | |||
| UL0 (LR-ORF2) | CGCATGCGCGAGCAGTTACTTT (F1) | 65 to 50 | (100517-100856) |
| CGAGAGCTCGGCGCAGAAGA (R1) | |||
| GCCTGCACTTGCCTTTGGCAT (F2) | 59 | 363 (100393-100756) | |
| TGGCGCCGCAGCCAGGCGGTC (R2) | |||
| UL11/tegument protein | ATGGGACAGGCGGCGTCGTGC (F) | 61 | 330 (84191-84520) |
| CAAATCGTGTAATTCTTTGCGTGT (R) | |||
| UL27/gB (N terminal) | TGGGCATGGGCGAGATCA (F) | 59 | 351 (56001-56352) |
| CGCTCGGTGGGTCGATGAG (R) | |||
| UL44/gC (N terminal) | CGCGCACCGCCAGTGGCGTTG (F1) | 59 | 508 (17701-18209) |
| TCCGCCATGGGCCCGCTGGGGCGA (R1) | |||
| GCGTGCTGTTGGTAACGGGCG (F2) | 60 | 323 (17924-18247) | |
| AGAGACCGCCAGCCCGAGACC (R2) |
BoHV-1 GenBank accession number AJ004801.
F, forward; R, reverse.
Tm, melting temperature.
Sequence analysis.
Primary DNA sequence assembly and analysis were performed using ClustalW (45a) to locate useful SNPs. All specified sequence positions of SNPs correspond to nucleotide positions in BoHV-1 reference strain Cooper (GenBank accession number AJ004801). Short stretches of BoHV-1.2 strains K22 (38) (accession numbers AF078725, AF078731, and EF624466) and ST (23) (accession number Z23068) were included in the analysis where they were available online.
SNP genotyping assay.
According to the localization of the SNPs enabling the differentiation between BoHV-1.1 and BoHV-1.2, primers and minor groove binder probes for TaqMan assay were designed using Primer Express v 2.0 (Applied Biosystems, Applera Corporation, CA). Sequences for the primers and probes used are summarized in Table 3. Probes specific for BoHV-1.1 SNPs were labeled with VIC dye, and those specific for BoHV-1.2 SNPs were labeled with 6-carboxyfluorescein (FAM). PCRs were carried out using 10 ng of dried cellular/viral DNA in a final volume of 5 μl composed of 2.5 μl of TaqMan PCR master mix (Applied Biosystems), 0.125 μl of one SNP genotyping assay mixture containing the two primers and the two probes designed for one target SNP, and 2.375 μl of nuclease-free water. The PCR profile was 4 min at 50°C, 15 min at 95°C (activation of the polymerase), and 45 cycles of 15 s at 95°C and 1 min at 60°C. Endpoint reading of the fluorescence generated during PCR amplification was made using the ABI Prism 7900HT apparatus, and genotype assignments were obtained with Sequence Detection System (SDS) software. For each SNP locus, results were plotted on a two-dimensional scatter plot of the BoHV-1.1 “allele” versus the BoHV-1.2 “allele.”
TABLE 3.
Primers and probes used in TaqMan assays to discriminate BoHV-1.1 and BoHV-1.2 at seven SNP positions
| SNP | SNP position in BoHV-1a | Nucleotide identity
|
TaqMan probec | TaqMan primerb | |
|---|---|---|---|---|---|
| BoHV-1.1 | BoHV-1.2 | ||||
| UL44/gC | 17983 | A | C | VIC-CGCGTCCCCACGGG | GGCGTGCTGTTGGTAACG (F) |
| FAM-CGCGTCCCCCCGGG | ACGCCCGGTGCTACTAC (R) | ||||
| UL27/gB | 56095 | T | C | VIC-TGAAGCCTGCGCGGC | ACGACGACCCCTGGGA (F) |
| FAM-AAGCCCGCGCGGC | CGTCCGTCGTGTGCCA (R) | ||||
| UL11 | 84380 | G | A | VIC-ACTGGTCCGACGTCAT | GGGCGGCGAGGACAA (F) |
| FAM-ACTGGTCCAACGTCAT | GCGGCCGGTACGGTTTTA (R) | ||||
| UL0/LRT | 100561 | A | G | VIC-TCGAGCCGCAGTCG | CGAGCAGTTACTTTCGGTTTGG (F) |
| FAM-TCGAGCCGCAGCCG | CCCCAGCCTGCATACTTAACTT (R) | ||||
| US2 | 115577 | A | G | VIC-CTCGGCAAGGAAA | AGCGCCCGGCTCTG (F) |
| FAM-CTCGGCGAGGAAA | GGGCAAGCCAGACGT (R) | ||||
| US6/gD | 119103 | A | G | VIC-CTACGCGACGAGCG | GCAGCCCGTCGAGGT (F) |
| FAM-TACGCGGCGAGCG | GATCAGCGCCAGCATGTC (R) | ||||
| US8/gE | 121768 | T | C | VIC-CTACGCGACGAGCG | CTGCCGCTGCTGCTG (F) |
| FAM-TACGCGGCGAGCG | CGGCCATCAAGCCCGAAA (R) | ||||
BoHV-1 GenBank accession number AJ004801.
F, forward; R, reverse.
SNPs differentiating BoHV-1.1 and BoHV-1.2 are indicated in boldface type.
Statistical analysis of the progeny virus population.
The confidence intervals (α = 0.05) of the individual progeny configuration were calculated by establishing the lower and upper limits of each of the intervals in a binomial sampling distribution. To test the existence of interference in the three-factor crosses, Fisher's exact test was used. The number of observed double recombinants was compared to the number of expected recombinants under a null hypothesis of independence of two neighboring recombination events. The P value represented the sum of the P values calculated from the hypergeometric distribution in the contingency tables of double-recombination events equal to or higher than the observed frequency.
RESULTS
Selection of two wild-type BoHV-1 strains as parental strains for recombination experiments.
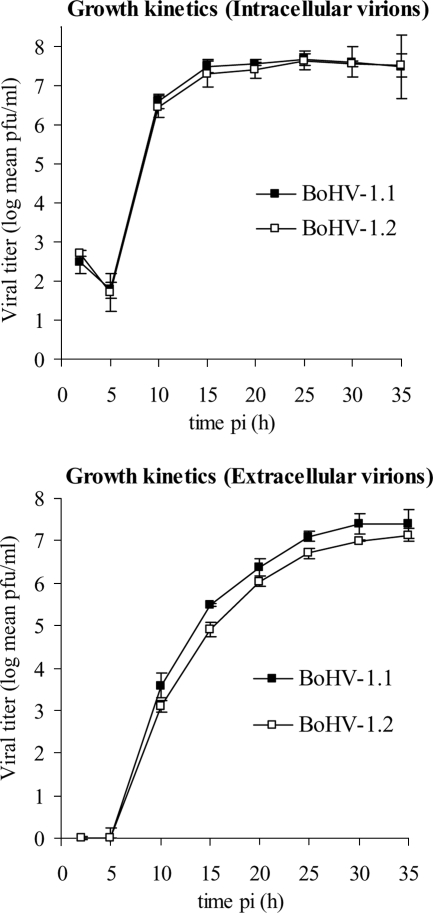
In order to overcome the biases introduced by recombination markers artificially introduced into the parental strains previously used in the study of recombination in herpesviruses (3, 4, 17, 28, 40), we used point mutation markers differentiating two wild-type BoHV-1 field strains, namely, Ciney (BoHV-1.1) and K22 (BoHV-1.2). Several advantages favor the use of two wild-type strains for recombination experiments. First, differential growth phenotypes can be discounted. As illustrated in Fig. 1, no significant difference was evidenced here in the single-cycle growth kinetics for the intracellular virus titers. However, BoHV-1.1 strain Ciney released slightly more virions into the supernatant, as evidenced by the higher-level titer of extracellular BoHV-1.1 Ciney from 12 to 35 h postinfection. Second, the high degree of homology between the two selected parental strains allowed us to overcome restrictions on the homologous recombination imposed by nonhomology (27).
FIG. 1.
Growth kinetic curves of two wild-type BoHV-1 strains selected as parental strains in coinfection experiments. Data for intra- and extracellular growth kinetics of BoHV-1.1 (strain Ciney) and BoHV-1.2 (strain K22) were obtained after infection of MDBK cells at an MOI of 10. Titers, expressed as PFU, were recorded after 2, 5, 10, 15, 20, 25, 30, and 35 h postinfection (pi). The data presented are mean values ± 1.96 standard deviations of triplicates (α = 0.05).
Stable SNPs discriminated by TaqMan PCR assays are suitable as recombination markers.
In order to detect point mutations differentiating the two wild-type BoHV-1 strains, seven fragments were amplified and sequenced in the UL (four fragments) and the US (three fragments) sequences from BoHV-1.1 and BoHV-1.2 genomic DNA. The target sequences were localized in the US8, US6, US2, UL0, UL11, UL27, and UL44 BoHV-1 genes, encoding glycoprotein E (gE), gD, US2 tegument protein, latency-related product, UL11 tegument protein, gB, and gC, respectively (Table 1). Expressed as the relative percentage of the genome length, the locations of the amplified loci are at 11% (US8), 13% (US6), 15% (US2), 26% (UL0), 38% (UL11), 59% (UL27), and 87% (UL44) of the BoHV-1 genome. Sequencing data were analyzed in the seven target sequences obtained from BoHV-1.1 and BoHV-1.2. The alignment of the nucleotide sequences showed a mean degree of homology of 99.2% between BoHV-1.1 and BoHV-1.2. Twenty-one point mutations were identified in the seven regions that were targeted along the BoHV-1.1 and BoHV-1.2 genomes (Table 2). With the exception of two mutations localized in US2- and gC (UL44)-encoding genes, all mutations were transitions. Each mutation was confirmed by the alignment of six sequence analyses (three on the forward strand and three on the reverse strand) of PCR products obtained independently. The stability of the mutations was demonstrated by sequencing of PCR products obtained from BoHV-1.1 and BoHV-1.2 DNA extracted after five successive passages in cell culture.
TABLE 2.
Mutations differentiating BoHV-1.1 Ciney and BoHV-1.2 K22 obtained by sequence alignment of PCR products in seven less conserved regions of BoHV-1 genome
| Gene (sequenced region) | Length of the sequenced region (bp) | SNP position(s) in the ORFa | Codon modification [BoHV-1.1]aa→[BoHV-1.2]aac | Mutation typeb |
|---|---|---|---|---|
| US8 (N terminal) | 258 | 55 | [TTA]Leu→[CTA]Leu | Ts S |
| US6 (N terminal) | 511 | 208 | [ACG]Thr→[GCG]Ala | Ts NS |
| 426 | [TTT]Phe→[TTC]Phe | Ts S | ||
| 462 | [ACG]Thr→[ACA]Thr | Ts S | ||
| US2 (N terminal) | 273 | 87 | [CTT]Leu→[CTC]Leu | Ts S |
| 149 | [CAC]His→[CGC]Arg | Ts NS | ||
| 169, 170 | [AAG]Lys→[GCG]Ala | Ts-Tv NS | ||
| 265 | [CGC]Arg→[TGC]Cys | Ts NS | ||
| LR ORF2 | 321 | 73 | [ACT]Thr→[GCT]Ala | Ts NS |
| UL11 | 288 | 165 | [AAA]Lys→[AAG]Lys | Ts S |
| 166, 167 | [AAA]Lys→[GGA]Gly | Ts NS | ||
| 169 | [AAC]Asn→[GAC]Asp | Ts NS | ||
| 187 | [GAC]Asp→[AAC]Asn | Ts NS | ||
| UL27 (N terminal) | 314 | 702 | [TTT]Phe→[TTC]Phe | Ts S |
| 740 | [CTG]Leu→[CCG]Pro | Ts S | ||
| UL44 (N terminal) | 280 | 144 | [GCT]Ala→[GCC]Ala | Ts S |
| 227 | [GTG]Val→[GGG]Gly | Tv NS | ||
| 303 | [CCC]Pro→[CCT]Pro | Ts S | ||
| 372 | [CCT]Pro→[CCC]Pro | Ts S |
SNPs selected for TaqMan genotyping assay are indicated in boldface type. ORF, open reading frame.
Ts, transition; Tv, transversion; S, synonymous; NS, nonsynonymous.
aa, amino acid.
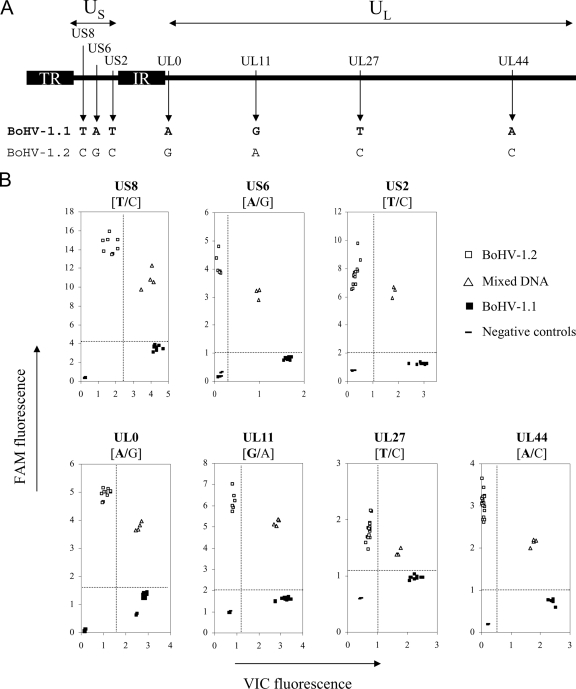
One mutation per investigated locus was selected for setting up seven BoHV-1.1/BoHV-1.2-discriminative TaqMan PCR assays (Table 2). Primers and hybridization probes spanning the selected SNPs were designed using Primer Express v.2.0 (Applied Biosystems) (Table 3). Endpoint reading of the fluorescence generated during PCR amplification demonstrated that the TaqMan assays were efficacious at specifically discriminating BoHV-1.1 and BoHV-1.2 at the seven loci. Figure 2 illustrates the genotyping results for the two allele versions of the UL44, UL27, UL11, UL0, US2, US6, and US8 genes. Through the specific degradation of the VIC probes during PCR elongation in the presence of BoHV-1.1 alleles, an intense fluorescence was emitted and was measured along the x axis. DNA from cell cultures infected by BoHV-1.2 K22 hybridized with the FAM-labeled probes, resulting in a high fluorescence signal along the y axis. When different ratios of BoHV-1.1 and -1.2 DNA (1/10, 1/1, and 10/1) were mixed together and submitted for analysis, these samples were hybridized with both the FAM- and VIC-labeled probes, resulting in increased fluorescence signals along both axes. This result indicated that the TaqMan genotyping assays enabled the detection of a mixed viral population. Therefore, this provides a useful prerequisite condition for validating the clonality of isolated plaque-purified progeny viruses. The specificity was further demonstrated by the lack of fluorescence signals when DNA from mock-infected bovine cells was submitted to the seven TaqMan assays (Fig. 2). The sensitivity of the discrimination between BoHV-1.1 and -1.2 in the seven target SNP positions was investigated. Total DNA quantities ranging from 1 to 20 ng of DNA extracted from infected cell cultures gave a similar signal, enabling the discrimination (Fig. 2). Additional controls included a second strain for each BoHV-1 subtype and DNA extracted after five passages in cell culture (data not shown). BoHV-1 strain Cooper displayed the BoHV-1.1 genotype at the seven positions; BoHV-1 strain ST displayed the BoHV-1.2 genotype. Thus, the seven point mutations discriminated by genotyping assays represent seven SNPs segregating the BoHV-1.1 and BoHV-1.2 subtypes. Furthermore, the TaqMan assays showed similar discriminative results when performed on DNA obtained after five successive passages of the four BoHV-1 strains in cell culture (data not shown). Viral strains Ciney and Cooper were still assigned to the BoHV-1.1 subtype at the seven SNPs loci; strains K22 and ST were still assigned to the BoHV-1.2 subtype. Taken together, these results indicate that seven SNPs spanning and differentiating the BoHV-1.1 and BoHV-1.2 genomes are stable. They represent good recombination markers, which are discriminated rapidly and accurately in high-throughput TaqMan PCR assays.
FIG. 2.
TaqMan genotyping assays discriminate seven SNPs differentiating BoHV-1.1 and BoHV-1.2 genomes. (A) Organization of the BoHV-1 genome including two unique sequences, a long one (UL) and a short one (US). The latter is flanked by two repeated and inverted sequences (IR, internal repeat; TR, terminal repeat). Shown are the localizations of the seven point mutations detected in seven BoHV-1 genes that are targeted by TaqMan genotyping assays. The nucleotides indicate the allelic version discriminating BoHV-1.1 (in boldface type) and BoHV-1.2 at each position. (B) Endpoint reading of the fluorescence generated during PCR amplification of seven target genes discriminating BoHV-1.1 (▪) and BoHV-1.2 (□). BoHV-1.1 DNA hybridized with the VIC-labeled probes, giving rise to an intense fluorescence along the x axis during amplification. BoHV-1.2 alleles hybridized with the FAM-labeled probes, resulting in a high fluorescence signal along the y axis. Different ratios of BoHV-1.1 and -1.2 DNA (1/10, 1/1, and 10/1) were mixed together and submitted to the analysis; these samples (▵) were hybridized with both the FAM- and VIC-labeled probes, resulting in increased fluorescence signals along both axes. Dot plot data were generated from endpoint readings of the fluorescence recorded after TaqMan PCR assays were performed on total DNA quantities ranging from 1 to 20 ng.
Analysis of BoHV-1 progeny generated by coinfection between BoHV-1.1 and -1.2.
Wild-type BoHV-1.1 Ciney (P1) and BoHV-1.2 K22 (P2) were used as the two parents in a large-scale recombination experiment. Cells were coinfected at a total MOI of 20 at a P1-to-P2 ratio of 1:1. Control cell cultures were infected separately with P1 or P2 at an MOI of 10. After 24 h of incubation, the cells and supernatants were harvested, the virus was released by sonication, and the progeny was titrated (data not shown). The range of differences between progeny virus titers for BoHV-1.1 (5 × 107 PFU/ml) and BoHV-1.2 (1.1 × 107 PFU/ml) grown alone was equivalent to that shown in Fig. 1, which indicates that BoHV-1.1-infected cells released more infectious virus. The progeny titer of coinfection supernatant (4.3 × 107 PFU/ml) was slightly lower than the BoHV-1.1 titer grown alone.
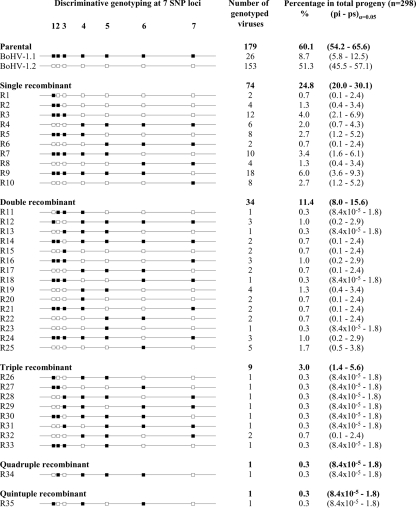
A total of 304 randomly selected progeny viruses were plaque purified and propagated individually from the coinfection supernatant. DNAs were individually extracted and submitted to the seven SNP genotyping assays capable of discriminating the allelic versions specific for BoHV-1.1 and BoHV-1.2 in the US8, US6, US2, UL0, UL11, UL27, and UL44 genes. With seven SNP sites that could be counted as originating from BoHV-1.1 or BoHV-1.2, there were 27, i.e., 128, possible progeny genomes (2 parental genomes and 126 recombinants). With regard to the signal obtained by allelic discrimination, each of the 304 progeny was counted as either P1, P2, or belonging to 1 of the 126 recombinant classes (Fig. 3). Of the progeny, six BoHV-1 virions were removed from the analysis because both genotypes were assigned to at least one SNP position. These viruses were considered as mixes of either the two parental (n = 1) or the two recombinant (n = 5) BoHV-1 populations. When both genotypes were detected at the seven SNP positions, the population was considered to be a mix of the parental virus. When both genotypes were assigned to six of the SNP positions at maximum, the population was considered to be a mix of two recombinants. Of the 298 monoclonal progeny viruses, 119 were characterized as recombinants, giving an overall frequency of 39.9% of recombinants, and 179 were defined as parental (60.1% of the progeny) (Fig. 3). From the binomial sampling distribution, any configuration that was not represented in the sample (n = 0 in 298 events) had a confidence interval ranging from 0 to 1.2%. Among the parental structures, BoHV-1.2 was five times more frequent than BoHV-1.1 (Fig. 3). Among the recombinant configurations, a high level of diversity was observed, since 35 of the 126 possible configurations were represented. The class of single recombinants was the most abundant, representing 24.8% of the progeny, where recombinants generated from double, triple, quadruple, and quintuple recombination events represented 11.4, 3.0, 0.3, and 0.3% of the progeny, respectively. Taken together, these results show that several mosaic structures combining up to six different segments were generated following the simultaneous in vitro infection with two wild-type BoHV-1 strains. It is also obvious that recombination events occurred along the entire genome, since at least one configuration combining two neighboring SNP markers was detected within both US and UL segments.
FIG. 3.
Characterization and division of the progeny virus genotypes issued from in vitro coinfection with BoHV-1.1 and BoHV-1.2. A total of 298 randomly plaque-purified virions issued from a BoHV-1.1 and BoHV-1.2 coinfection experiment were submitted to TaqMan genotyping assays at seven selected SNP sites in BoHV-1, the US8 (1), US6 (2), US2 (3), UL0 (4), UL11 (5), UL27 (6), and UL44 (7) genes. The allelic discrimination data are represented by small squares at each SNP position. Filled squares (▪) indicate the alleles inherited from BoHV-1.1; empty squares (□) indicate the alleles inherited from BoHV-1.2. Progeny virus genotypes were distributed in 37 different SNP combinations: 2 parental (P1 and P2) and 35 recombinants (R1 to R35). The configurations of recombinants were further classified as single, double, triple, quadruple, and quintuple recombinants by counting the recombination events present in the virus genotype. Quantitative data are presented as the absolute number of genotyped viruses classified in each configuration and as the relative frequency in the total progeny. The confidence intervals (α = 0.05) were calculated by determining the lower (pi) and upper (ps) limits of frequency in a binomial sampling distribution.
Negative genetic interference was observed in three-factor crosses of the BoHV-1 progeny.
Several multiple recombinants were observed in the 298 progeny viruses since 45 recombinants combined more than two SNPs of different origin (Fig. 3). In order to investigate the influence of a first recombination event on the occurrence of a second recombination event, we decided to compare the number of observed double recombinants in the progeny with the number of expected double recombinants. Under a null hypothesis of independent occurrence, the double-recombinant frequency (rF) is the product of the single-recombinant frequency (Table 4). From the ratio of the number of observed double recombinants to the number of expected double recombinants (a ratio defined as the coefficient of coincidence [c]), the genetic interference (I) was calculated as follows: I = 1 − c (Table 4). I values ranging from 0 to 1 indicate that the occurrence of one crossing over (CO) decreases the probability of a second CO occurring nearby. Negative I values are obtained when the number of observed double recombinants is higher than the expected number. In the BoHV-1 progeny generated by the coinfection between BoHV-1.1 and BoHV-1.2, negative interference was observed in the five situations of three-factor crosses (Table 4). Significant negative interference was observed in two situations involving US6, US2, and UL0 (I = −2.11) and UL11, UL27, and UL44 (I = −1.09). Thus, BoHV-1 recombination generated more double recombinants than expected in these situations. This means that double-recombination events were interdependent in these situations and indicates that a first recombination event enhanced the occurrence of a second recombination in the neighboring regions. To our knowledge we provide here the first evidence of genetic interference in the recombination process in herpesviruses.
TABLE 4.
Interference in the three-factor crossesc
| SNP as recombination marker:
|
Single rF (%)
|
% of expected double rF | No. of expected double recombinants | No. of observed double recombinants | Σ of the P valuea | Interference calculationb | |||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | A/B | B/C | |||||
| US8 | US6 | US2 | 3 | 5 | 0.15 | 0.45 | 1 | 0.38 | −1.24 |
| US6 | US2 | UL0 | 5 | 15.1 | 0.76 | 2.25 | <7 | <0.01 | −2.11 |
| US2 | UL0 | UL11 | 15.1 | 7.4 | 1.12 | 3.33 | 5 | 0.22 | −0.50 |
| UL0 | UL11 | UL27 | 7.4 | 12.4 | 0.92 | 2.73 | 4 | 0.28 | −0.46 |
| UL11 | UL27 | UL44 | 12.4 | 16.8 | 2.08 | 6.21 | <13 | <0.01 | −1.09 |
Σ of the P value is the sum of the P values obtained from Fisher's exact test as described in Materials and Methods.
Determined as 1 − (number of observed double recombinants divided by the number of expected double recombinants).
Three-factor crosses where significant negative interference was observed are indicated in boldface type.
Overcoming the bias of invisible recombinants in the assessment of RF.
The use of seven recombination markers spanning BoHV-1 genomes enabled us to address the question of invisible recombinants between distantly separated markers (Table 5). When recombination was assessed between two nonneighboring markers (e.g., US8-US2, UL0-UL27, and US8-UL44), a difference was consistently observed between the number of apparent recombinants and the number of recombination events that had occurred between the two distant markers. The gap between these two counts highlighted the existence of recombinants that cannot be detected when the investigation of recombination relies only on distant markers. We define these recombinants as invisible recombinants. To assess the bias induced by invisible recombinants in the quantitative evaluation of recombination, we compared the frequency obtained from apparent recombinant counts (r) and recombination events (R) (Table 5). Since invisible recombinants have a pronounced effect on the quantitative evaluation of recombination frequency (RF), we further based our analysis on the RF between any two SNP sites of BoHV-1 in relationship to the relative distance separating these SNPs and their respective localizations.
TABLE 5.
rF and RF values between any SNP site calculated from the progeny virus genotypes generated after BoHV-1.1 and BoHV-1.2 coinfectionb
| Event | Coposition | Distance (kbp) | No. of events
|
% in the total progenya
|
%/kb
|
|||
|---|---|---|---|---|---|---|---|---|
| r | R | r | R | rF | RF | |||
| Within US | US8 × US6 | 2.67 | 9 | 9 | 3.0 | 3.0 | 1.13 | 1.13 |
| US6 × US2 | 3.53 | 15 | 15 | 5.0 | 5.0 | 1.43 | 1.43 | |
| US8 × US2 | 6.19 | 22 | 24 | 7.4 | 8.1 | 1.19 | 1.30 | |
| Within UL | UL0 × UL11 | 16.18 | 22 | 22 | 7.4 | 7.4 | 0.46 | 0.46 |
| UL11 × UL27 | 28.29 | 37 | 37 | 12.4 | 12.4 | 0.44 | 0.44 | |
| UL27 × UL44 | 38.11 | 50 | 50 | 16.8 | 16.8 | 0.44 | 0.44 | |
| UL0 × UL27 | 44.47 | 51 | 59 | 17.1 | 19.8 | 0.38 | 0.45 | |
| UL0 × UL44 | 82.58 | 69 | 109 | 23.2 | 36.6 | 0.28 | 0.44 | |
| UL11 × UL44 | 66.40 | 61 | 87 | 20.5 | 29.2 | 0.31 | 0.44 | |
| Between US and UL | US2 × UL0 | 15.02 | 45 | 45 | 15.1 | 15.1 | 1.01 | 1.01 |
| US2 × UL11 | 31.20 | 57 | 67 | 19.1 | 22.5 | 0.61 | 0.72 | |
| US2 × UL27 | 59.48 | 74 | 104 | 24.8 | 34.9 | 0.42 | 0.59 | |
| US2 × UL44 | 97.59 | 80 | 154 | 26.8 | 51.7 | 0.28 | 0.53 | |
| US6 × UL0 | 18.54 | 46 | 60 | 15.4 | 20.1 | 0.83 | 1.09 | |
| US6 × UL11 | 34.72 | 58 | 82 | 19.5 | 27.5 | 0.56 | 0.79 | |
| US6 × UL27 | 63.01 | 71 | 119 | 23.8 | 39.9 | 0.38 | 0.63 | |
| US6 × UL44 | 101.12 | 85 | 169 | 28.5 | 56.7 | 0.28 | 0.56 | |
| US8 × UL0 | 21.21 | 45 | 69 | 15.1 | 23.2 | 0.71 | 1.09 | |
| US8 × UL11 | 37.39 | 57 | 91 | 19.1 | 30.5 | 0.51 | 0.82 | |
| US8 × UL27 | 65.67 | 70 | 128 | 23.5 | 43.0 | 0.36 | 0.65 | |
| US8 × UL44 | 103.79 | 84 | 178 | 28.2 | 59.7 | 0.27 | 0.58 | |
The total number of progeny was 298.
r, recombinant; R, recombination. Values calculated from the number of recombination events are indicated in boldface type.
The highest levels of RF were detected within the unique short segment of BoHV-1.
Based on the results presented above, RF levels between any two SNP sites of the BoHV-1 genome led to the following observations (Table 5): (i) the average RF between any markers localized in the UL segment was around 0.45%; (ii) the recombination observed between any US markers was three times higher, with a range from 1.13% to 1.43%; and (iii) the RF between any US and any UL marker decreased from 1.09% to 0.53% when the distance between the UL marker and the US marker increased, i.e., when the proportion of the US region decreased in the physical distance separating the two markers. These findings support the idea that the localization of a gene in the BoHV-1 genome influences its recombination rate. These observations also reveal that recombination occurs at a very high rate within the BoHV-1 US segment.
DISCUSSION
Through the analysis of the progeny obtained in an in vitro coinfection experiment involving two wild-type BoHV-1 strains, we report the following original results. A significant genetic diversification is generated by the multiple distributions of recombination events. Negative genetic interference is demonstrated in the recombination process in an animal DNA virus. Another salient finding is that the localization of a gene in the BoHV-1 genome influences its recombination rate. The use of SNPs is also validated as reliable recombination markers for a prospective approach regarding recombination in a DNA virus population.
The high frequency of multiple recombinants observed in the progeny provided the first evidence of negative genetic interference in herpesviruses. In the two BoHV-1 regions where significant negative interferences were measured, the occurrence of a first recombination event positively influenced the occurrence of a second recombination event in the neighboring region. This fundamental observation would need further investigations both to determine its influence on the generation of natural recombinants in herpesvirus populations and to characterize the mechanism involved in this interference. Recombination analyses suggest that varicella-zoster virus strains belonging to the M1 and M2 genotypes are mosaic recombinant strains originating from ancestral isolates belonging to the E and J genotypes through recombination on multiple occasions (35). Similar conclusions were drawn from recombination analyses performed using other natural recombinant isolates of varicella-zoster virus (37), human simplex virus 1 (HSV-1), and HSV-2 (1, 33, 34). From the negative interference obtained in our recombination assay, it would be interesting to investigate whether these recombinants were generated from a single coinfection event producing multiple recombinant genomes or through successive occasions of single recombination events.
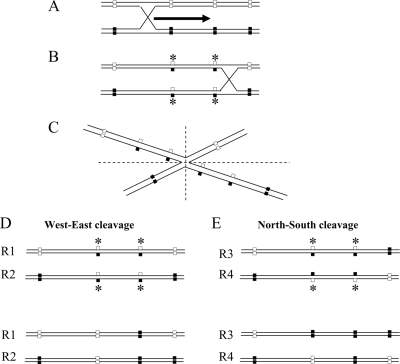
Two main hypothesizes could explain the rise of an excess of multiple recombinants in our analyses. This could result either from double-CO events, whose cooccurrence would have been favored by an unidentified factor, or from gene conversion, a process occurring in the heteroduplex region formed during homologous recombination (5). The relatively short distance separating the recombination markers used in our analysis argues in favor of the latter hypothesis. Indeed, recombination analysis of closely linked markers in both prokaryotic and eukaryotic organisms has generally revealed a value of c much larger than 1 and produced a large negative value of I. This type of result has been named high negative interference (50). In these situations, double recombinants have been shown to appear much more frequently than expected. The high frequency of apparent double recombinants is likely to be the result of a gene conversion occurring at mismatched positions in the region of the heteroduplex expanding from the initial recombination process (19, 39) (Fig. 4). Following a single homologous recombination, repairs convert the nucleotide at the neighboring SNP included in the heteroduplex of the initial CO, giving an “artifactual” result of double recombinant (5).
FIG. 4.
Schematic representation of gene conversion occurring in the heteroduplex region formed during homologous recombination. (A) The two parental DNA molecules (P1 and P2) are aligned before recombination. The initial CO takes place by an exchange of single strands between the parental DNAs. (B) Once the cross-bridge has formed, its position can diffuse along the paired molecules. This process can create long regions of heteroduplex DNA. Mismatched positions in the heteroduplex are indicated by asterisks. (C) The Holliday structure can be redrawn, as indicated in B, after a rotation of 180° around the cross-bridge. The Holliday structure might be resolved either by West-East or by North-South cuts. (D) West-East cut yields linear molecules that are not recombinant for external genetic markers. However, repairs of mismatches within heteroduplex-combining alleles of the two parental DNAs might create apparent double recombinants through gene conversion in favor of one of the parental templates. (E) North-South cut yields linear molecules that are recombinant for parental genetic markers on either side of heteroduplex. Repairs of mismatches might also create multiple recombinants (R4) through gene conversion.
Another result of the present study is the demonstration of differences in RF between the US and UL subunits of BoHV-1. A high RF was observed between the three investigated loci of the S subunit. This is the first prospective demonstration of the high plasticity of the short segment of the herpesvirus genome. The numerous sequence exchanges that we observed in vitro during a very short time window of interaction between two wild-type BoHV-1 strains could correlate with the intriguing recombination mosaics that have been revealed by the retrospective phylogenetic analysis of three neighboring genes (gG, gI, and gE) in the US segment in both HSV-1 (33) and HSV-2 (34). From these field data, it was postulated that the genomes of HSV-1 and HSV-2 isolates consist of a mosaic of segments from different genetic groups.
Our analysis of 300 progeny BoHV-1 strains issued from coinfection between two wild-type BoHV-1 strains differentiated by seven SNP markers revealed an overall rF of 40%. Compared to previous results obtained with nonselected recombination markers, this is twice as high as the overall rF observed in 1,019 HSV-2 progeny (21.8%) by Brown and collaborators (4), who used two viruses originating from the same strain background but differing by five RE sites as unselected markers. However, our result is lower than the rF observed for 93 HSV-1 progeny (54.8%) by Umene (47), who used two strains differing in eight RE sites. More interesting comparisons can be made at the level of RF expressed as a percentage of recombination per kbp. In the present study, we found an overall frequency of 0.58% per kbp in BoHV-1, with significant differences between the L and S segments. This frequency is lower than the 0.7% per kbp previously observed in HSV-1 by Umene (47), who did not obtain any evidence to support the existence of enhanced recombination events in any region of HSV-1. Brown and collaborators (4) previously observed lower overall frequencies of around 0.43% per kbp in HSV-2 and 0.45% per kbp in HSV-1, although they used parental strains that were assumed to be highly homologous. The discrepancies among these three observations could be due to (i) virus species variations (HSV-1 and -2 belong to the genus Simplexvirus and have a class E genome structure, and BoHV-1 belongs to the genus Varicellovirus and has a class D genome structure), (ii) technical limits in the analyses based on RE sites, (iii) the effect of the strains involved in the respective studies, and (iv) an effect of the cell line on which the coinfections have been performed. The latter factor was previously identified as having a deep impact on the recombination process efficiency by Brown et al. (4). Nonetheless, the RF levels found in all the above-mentioned studies are in relatively good agreement and correlate with the RF of 0.5% observed in another DNA virus, human adenovirus type 5 (51).
The present study describes the use of isolated nucleotide mutations as markers in investigating recombination dynamics in herpesviruses. The use of SNPs as recombination markers is advantageous, as large numbers of markers can be detected in any kind of gene, essential or nonessential. Moreover, this is a robust method, as the presence of one allele version at one SNP locus is an all-or-nothing condition. Reversion is also expected to be a rare event, and it was not observed under our conditions. Another benefit of the tool developed in this study is its capacity to detect viruses showing “heterozygosis.” Six plaque-purified viruses were withdrawn from our analysis, as they were considered to be mixes of either the two parental (n = 1) or the two recombinant BoHV-1 (n = 5) populations. However it can be reasonably postulated that some of these configurations were originally produced from a monoclonal recombinant virus harboring a “nonrepaired” mismatched position(s) in the heteroduplex region involved in recombination. Under this hypothesis, the packaging and the release of viral genomes harboring heterozygosis would participate in the genetic diversification following virus replication.
SNP genotyping is also a useful method that allows the investigation of recombination at multiple positions. Although we restricted our investigation to seven loci, we were able to detect several recombination events and recombinant structures that were invisible by the use of two deletion markers (27, 28, 30, 40, 41). In addition, the use of seven SNP markers enabled us to partially answer this question of “invisible recombinants,” but an increased number of target SNPs along the BoHV-1 genome would be needed to resolve this definitively. The better accuracy of this method for investigating recombination is demonstrated by the differences observed between rF and RF in the quantitative assessment of recombination between widely separated markers (such as US8 and UL44). It is obvious that the underestimation provided by the rF results from the invisible recombinants that cannot be detected when distantly separated markers are used.
Regarding the numerous genetic structures (35 out of the 126 possible recombinant combinations) obtained in vitro, the question of the in vivo rise of such recombinants needs to be addressed. A previous study showed that recombinant viruses represent up to 30% of the progeny population isolated from cattle infected with two BoHV-1 mutants (40). Therefore, the rise and the evolution of a recombination mosaic could be monitored following the coinoculation of cattle with the two parental BoHV-1 strains used in this study. The in vivo replication context is much more complex. First, multiple growth cycles are observed during primary BoHV-1 infection. If recombinants are present in the first progeny, they will have the opportunity to coinfect new target cells, giving rise to new gene combinations. Second, several selection factors (the presence of immune effectors and the adaptability of the strains to cattle) may influence the fitness of progeny viruses arising from these coinfections. In vitro, BoHV-1.2 seemed to be favored when cells were coinfected by both strains. The situation could be totally different in vivo. A BoHV-1.1 epidemiological overdominance was observed in the 1970s when BoHV-1.1 strains were introduced into European cattle herds infected hitherto by BoHV-1.2 strains (12). Finally, the establishment of a latent infection with both strains increases the likelihood of cellular coinfection following a reactivation stimulus acting as a virus synchronizer. It was previously demonstrated that cattle can support the latent infection of two distinguishable BoHV-1 strains, namely, BoHV-1.2 K22 and BoHV-1.1 Cooper (49).
In conclusion, the use of SNPs as recombination markers detected by TaqMan assays provides an accurate tool for the further investigation of recombination in herpesvirus both in vitro and in vivo. The data obtained in this study demonstrate that in vitro, (i) BoHV-1 recombines at a high rate, (ii) multiple recombination events occur within one BoHV-1 growth cycle, (iii) BoHV-1 displays a negative interference, and (iv) levels of RF are different within the US and UL sequences.
Acknowledgments
We are grateful to Wojtek Socha, Dominique Ziant, and Lorène Dams for their enthusiastic technical assistance and to Corinne Fasquelle and Bernard Grisart for their support in the sequencing and the TaqMan genotyping processes.
This study was supported by grants from the Fonds National de la Recherche Scientifique (grants FRFC 2.4532.98, FRFC 2.4508.02, and FRFC 1.5.105.03). B. Muylkens is a postdoctoral researcher for the FNRS.
Footnotes
Published ahead of print on 19 January 2009.
REFERENCES
- 1.Bowden, R., H. Sakaoka, P. Donnelly, and R. Ward. 2004. High recombination rate in herpes simplex virus type 1 natural populations suggests significant infection. Infect. Genet. Evol. 4115-123. [DOI] [PubMed] [Google Scholar]
- 2.Brown, G. R., G. P. Gill, R. J. Kuntz, C. H. Langley, and D. B. Neale. 2004. Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc. Natl. Acad. Sci. USA 10115255-15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S. M., D. A. Ritchie, and J. H. Subak-Sharpe. 1973. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J. Gen. Virol. 18329-346. [DOI] [PubMed] [Google Scholar]
- 4.Brown, S. M., J. H. Subak-Sharpe, J. Harland, and A. R. MacLean. 1992. Analysis of intrastrain recombination in herpes simplex virus type 1 strain 17 and herpes simplex virus type 2 strain HG52 using restriction endonuclease sites as unselected markers and temperature-sensitive lesions as selected markers. J. Gen. Virol. 73293-301. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. M., D. N. Cooper, N. Chuzhanova, C. Ferec, and G. P. Patrinos. 2007. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8762-775. [DOI] [PubMed] [Google Scholar]
- 6.Crute, J. J., and I. R. Lehman. 1989. Herpes simplex-1 DNA polymerase. Identification of an intrinsic 5′-3′ exonuclease with ribonuclease H activity. J. Biol. Chem. 26419266-19270. [PubMed] [Google Scholar]
- 7.Delhon, G., M. P. Moraes, Z. Lu, C. L. Afonso, E. F. Flores, R. Weiblen, G. F. Kutish, and D. L. Rock. 2003. Genome of bovine herpesvirus 5. J. Virol. 7710339-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dewals, B., M. Thirion, N. Markine-Goriaynoff, L. Gillet, K. de Fays, F. Minner, V. Daix, P. M. Sharp, and A. Vanderplasschen. 2006. Evolution of bovine herpesvirus 4: recombination and transmission between African buffalo and cattle. J. Gen. Virol. 871509-1519. [DOI] [PubMed] [Google Scholar]
- 9.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 722010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drake, J. W., and C. B. Hwang. 2005. On the mutation rate of herpes simplex virus type 1. Genetics 170969-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutch, R. E., V. Bianchi, and I. R. Lehman. 1995. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J. Virol. 693084-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, S., H. White, and P. Nixon. 1990. A study of the predominant genotypes of bovid herpesvirus 1 found in the U.K. Vet. Microbiol. 22213-223. [DOI] [PubMed] [Google Scholar]
- 13.Engels, M., C. Giuliani, P. Wild, T. M. Beck, E. Loepfe, and R. Wyler. 1986. The genome of bovine herpesvirus 1 (BHV-1) strains exhibiting a neuropathogenic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridization. Virus Res. 657-73. [DOI] [PubMed] [Google Scholar]
- 14.Faga, B., W. Maury, D. A. Bruckner, and C. Grose. 2001. Identification and mapping of single nucleotide polymorphisms in the varicella-zoster virus genome. Virology 2801-6. [DOI] [PubMed] [Google Scholar]
- 15.Glazenburg, K. L., R. J. Moormann, T. G. Kimman, A. L. Gielkens, and B. P. Peeters. 1994. In vivo recombination of pseudorabies virus strains in mice. Virus Res. 34115-126. [DOI] [PubMed] [Google Scholar]
- 16.Halliburton, I. W. 1980. Intertypic recombinants of herpes simplex viruses. J. Gen. Virol. 481-23. [DOI] [PubMed] [Google Scholar]
- 17.Henderson, L. M., R. L. Levings, A. J. Davis, and D. R. Sturtz. 1991. Recombination of pseudorabies virus vaccine strains in swine. Am. J. Vet. Res. 52820-825. [PubMed] [Google Scholar]
- 18.Hughes, A. L., and P. Rivailler. 2007. Phylogeny and recombination history of gallid herpesvirus 2 (Marek's disease virus) genomes. Virus Res. 13028-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huisman, O., and M. S. Fox. 1986. A genetic analysis of primary products of bacteriophage lambda recombination. Genetics 112409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javier, R. T., F. Sedarati, and J. G. Stevens. 1986. Two avirulent herpes simplex viruses generate lethal recombinants in vivo. Science 234746-748. [DOI] [PubMed] [Google Scholar]
- 21.Kaashoek, M. J., P. H. Straver, E. M. Van Rooij, J. Quak, and J. T. van Oirschot. 1996. Virulence, immunogenicity and reactivation of seven bovine herpesvirus 1.1 strains: clinical and virological aspects. Vet. Rec. 139416-421. [DOI] [PubMed] [Google Scholar]
- 22.Lemaire, M., F. Schynts, G. Meyer, and E. Thiry. 1999. Antibody response to glycoprotein E after bovine herpesvirus type 1 infection in passively immunised, glycoprotein E-negative calves. Vet. Rec. 144172-176. [DOI] [PubMed] [Google Scholar]
- 23.Leung-Tack, P., J. C. Audonnet, and M. Riviere. 1994. The complete DNA sequence and the genetic organization of the short unique region (US) of the bovine herpesvirus type 1 (ST strain). Virology 199409-421. [DOI] [PubMed] [Google Scholar]
- 24.Loparev, V., E. Martro, E. Rubtcova, C. Rodrigo, J. C. Piette, E. Caumes, J. P. Vernant, D. S. Schmid, and A. M. Fillet. 2007. Toward universal varicella-zoster virus (VZV) genotyping: diversity of VZV strains from France and Spain. J. Clin. Microbiol. 45559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loparev, V. N., A. Gonzalez, M. Deleon-Carnes, G. Tipples, H. Fickenscher, E. G. Torfason, and D. S. Schmid. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 788349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markine-Goriaynoff, N., J.-P. Georgin, M. Goltz, W. Zimmermann, H. Broll, H. M. Wamwayi, P.-P. Pastoret, P. M. Sharp, and A. Vanderplasschen. 2003. The core 2 β-1,6-N-acetylglucosaminyltransferase-mucin encoded by bovine herpesvirus 4 was acquired from an ancestor of the African buffalo. J. Virol. 771784-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meurens, F., G. M. Keil, B. Muylkens, S. Gogev, F. Schynts, S. Negro, L. Wiggers, and E. Thiry. 2004. Interspecific recombination between two ruminant alphaherpesviruses, bovine herpesviruses 1 and 5. J. Virol. 789828-9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meurens, F., F. Schynts, G. M. Keil, B. Muylkens, A. Vanderplasschen, P. Gallego, and E. Thiry. 2004. Superinfection prevents recombination of the alphaherpesvirus bovine herpesvirus 1. J. Virol. 783872-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muir, W. B., R. Nichols, and J. Breuer. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 761971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muylkens, B., F. Meurens, F. Schynts, F. Farnir, A. Pourchet, M. Bardiau, S. Gogev, J. Thiry, A. Cuisenaire, A. Vanderplasschen, and E. Thiry. 2006. Intraspecific bovine herpesvirus 1 recombinants carrying glycoprotein E deletion as a vaccine marker are virulent in cattle. J. Gen. Virol. 872149-2154. [DOI] [PubMed] [Google Scholar]
- 31.Muylkens, B., J. Thiry, P. Kirten, F. Schynts, and E. Thiry. 2007. Bovine herpesvirus 1 infection and infectious bovine rhinotracheitis. Vet. Res. 38181-209. [DOI] [PubMed] [Google Scholar]
- 32.Nishiyama, Y., H. Kimura, and T. Daikoku. 1991. Complementary lethal invasion of the central nervous system by nonneuroinvasive herpes simplex virus types 1 and 2. J. Virol. 654520-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norberg, P., T. Bergstrom, E. Rekabdar, M. Lindh, and J. A. Liljeqvist. 2004. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J. Virol. 7810755-10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norberg, P., M. J. Kasubi, L. Haarr, T. Bergstrom, and J. A. Liljeqvist. 2007. Divergence and recombination of clinical herpes simplex virus type 2 isolates. J. Virol. 8113158-13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norberg, P., J. A. Liljeqvist, T. Bergstrom, S. Sammons, D. S. Schmid, and V. N. Loparev. 2006. Complete-genome phylogenetic approach to varicella-zoster virus evolution: genetic divergence and evidence for recombination. J. Virol. 809569-9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagamjav, O., T. Sakata, T. Matsumura, T. Yamaguchi, and H. Fukushi. 2005. Natural recombinant between equine herpesviruses 1 and 4 in the ICP4 gene. Microbiol. Immunol. 49167-179. [DOI] [PubMed] [Google Scholar]
- 37.Peters, G. A., S. D. Tyler, C. Grose, A. Severini, M. J. Gray, C. Upton, and G. A. Tipples. 2006. A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J. Virol. 809850-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ros, C., and S. Belak. 1999. Studies of genetic relationships between bovine, caprine, cervine, and rangiferine alphaherpesviruses and improved molecular methods for virus detection and identification. J. Clin. Microbiol. 371247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruksc, A., P. L. Bell-Rogers, J. D. Smith, and M. D. Baker. 2008. Analysis of spontaneous gene conversion tracts within and between mammalian chromosomes. J. Mol. Biol. 377337-351. [DOI] [PubMed] [Google Scholar]
- 40.Schynts, F., F. Meurens, B. Detry, A. Vanderplasschen, and E. Thiry. 2003. Rise and survival of bovine herpesvirus 1 recombinants after primary infection and reactivation from latency. J. Virol. 7712535-12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schynts, F., A. Vanderplasschen, E. Hanon, F. A. Rijsewijk, O. J. van, and E. Thiry. 2001. Use of PCR and immunofluorescence to detect bovine herpesvirus 1 recombinants. J. Virol. Methods 9299-104. [DOI] [PubMed] [Google Scholar]
- 42.Smith, G. A., P. L. Young, and K. C. Reed. 1995. Emergence of a new bovine herpesvirus 1 strain in Australian feedlots. Arch. Virol. 140599-603. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 785856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiry, E., F. Meurens, B. Muylkens, M. McVoy, S. Gogev, J. Thiry, A. Vanderplasschen, A. Epstein, G. Keil, and F. Schynts. 2005. Recombination in alphaherpesviruses. Rev. Med. Virol. 1589-103. [DOI] [PubMed] [Google Scholar]
- 45.Thiry, E., B. Muylkens, F. Meurens, S. Gogev, J. Thiry, A. Vanderplasschen, and F. Schynts. 2006. Recombination in the alphaherpesvirus bovine herpesvirus 1. Vet. Microbiol. 113171-177. [DOI] [PubMed] [Google Scholar]
- 45a.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45191-223. [DOI] [PubMed] [Google Scholar]
- 47.Umene, K. 1985. Intermolecular recombination of the herpes simplex virus type 1 genome analysed using two strains differing in restriction enzyme cleavage sites. J. Gen. Virol. 662659-2670. [DOI] [PubMed] [Google Scholar]
- 48.Umene, K. 1999. Mechanism and application of genetic recombination in herpesviruses. Rev. Med. Virol. 9171-182. [DOI] [PubMed] [Google Scholar]
- 49.Whetstone, C. A., and J. M. Miller. 1989. Two different strains of an alphaherpesvirus can establish latency in the same tissue of the host animal: evidence from bovine herpesvirus 1. Arch. Virol. 10727-34. [DOI] [PubMed] [Google Scholar]
- 50.White, R. L., and M. S. Fox. 1974. On the molecular basis of high negative interference. Proc. Natl. Acad. Sci. USA 711544-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young, C. S., and S. J. Silverstein. 1980. The kinetics of adenovirus recombination in homotypic and heterotypic genetic crosses. Virology 101503-515. [DOI] [PubMed] [Google Scholar]