Abstract
Both initial infection and cell-to-cell spread by herpes simplex virus type 1 (HSV-1) require the interaction of the viral glycoprotein D (gD) with an entry receptor on the cell surface. The two major HSV entry receptors, herpesvirus entry mediator (HVEM) and nectin-1, mediate infection independently but are coexpressed on a variety of cells. To determine if both receptors are active in these instances, we have established mutant viruses that are selectively impaired for recognition of one or the other receptor. In plaque assays, these viruses showed approximately 1,000-fold selectivity for the matched receptor over the mismatched receptor. Separate assays showed that each virus is impaired for both infection and spread through the mismatched receptor. We tested several human tumor cell lines for susceptibility to these viruses and observed that HT29 colon carcinoma cells are susceptible to infection by nectin-1-restricted virus but are highly resistant to HVEM-restricted virus infection, despite readily detectable HVEM expression on the cell surface. HVEM cDNA isolated from HT29 cells rendered HSV-resistant cells permissive for infection by the HVEM-restricted virus, suggesting that HT29 cells lack a cofactor for HVEM-mediated infection or express an HVEM-specific inhibitory factor. Passaging of HVEM-restricted virus on nectin-1-expressing cells yielded a set of gD missense mutations that each restored functional recognition of nectin-1. These mutations identify residues that likely play a role in shaping the nectin-1 binding site of gD. Our findings illustrate the utility of these receptor-restricted viruses in studying the early events in HSV infection.
Entry of herpes simplex virus type 1 (HSV-1) into cells requires the envelope glycoprotein D (gD), gB, gH, and gL (5, 13, 21, 27). After virion adsorption to cell surface proteoglycans, gD binds to one of its receptors, herpesvirus entry mediator (HVEM, or HveA), nectin-1 (HveC), or 3-O-sulfated heparan sulfate (3-O-S HS) (19, 32, 43). Receptor binding results in a conformational change in gD, which is believed to activate either or both gB and gH as mediators of fusion between the viral envelope and the plasma membrane or an endosomal membrane (7, 18, 20, 26, 31, 34, 35). Fusion leads to nucleocapsid release into the cytoplasm for subsequent viral genome delivery to the nucleus. Since productive HSV-1 infection is set in motion by the gD-receptor interaction, cellular susceptibility to HSV-1 is initially determined by the presence of one or more functional gD receptors on the cell surface.
In addition to supporting HSV-1 entry, gD receptors also support HSV-1 cell-to-cell spread (8, 41). However, it is not clear whether they act by the same mechanism in entry and spread. For example, gD mutants that allow gD-receptor-independent virus spread but not receptor-independent entry have been isolated (39). Likewise, gD is required for entry of the related alphaherpesvirus pseudorabies virus (PRV) but not for PRV lateral spread (2, 36, 37, 40). Two additional glycoproteins, gE and gI, are important for HSV-1 spread but are dispensable for entry (reviewed in reference 23). Further studies of the effects of gD mutations on HSV-1 spread will help distinguish the roles of this glycoprotein in virus infection and spread.
The individual contributions of HVEM and nectin-1 to infection in vitro and in vivo are difficult to assess since cells often express both receptors (24). Receptor-specific blocking antibodies have been used to separate the entry-mediating activities of the two receptors for primary cell cultures derived from human dorsal root ganglia (44). This study showed that infection of sensory neurons was blocked by antibodies to nectin-1, while virus entry into fibroblasts was not inhibited by antibodies to either nectin-1 or HVEM. Taylor et al. have described the use of knockout mice to distinguish the contributions of the two receptors to natural infection and to the pathogenesis of HSV-2 (48). In addition, virus mutants have been isolated that are severely impaired for infection through HVEM and 3-O-S HS but not nectin-1 (51). While some of these isolates have the additional ability to infect through another nectin family member, nectin-2, others appear to be genuinely selective for nectin-1 (51). To complement these tools and facilitate the rapid examination of cell lines for receptor usage in both entry and spread, we have sought to generate virus mutants that are deficient for the recognition of nectin-1 but not of HVEM. We refer to viruses and mutant gD molecules that are reactive with one but not the other major HSV-1 entry receptor as HVEM-restricted or nectin-1-restricted, regardless of their reactivities with the less prominent entry receptors 3-O-S HS and nectin-2. Receptor-restricted viruses offer the benefit that they can be tested in animal models in the absence of physiological defects inherent in knockout mice.
It is well documented that point mutations and deletions near the amino terminus of gD frequently abolish gD's ability to interact with HVEM without diminishing productive interaction with nectin-1 (11, 45, 52). Additional studies have identified various types of mutations throughout the gD ectodomain that eliminate gD's activity with both receptors, but mutations that specifically interfere with the nectin-1 interaction have been elusive. Using site-directed saturation mutagenesis and a transient complementation assay that measures the rescue of a gD-null virus by gD expression in trans, we previously identified the first HVEM-restricted gD mutant protein, gD:R222N/F223I (Q. Bai, W. A. Shah, J. B. Cohen, R. J. Eisenberg, G. H. Cohen, and J. C. Glorioso, J. C. Abstr. 2.10. 26th International Herpesvirus Workshop, Regensberg, Germany, 28 July to 3 August 2001). Further characterization by others has confirmed the nectin-1-specific defect of gD:R222N/F223I by using both transient complementation and cell-cell fusion assays and has provided evidence that this mutant protein is selectively impaired for binding to nectin-1 (29). A second HVEM-restricted gD mutant, gD:A3C/Y38C, was identified in a study designed to lock the HVEM-binding N-terminal domain of gD in the hairpin conformation it assumes upon HVEM binding (10). Surprisingly, this mutant protein showed cell fusion, complementation, and receptor-binding characteristics similar to those of gD:R222N/F223I. However, it is not known whether these mutant proteins restrict virus infection when expressed from the viral genome.
Here we describe the establishment and characterization of gD:R222N/F223I and gD:A3C/Y38C mutant viruses. We demonstrate that both viruses are substantially impaired for nectin-1-dependent infection and spread. We isolated phenotypic revertants of one of the viruses by passaging it through nectin-1-bearing cells and identified distal gD mutations that suppress the effects of the receptor-restricting mutations. We provide an example illustrating the potential of our vectors to expose additional requirements for HVEM-dependent HSV infection.
MATERIALS AND METHODS
Cells.
Baby hamster kidney J1.1-2 cells and murine melanoma B78H1 cells, kindly provided by Gabriella Campadelli-Fiume (University of Bologna, Italy) and Gary Cohen (University of Pennsylvania, PA), respectively, were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) (J1.1-2) or with 5% FBS (B78H1). African green monkey kidney Vero cells (ATCC CCL-81) and gD-complementing VD60 cells (a kind gift from David Johnson, Oregon Health and Science University, OR) were cultured in DMEM supplemented with 5% FBS. Human colon adenocarcinoma HT29 cells (ATCC HTB-38) and human pancreatic adenocarcinoma BxPC3 cells (ATCC CRL-1687) were cultured in DMEM supplemented with 10% FBS. J/A and B78/A cells were established by transfection of J1.1-2 and B78H1 cells with pBEC14 (32) (kindly provided by Patricia Spear, Northwestern University, IL), which expresses human HVEM from the human cytomegalovirus (HCMV) major immediate early (IE) promoter in pcDNA3. Likewise, B78/C cells were established by transfection of B78H1 cells with pBG38, a pcDNA3-based expression plasmid for human nectin-1 (also from Patricia Spear) (19). Transfections were carried out with Lipofectamine2000 (Invitrogen), and cells were selected with G418 (Sigma, St. Louis, MO) at 400 μg/ml (J1.1-2) or at 800 μg/ml (B78H1). J/C cells were established and cultured as described previously (16).
Plasmids.
Targeting plasmids for the construction of gD mutant viruses were derived from a previously described expression plasmid for wild-type gD, pgDSac, which contains the complete gD coding region and regulatory flanking sequences (49). The R222N/F223I mutant version of pgDSac, designated pgD:R222N/F223I, was also previously described (49). The A3C/Y38C derivative of pgDSac, termed pgD:A3C/Y38C, was constructed in several steps. A 122-bp BamHI-BstBI restriction fragment was generated by PCR using pgDSac as template and a pair of primers specifying the desired mutations (hu5, 5′-CCGGATCCCCATGGGGTCCGCGGCAAATATTGTTTGGCGGATGCCTCTC TCAAGATG-3′; and hu2, 5′-ACACTCTTCGAACCCCCGGAGGGTCGGTCAGCTG-3′). In parallel, a 168-bp BstBI-EcoRI fragment was generated by PCR using the same template and a pair of primers matching the wild-type gD sequence (hu3, 5′-CGGGGGTTCGAAGAGTGTGTCACATCCAGGCGGGCCTACCGG-3′; and hu4, 5′-CCGAATTCTCCGGACGTCTTCGGAGGCCCC-3′). The two fragments were inserted between the BamHI and EcoRI sites of cloning vector pSP72 (Promega, Madison, WI), resulting in pSP72-A3C/Y38C. The insert was isolated as a 278-bp NcoI-BspEI fragment and used to replace the corresponding fragment of pgDSac, yielding pgD:A3C/Y38C. Targeting plasmid pgD:d5-28V was generated by annealing two complementary oligonucleotides, yielding 5′-GATC overhangs (VBPgD1.2, 5′-GATCCGGGTCAGGTTCCGCGACTTGGTTACCTCCTCGTGGATCTA-3′, and VBPgD2.2, 5′-GATCTAGATCCACGAGGAGGTAACCAAGTCGCGGAACCTGACCCG-3′), and ligation into the unique BamHI site of plasmid pgDΔ5-28Bam, a derivative of pgDSac containing a short BamHI linker in place of codons 5 to 28 for mature gD (M. Tsvitov and J. C. Glorioso, unpublished results).
Mutant gD expression plasmids for transient complementation assays were constructed by replacement of the BspEI-BglII fragment of pgDSac with 1,410-bp BspEI-BamHI mutant fragments obtained by PCR with DNA from viral isolates selected for the loss of the receptor restriction of K26-gD:R222N/F223I (the D139N/R222N/F223I, R222K/F223I, R222N/F223N, and R222N “revertant” isolates). The gD sequence of the L25P/R222N/F223I revertant isolate was obtained as a PCR-derived 1,779-bp AflII-BamHI fragment and cloned between the AflII and BglII sites of pgDSac. The PCR primers flanking the gD coding sequence and providing a 3′ BamHI site were hu13, 5′-TCGGTACCCGGCCGTGTGACACTATCGTCC-3′, and hu14, 5′-ATGGATCCACGAATTCGAGCTCCAGGGTGGGATATGCG-3′.
The HVEM-V241I expression plasmid was derived from pBEC14 (32) by the replacement of an 821-bp NheI-BspEI fragment with a corresponding reverse transcription-PCR fragment generated from HT29 mRNA, using a primer pair straddling the HVEM coding region (hu37, 5′-CTGGAGTTCATCCTGCTAGCTGG-3′, and hu38, 5′-GCTGGGTGTGTGGTCTGTGAGC-3′).
All plasmid constructs were fully confirmed by DNA sequencing.
Recombinant viruses.
Parental virus K26GFP, kindly provided by Prashant Desai (Johns Hopkins University, MD), is an HSV-1 (KOS) recombinant virus carrying green fluorescent protein (GFP) in its capsid as a fusion with the viral VP26 capsid protein (12). Derivatives carrying specific mutations in the gD gene were generated by homologous recombination as detailed below.
K26GFP viral DNA was isolated from infected Vero cells by phenol-chloroform extraction. K26-gD:R222N/F223I was generated by cotransfection of J/A cells with K26GFP DNA and targeting plasmid pgD:R222N/F223I by using Lipofectamine 2000 reagent. Progeny viruses were separated by limiting dilution on J/A cells and screened for the failure to infect J/C cells. One isolate was purified by an additional three rounds of limiting dilution on J/A cells and sequenced through the entire gD open reading frame. K26-gD:d5-28V was then generated by cotransfection of J/C cells with K26-gD:R222N/F223I viral DNA and targeting plasmid pgD:d5-28V, limiting dilution of progeny viruses on J/C cells, and screening for the failure to infect J/A cells. Following additional rounds of limiting dilution, the anticipated sequence of the gD gene was confirmed for one clonal isolate. Last, K26-gD:A3C/Y38C was generated by cotransfecting J/A cells with K26-gD:d5-28V viral DNA and pgD:A3C/Y38C, screening of progeny viruses for the failure to infect J/C cells, and sequencing of the gD gene of a clonal isolate. Rescued viruses were established by cotransfection of J/C cells with pgDSac and K26-gD:R222N/F223I or K26-gD:A3C/Y38C viral DNA or by cotransfection of J/A cells with pgDSac and K26-gD:d5-28V viral DNA. Isolates were purified by three rounds of limiting dilution and sequenced through the entire gD gene.
Virus stocks were prepared by infection of six T-150 flasks containing confluent cells at a multiplicity of infection (MOI) of 0.05. K26-gD:d5-28V was grown on J/C cells, K26-gD:A3C/Y38C and K26-gD:R222N/F223I were grown on J/A cells, and K26GFP was grown separately on both J/A and J/C cells, all at 37°C; the three rescued viruses were grown on J/A cells. Sixteen hours later, media were refreshed, and cultures were shifted to 33°C. When 100% cytopathic effect was observed, the cells were incubated with 0.45 M NaCl at room temperature (RT) for 30 min. The culture media were collected, filtered through an 0.8-μm-pore filter, and centrifuged at 48,400 × g for 30 min at 4°C. Virus pellets were resuspended in phosphate-buffered saline (PBS) and incubated with 300 U/ml of Benzonase nuclease (Sigma) for 1 h at RT in the presence of 2 mM MgCl2. The virus particles were recentrifuged as described above, resuspended in PBS, and stored at −80°C with 10% glycerol. Biological titers, expressed in PFU per ml, were determined by standard procedures in triplicate. Genome titers, expressed as genome copies (gc) per ml, were established by real-time quantitative PCR (qPCR) for the IE gene ICP47 exactly as described previously (49).
The dual-marker gD-null virus KΔUs3-8Z41HG was derived from KΔUs3-8Z (1) by insertion of an HCMV IE promoter-enhanced GFP cassette into UL41 (unpublished results). KΔUs3-8Z contains lacZ controlled by the HCMV IE promoter in place of the unique short (Us) region genes 3 through 8. KΔUs3-8Z41HG was grown on gD-complementing VD60 cells (27).
CELISA.
Cellular enzyme-linked immunosorbent assay (CELISA) was performed as described by Walker et al. (50) with modifications. Cells were incubated in triplicate with a 1:40 dilution of CW10 anti-HVEM mouse monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or a 1:40 dilution of R1.302.12 anti-nectin-1 mouse monoclonal antibody (Santa Cruz Biotechnology) in 1% horse serum (Invitrogen) in PBS (HS-PBS) at RT for 30 min. The cells were washed with PBS, fixed with 2% paraformaldehyde (Electron Microscopy Science, Hatfield, PA) and 0.2% glutaraldehyde (Sigma) at RT for 30 min, washed with 10% HS-PBS, and incubated with a 1:500 dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) (Santa Cruz Biotechnology) in 1% HS-PBS at RT for 30 min. The cells were washed again with PBS, rinsed with 20 mM citrate buffer (pH 4.5), and incubated with TMB One solution (Promega) at RT for 10 min. The reaction was terminated with 1 N H2SO4, and absorbance at 405 nm was measured. For normalization, the value obtained by incubation with the secondary antibody alone was subtracted from each experimental value, and the difference was divided by the cell number determined with parallel cultures.
Immunofluorescence.
For evaluation of gD receptor expression, test cells were plated onto 35-mm glass-bottom culture dishes (MatTek, Ashland, MA), fixed with 4% paraformaldehyde at RT for 30 min, and permeabilized with 0.1% Triton X-100 (Packard, Downers Grove, IL) at RT for 10 min. The cells were sequentially incubated with (i) 10% HS-PBS at 37°C for 1 h, (ii) a 1:400 dilution of R140 anti-HVEM rabbit polyclonal antibody (a gift from Gary Cohen) or a 1:100 dilution of H-62 anti-nectin-1 rabbit polyclonal antibody (Santa Cruz Biotechnology) in 1% HS-PBS at RT for 1 h, and (iii) a 1:200 dilution of Cy3-conjugated sheep anti-rabbit IgG (Sigma) in 1% HS-PBS at RT for 1 h. The cell nuclei were stained by incubation with 1 μM Hoechst 33342 (Invitrogen) at RT for 10 min. Images were obtained with a FluoView 1000 confocal microscope (Olympus, Center Valley, PA) at a magnification of ×60.
Primary infection assay.
Cells were infected with 100 or 300 gc/cell at 37°C for 8 h and fixed in 100% methanol (VWR, West Chester, PA) at −20°C for 10 min. Immunofluorescence was performed as described above using a 1:400 dilution of 1-21 mouse monoclonal anti-VP16 primary antibody (Santa Cruz Biotechnology) and a 1:400 dilution of Cy3-conjugated sheep anti-mouse IgG (Sigma). Images were obtained with a Nikon Diaphot fluorescence microscope (Nikon, Melville, NY).
Infectious center assay.
The infectious center assay was performed as described by Roller and Rauch (41) with minor modifications. Donor cells (J/A or J/C) were infected at an MOI of 10 at 37°C for 2 h and treated with 0.1 M glycine (pH 3.0) at RT for 2 min. The cells were incubated at 37°C for 1 h, trypsinized, and suspended in culture medium. Equal numbers of donor cells were seeded onto monolayers of different test (acceptor) cells in a 48-well plate. The plate was incubated for 3 h at 37°C, and the cells were overlaid with medium containing 1% methylcellulose (Sigma). Two days later, the methylcellulose overlay was removed, and the cells were fixed with 100% methanol and immunostained with anti-VP16 antibody, as described above.
Confocal imaging of virus infection.
HT29 cells were plated onto 35-mm glass bottom culture dishes, incubated with the wild-type or the mutant viruses at 1,000 gc/cell for 45 min at 4°C, and then incubated for 2 h at 37°C. The cells were fixed with 4% paraformaldehyde at 4°C for 30 min and incubated with 5 μg/ml wheat germ agglutinin-Alexa Fluor 647 conjugate (Invitrogen) and 1 μM Hoechst 33342 at RT for 10 min. Confocal microscopy images were obtained at a magnification of ×60.
Selection of phenotypic revertants.
J/C cells were separately inoculated with two different stocks of K26-gD:R222N/F223I at 100 J/A-based PFU/cell. Sixteen hours postinfection (p.i.), the cells were incubated with 0.1 M glycine (pH 3.0) at RT for 2 min and cultured until infectious foci were visible. Medium was collected, and cell lysate was prepared by repeated sonication on ice water, followed by centrifugation to remove cellular debris. A portion of the combined medium and lysate was used directly for virus cloning by limiting dilution on J/C cells. A second portion was passaged once more through J/C cells prior to cloning. Selected clones were analyzed by sequencing of the entire gD open reading frame. All mutant gD sequences reported here were unambiguous, confirming the purity of the isolates and the absence of wild-type virus.
Transient complementation assay.
Transient complementation of gD-null virus was performed essentially as described previously (1). Briefly, monolayers of Vero cells were transfected with pgDSac or mutant derivatives by using Lipofectamine LTX reagent (Invitrogen). Twenty-four hours posttransfection, the monolayers were infected with KΔUs3-8Z41HG at an MOI of 5 at 37°C for 2 h and treated with 0.1 M glycine (pH 3.0) at RT for 2 min to inactivate extracellular virus. Twenty-four hours p.i., cell lysates were prepared by repeated sonication on ice water, followed by centrifugation to remove cellular debris. Equal lysate volumes were incubated with test cells at 37°C for 16 h. The cells were fixed with 0.5% glutaraldehyde and stained with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal; Sigma).
RESULTS
Establishment of cell lines expressing HVEM or nectin-1.
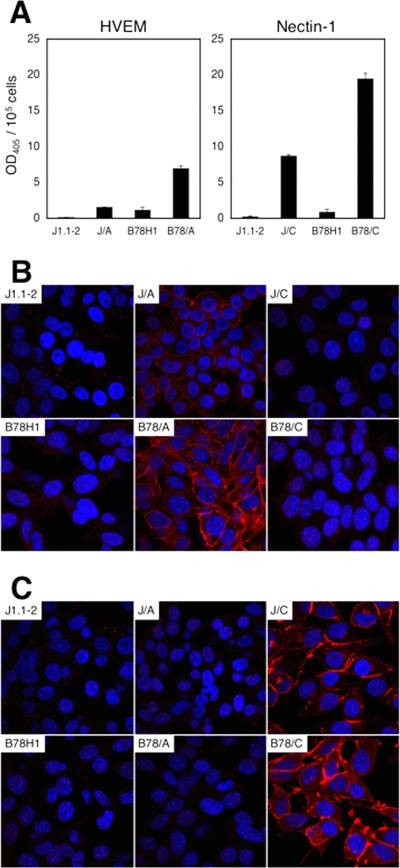
Baby hamster kidney J1.1-2 and murine melanoma B78H1 cells are resistant to HSV-1 infection due to the absence of gD receptors (9, 25, 30). To facilitate the selection, growth, and characterization of gD-mutant viruses that may preferentially infect through either HVEM or nectin-1, clonal lines expressing one or the other receptor were derived from J1.1-2 and B78H1 cells by stable transfection. One representative line of each kind, designated J/A or B78/A for HVEM (HveA)-expressing cells and J/C or B78/C for nectin-1 (HveC)-expressing cells, was used in the remainder of this study. Quantification of the antibody binding to cells (CELISA) (50) showed that the two B78 lines expressed more receptor than their J counterparts (Fig. 1A). Immunofluorescence analysis demonstrated that HVEM was distributed evenly along the cell surface (Fig. 1B), whereas nectin-1 was particularly abundant at cell-cell contact sites (Fig. 1C), consistent with published observations (17, 47).
FIG. 1.
Expression of HVEM and nectin-1 on J1.1-2- and B78H1-derived cell lines. (A) Cells were reacted with anti-HVEM (left panel) or anti-nectin-1 (right panel) mouse monoclonal antibodies, fixed, and incubated with HRP-conjugated secondary antibody. Substrate was added, and HRP activity was measured as absorbance at 405 nm. Background reading values for secondary antibody alone were subtracted from the experimental values and divided by the cell number for normalization. The mean values ± standard deviations per 105 cells from three determinations are plotted. (B and C) Cells were reacted with anti-HVEM (B) or anti-nectin-1 (C) rabbit polyclonal antibodies and stained with Cy3-conjugated secondary antibody. Nuclei were stained with Hoechst 33342. The cells were observed with a FluoView 1000 confocal microscope.
Construction of receptor-restricted gD-mutant viruses.
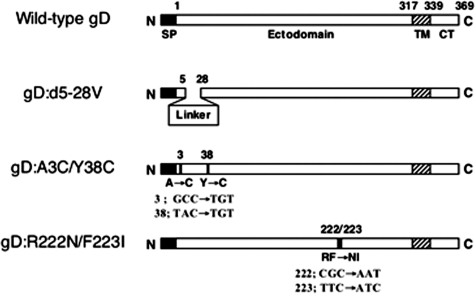
Targeting plasmids containing the gene for HVEM-selective (R222N/F223I or A3C/Y38C) or nectin-1-selective (d5-28V) gD were used to establish the virus mutants studied in this work; the gD:d5-28V gene contains a 20-codon linker sequence that replaces gD codons 5 to 28 (Fig. 2) and failed to enable the entry of a gD-deficient virus into J/A cells, but not J/C cells, in a transient complementation assay (data not shown). Following cotransfection of the targeting plasmids with viral DNA, recombinant viruses were identified by screening or selection on cell lines expressing one or the other receptor. Virus mutant K26-gD:R222N/F223I was derived directly from K26GFP, an HSV-1 KOS recombinant virus expressing VP26 as a fusion with GFP (12), and was identified by screening for infection of J/A but not of J/C cells. K26-gD:d5-28V was then derived from K26-gD:R222N/F223I and selected on J/C cells. Finally, K26-gD:A3C/Y38C was derived from K26-gD:d5-28V and selected on J/A cells. In addition, we rescued each of these mutant viruses by homologous recombination with the wild-type gD gene; these rescued (R) virus constructs are referred to as R-gD:R222N/F223I, R-gD:d5-28V, and R-gD:A3C/Y38C, respectively. All recombinant viruses were purified by at least three rounds of limiting dilution and confirmed by DNA sequencing through the gD gene. Stocks of the recombinant viruses were prepared on J/A or J/C cells, and for comparison, stocks of K26GFP were prepared separately on both cell lines.
FIG. 2.
Schematic representation of receptor-restricting gD mutations. In gD:d5-28V, gD amino acids Ala-5 to Leu-28 were replaced with a linker sequence of 20 residues. In gD:A3C/Y38C, cysteines were substituted for Ala-3 and Tyr-38 (codon changes are shown underneath), and in gD:R222N/F223I, Arg-222 and Phe-223 were exchanged for asparagine and isoleucine, respectively. SP, signal peptide; TM, transmembrane domain; CT, cytoplasmic tail.
Tropism of the gD mutant viruses.
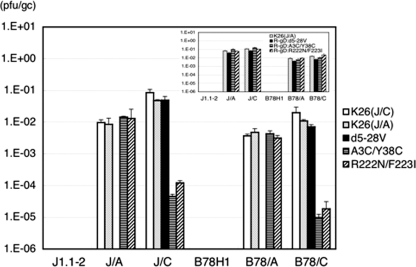
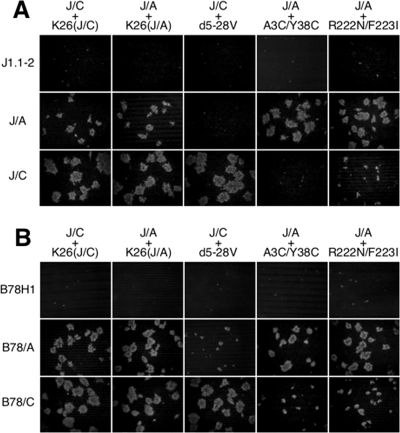
We determined the titers of the different viral preparations on J1.1-2 and B78H1 cells and their receptor-bearing derivatives by using a standard plaque formation assay and normalized the results, expressed as PFU/ml, to genome titers (gc per ml) as determined by real-time qPCR for the viral ICP47 gene (22). As shown in Fig. 3, the PFU/gc values (specific infectious activities) for all of the stocks on receptor-negative J1.1-2 and B78H1 cells were below 10−6. The values for the two K26GFP stocks prepared separately on J/C and J/A cells [K26(J/C) and K26(J/A), respectively] were similar for all of the receptor-bearing cell lines, indicating that the growth of this wild-type gD virus in the presence of one or the other receptor did not select for enhanced activity toward that receptor. For all of the viruses, the PFU/gc values on receptor-transduced J cells were higher than those on the corresponding B78-derived cells (1.8-fold to 7.1-fold). This may reflect differences in virus replication or in later events occurring on the two cell types, as it is opposite to the differences in receptor abundance noted between the J- and B78-derived cell lines (Fig. 1) and as similar differences were not consistently observed for the single-cell infection assays presented below (Fig. 4). The PFU/gc values for K26-gD:d5-28V on J/A and B78/A cells were below 10−6, at least 3 to 4 orders of magnitude lower than the values for the wild-type K26GFP stocks on these cells, while on J/C and B78/C cells, the PFU/gc values for K26-gD:d5-28V were similar to those of the K26GFP stocks. Thus, nectin-1 appeared to be an effective receptor for this mutant gD virus, whereas HVEM was not. Conversely, the PFU/gc values for K26-gD:A3C/Y38C and K26-gD:R222N/F223I on J/A and B78/A cells were similar to those for the K26GFP stocks on these cells, but on J/C and B78/C cells, the values for the two mutant viruses were some 2 to 3 orders of magnitude lower than those of K26GFP; compared to the PFU/gc values for K26(J/C) virus, the values for the K26-gD:A3C/Y38C mutant virus on J/C and B78/C cells were 1,842-fold and 2,054-fold reduced, respectively, and 690-fold and 1,089-fold reduced for the K26-gD:R222N/F223I mutant virus. These results demonstrated that our mutant viruses are highly selective in their plaque-forming ability for either nectin-1- or HVEM-bearing cells. Since the PFU/gc values for the R-gD:d5-28V, R-gD:R222N/F223I, and R-gD:A3C/Y38C viruses were similar to those of the K26(J/A) stock (Fig. 3, inset), it can be concluded that the receptor selectivity of the mutant viruses was due exclusively to the mutations in the gD genes of these viruses.
FIG. 3.
Specific infectious activities (PFU/gc) of the gD mutant viruses on different cell lines. Biological titers (PFU/ml) were divided by genome titers (gc/ml), and the mean values ± standard deviations from three determinations were plotted on a logarithmic scale. K26(J/C), K26GFP virus propagated on J/C cells; K26(J/A), K26GFP virus propagated on J/A cells. Inset, PFU/gc values for the three rescued viruses are compared to those of K26(J/A).
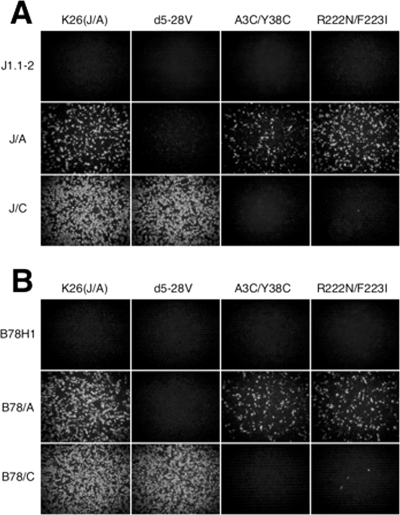
FIG. 4.
(A and B) Primary infection by the gD mutant viruses. Cells were infected at 100 gc/cell for 8 h and immunostained with anti-VP16 mouse monoclonal antibody and Cy3-conjugated secondary antibody. Images were obtained with a Nikon Diaphot fluorescence microscope.
Infection profiles of the receptor-restricted gD mutant viruses.
To determine whether the tropisms of the nectin-1- and HVEM-restricted viruses reflected selective receptor use during the initial round of infection prior to lateral spread, cells were infected with equal numbers of virions, based on genome titers (100 gc/cell), and single-cell infection was visualized by VP16 immunostaining at 8 h p.i. As shown in Fig. 4, K26-gD:d5-28V infected J/C and B78/C cells as efficiently as the wild-type gD virus [K26(J/A)] but failed to infect J/A or B78/A cells. Conversely, the gD:A3C/Y38C and gD:R222N/F223I viruses infected J/A and B78/A cells at efficiencies that were similar or somewhat reduced compared to the wild-type virus, but neither virus showed significant infection of J/C or B78/C cells. These profiles were consistent with the specific infectious activities (PFU/gc) of the three mutant viruses shown in Fig. 3, indicating that changes during the first round of infection, most likely at the entry stage, were at least in part responsible for the observed tropism alterations.
Spread profiles of the receptor-restricted mutant viruses.
Infectious center assays were carried out to determine if the gD mutant viruses were also altered in their lateral spread properties. J/C cells were infected with the J/C-grown wild-type gD virus or the nectin-1-restricted gD:d5-28V mutant virus, and J/A cells were infected with the J/A-grown wild-type gD virus or the HVEM-restricted gD:A3C/Y38C or gD:R222N/F223I mutant virus, each at 10 PFU/cell to achieve 100% infection. After residual extracellular virions were inactivated by acidic wash, equal numbers of infected (donor) cells were seeded onto monolayers of uninfected acceptor cells. The cultures were overlaid with 1% methylcellulose, and plaque formation was assessed 2 days later by VP16 immunostaining. As shown in Fig. 5, plaques formed by the wild-type viruses on each acceptor cell line were similar in number and size, regardless of the type of donor cells used (J/C or J/A), suggesting that the differences in spread described below were not caused by differences in the donor cell types. Plaque formation by K26-gD:d5-28V on J/C cells was similar to that of the two wild-type K26GFP stocks, but no plaques were observed for this mutant virus on J/A cells (Fig. 5A). Conversely, K26-gD:A3C/Y38C formed slightly larger plaques on J/A cells than the wild-type gD viruses, but none formed on J/C cells (Fig. 5A). Likewise, the spread of K26-gD:R222N/F223I on J/A cells was comparable to that of the wild-type viruses but was substantially impaired on J/C cells (Fig. 5A). Similar trends were observed for the B78 pair of cell lines, but in this case, small foci were observed for the nectin-1-restricted virus on B78/A cells and for both of the HVEM-restricted viruses on B78/C cells (Fig. 5B); this may reflect the higher receptor levels observed for the two B78 cell lines than for the two J lines (Fig. 1). Consistent with the results for J/C cells, the K26-gD:R222N/F223I foci on B78/C cells were somewhat larger and more numerous (approximately 2-fold) than the K26-gD:A3C/Y38C foci on these cells (Fig. 5B). This difference in spread properties between the two HVEM-restricted viruses was reminiscent of the difference in specific infectious activities between these viruses on both J/C and B78/C cells observed above (Fig. 3) and may be indicative of greater residual interaction of nectin-1 with gD:R222N/F223I than with gD:A3C/Y38C. Together, these results showed that our receptor-restricted gD mutant viruses are impaired not only for initial infection but also for spread on receptor-mismatched cells.
FIG. 5.
(A and B) Spread of the gD mutant viruses. J/C cells were infected with K26(J/C) or with K26-gD:d5-28V at an MOI of 10 based on the biological titer of each virus on J/C cells. Likewise, J/A cells were infected with K26(J/A), K26-gD:A3C/Y38C, or K26-gD:R222N/F223I, each at an MOI of 10 calculated from the biological titers of these viruses on J/A cells. After 2 h, extracellular virus was inactivated by acidic wash, and the cells were cultured for 1 h and trypsinized. Equal numbers of these infected (donor) cells (indicated above the panels) were added onto monolayers of the various cells indicated on the left (acceptor cells). The mixed cultures were incubated at 37°C for 3 h and overlaid with medium containing 1% methylcellulose. The cells were fixed 2 days later and immunostained with anti-VP16 primary antibody and Cy3-conjugated secondary antibody. Images were obtained with a Nikon Diaphot fluorescence microscope.
Functionality of gD receptors on human tumor lines.
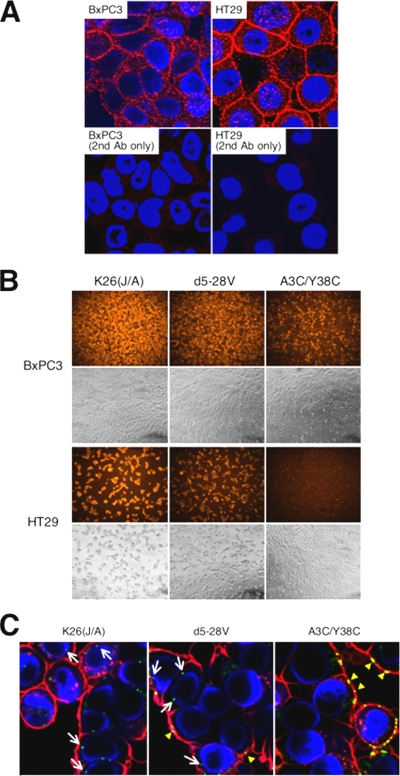
We tested a small panel of human tumor cell lines to evaluate the utility of our receptor-restricted viruses in assessing the use of naturally expressed receptors. Indirect immunofluorescence showed abundant peripheral HVEM expression on two of the cell lines, pancreatic adenocarcinoma BxPC3 and colon adenocarcinoma HT29 cells (Fig. 6A), and both of these lines also expressed nectin-1 (data not shown). We exposed both lines to wild-type gD virus [K26(J/A)] or to receptor-restricted mutant viruses at 300 gc/cell and examined infection 8 h later by VP16 staining. Remarkably, while K26(J/A) and nectin-1-restricted K26-gD:d5-28V infected the majority of cells of both lines under these conditions, HVEM-restricted K26-gD:A3C/Y38C infected only BxPC3 cells (Fig. 6B, first and third rows). Confirming the resistance of HT29 cells to K26-gD:A3C/Y38C infection, bright-field imaging showed a pronounced cytopathic effect (cell rounding and detachment) of K26(J/A) and K26-gD:d5-28V infection on these cells but no disruption of the monolayer by K26-gD:A3C/Y38C (Fig. 6B, fourth row). We used confocal microscopy to examine the location of virus particles at 2 h p.i. As shown in Fig. 6C, the A3C/Y38C virus remained at the cell periphery, whereas a sizable fraction of the K26(J/A) and K26-gD:d5-28V particles were found inside the cells near the nucleus. Thus, infection of HT29 cells by K26-gD:A3C/Y38C appeared to be blocked at the entry stage.
FIG. 6.
Receptor functionality on human tumor lines. (A) Cells were reacted with anti-HVEM rabbit polyclonal antibody and stained with Cy3-conjugated secondary antibody. Nuclei were stained with Hoechst 33342, and images were obtained with a FluoView 1000 confocal microscope. Cells reacted with secondary antibody alone (2nd Ab only) are shown as controls. (B) Cells were infected at 300 gc/cell for 8 h and immunostained with anti-VP16 primary antibody and Cy3-conjugated secondary antibody. Images were obtained with a Nikon Diaphot fluorescence microscope. Phase-contrast images of the same fields are shown in the lower panels. (C) HT29 cells were incubated with the indicated viruses at 1,000 gc/cell for 45 min at 4°C, followed by 2 h at 37°C, and were then fixed with 4% paraformaldehyde. Cell membranes and nuclei were stained with wheat germ agglutinin-Alexa Fluor 647 conjugate (red) and Hoechst 33342 (blue), respectively. Confocal images were obtained as described in the legend to panel A. Arrows point to internalized capsids; arrowheads indicate capsids remaining at the cell membrane (yellow).
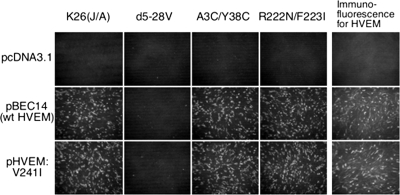
To determine whether the entry block could be due to mutations in HVEM, we sequenced PCR-amplified HVEM cDNA from HT29 cells. The predicted amino acid sequence differed at a single position from the published sequence of human HVEM (32), a valine-to-isoleucine substitution in the cytoplasmic domain at position 241 (V241I). The sequencing output was homogeneous without evidence of a V241 allele. Although the V241I substitution has been previously reported, along with evidence that it is a neutral polymorphism with respect to the HSV entry-mediating activity of HVEM (14, 46), we considered the possibility that it could selectively interfere with infection by gD mutant viruses. We tested this possibility by transfection of gD receptor-negative B78H1 cells with structurally identical HVEM-V241 and HVEM-V241I expression plasmids and infection 48 h later with gD wild-type or receptor-restricted mutant viruses at 100 gc/cell. HVEM expression at the time of infection was confirmed by anti-HVEM immunofluorescence on parallel cultures (Fig. 7). Infection was visualized by anti-VP16 staining at 8 h p.i. The results showed that the two HVEM plasmids yielded indistinguishable patterns of B78H1 infection by the panel of viruses (Fig. 7, first through fourth columns). In particular, both of the HVEM-restricted viruses showed infection of both sets of HVEM-transfected cells that was as abundant as that of the wild-type gD virus. These results demonstrated that HVEM-V241I has the intrinsic ability to function as an entry receptor for our HVEM-restricted mutant viruses. Thus, the resistance of HT29 cells to the A3C/Y38C virus suggests that these cells are deficient for an accessory function that is required for HVEM-mediated infection or that they express an inhibitory function that specifically blocks infection through HVEM.
FIG. 7.
Functionality of HT29-derived HVEM-V241I. B78H1 cells were transfected with pBEC14 expressing wild-type HVEM (32), a derivative expressing HVEM-V241I, or with pcDNA3.1. Two days later, the cells were infected with the indicated viruses at 100 gc/cell for 8 h and immunostained for VP16. A parallel set of transfected cells was immunostained for HVEM. Images were obtained with a Nikon Diaphot fluorescence microscope.
Phenotypic reversion of K26-gD:R222N/F223I.
We used K26-gD:R222N/F223I to select escape mutants on J/C cells as a strategy to identify gD residues that may be important for the integrity of the nectin-1 binding pocket. We passaged two separate stocks of K26-gD:R222N/F223I at high J/A-based MOI once or twice on J/C cells and characterized a number of progeny viruses that were able to grow and form plaques on these cells. Direct sequencing of the entire gD open reading frames of 17 cloned isolates derived from the first stock and 33 derived from the second revealed that each isolate carried one of seven different missense mutations compared to the parental gD:R222N/F223I allele (Table 1). Five of these missense mutations were found in multiple isolates and three were represented among derivatives of both parental virus stocks. All of the sequenced gD genes retained at least one of the two parental mutant codons. All of the sequences were homogeneous, arguing against significant contamination with wild-type or parent virus. The seven different mutations could be categorized as a simple reversion (I223F), as forward mutations (N222K, I223N), or as second-site mutations (all others). The simple reversion restored the wild-type amino acid at position 223, the forward mutations altered one or the other mutant amino acid of the parent virus to a different mutant amino acid, and the second-site mutations added a substitution elsewhere in the gD sequence to the parental R222N/F223I substitutions. At the nucleotide level, all of the second-site mutations were transitions, while the simple-reversion and forward mutations were transversions, suggesting a possible positional bias in the mechanisms that created these mutations.
TABLE 1.
gD mutations after passage of K26-gD:R222N/F223I virus on J/C cellsa
| Virus stock | Amino acid (nucleotide) mutationb | No. of isolates/total no. of isolates analyzed per group | Allelec |
|---|---|---|---|
| 1 | D139N (GAC→AAC) | 8/17 | D139N/R222N/F223I |
| I223F (ATC→TTC) | 7/17 | R222N | |
| I223N (ATC→AAC) | 2/17 | R222N/F223N | |
| 2 | L25P (CTG→CCG) | 11/33 | L25P/R222N/F223I |
| N222K (AAT→AAA) | 5/33 | R222K/F223I | |
| D139N (GAC→AAC) | 4/33 | D139N/R222N/F223I | |
| I223N (ATC→AAC) | 2/33 | R222N/F223N | |
| I223F (ATC→TTC) | 1/33 | R222N | |
| A5V (GCG→GTG) | 1/33 | A5V/R222N/F223I | |
| A98V (GCT→GTT) | 1/33 | A98V/R222N/F223I |
Two separate stocks of K26-gD:R222N/F223I virus were passaged once (stock no. 2) or twice (stock no. 1 and 2) through J/C cells, and progeny viruses were cloned by limiting dilution on J/C cells.
Amino acid and nucleotide changes are shown for individual isolates. All sequences retained the parental R222N and F223I mutations unless indicated otherwise.
Allele designations identify all amino acid substitutions in the encoded protein compared to wild-type gD.
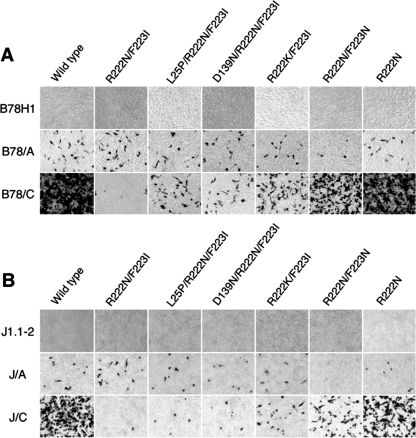
We used a transient complementation assay for functional characterization of several of the mutant gD alleles identified above. This assay examines the ability of mutant gD proteins expressed from transfected plasmids to render a gD-null virus introduced into the same cells competent for a single round of infection of test cells. We cloned the five most common gD alleles in plasmid vectors behind the gD promoter, confirmed the sequence of each, and transfected the recombinant plasmids into Vero cells. One day later, the cells were infected at an MOI of 5 with a gD-null virus, KΔUs3-8Z41HG, which had been grown on gD-complementing VD60 cells (27) to enable entry into Vero cells. Extracellular virus was inactivated after 2 h by acidic wash, virus was harvested the next day, and equal volumes were used to infect B78H1, B78/A, and B78/C cells. At 16 h p.i., the cells were stained for lacZ gene expression from the viral backbone; lacZ in this recombinant virus is controlled by the HCMV IE promoter. Representative results are shown in Fig. 8A. As expected, no infection was seen for any of the viral preparations on receptor-negative B78H1 cells (Fig. 8A, top row). The level of KΔUs3-8Z41HG complementation by transfected wild-type gD virus (Fig. 8A, first column) demonstrates that B78/C cells were infected more efficiently than B78/A cells under these experimental conditions. KΔUs3-8Z41HG harvested from cells transfected with the parent gD:R222N/F223I gene showed a dramatic reduction in the ratio of infected B78/C to B78/A cells (Fig. 8A, second column) compared to virus from cells that had been transfected with the wild-type gD gene, thus validating the assay. Each of the five R222N/F223I-derived mutant alleles (Fig. 8A, third through seventh columns) yielded increased infection of B78/C cells compared to that of the parental allele but only minor changes on B78/A cells. Similar results were obtained in an independent complementation experiment on J1.1-2, J/A, and J/C cells (Fig. 8B). Although we have seen some variation in the amounts of gD produced by the complementing plasmids (data not shown) and recognize the possibility that the new mutations may not be neutral with respect to entry via HVEM, these results were most consistent with various degrees of suppression of the nectin-1-specific defect of gD:R222N/F223I.
FIG. 8.
(A and B) Transient complementation analysis of gD genes isolated from K26-gD:R222N/F223I revertant viruses. Vero cells were transfected with expression plasmids for the gD genes indicated above the panels and infected 24 h later with KΔUs3-8Z41HG (MOI, 5) for 2 h, and extracellular virus was inactivated by acidic wash. Virus was collected at 24 h p.i., and equal volumes were used for infection of the cell lines listed at the left. Cells were fixed 16 h later and stained for lacZ expression. Images of representative fields are shown. Panels A and B show results from independent experiments.
DISCUSSION
The A3C/Y38C and R222N/F223I gD mutations used in this study were previously shown to selectively diminish the binding of purified soluble gD ectodomain to nectin-1, fusion of cells transduced for the four essential glycoproteins with nectin-1-expressing cells, and transient complementation of gD-deficient viruses for entry via nectin-1 (10, 29). We took advantage of these mutations to create HVEM-restricted viruses to facilitate the screening of cells and tissues for HVEM function in entry and spread. While we have not examined the ability of these viruses to use 3-O-S HS as an entry receptor, it has been reported that the R222N/F223I double mutation does not impair the reactivity of HSV-1 gD with 3-O-S HS (29).
We tested a series of human tumor cell lines to correlate HVEM expression with susceptibility to HVEM-restricted virus infection. We chose K26-gD:A3C/Y38C for this analysis because its selectivity in our assays on receptor-transduced J and B78 cells was tighter than that of K26-gD:R222N/F223I. We found that HT29 and BxPC3 cells expressed abundant HVEM on their surface, yet only BxPC3 cells were readily infected by K26-gD:A3C/Y38C; abundant HVEM expression on HT29 cells has previously been reported by others using flow cytometry and a different antibody than the one used in our imaging studies (53). Using confocal microscopy, we observed that infection of HT29 cells by the HVEM-restricted virus was blocked at the entry stage, whereas BxPC3 cells were permissive for entry by the same virus. These findings are reminiscent of previous observations that HVEM detected on human dorsal root ganglion neurons, human IMR-5 neuroblastoma cells, and human A431 epidermoid carcinoma cells is not functional as a mediator of HSV-1 infection (29, 44). However, the integrity of the expressed HVEM was not examined in those studies. Our analysis of HT29 HVEM cDNAs identified a single nucleotide difference from the published human HVEM sequence (32), resulting in an amino acid change (V241I) in the cytoplasmic domain. This substitution was previously identified in HVEM cDNAs isolated from chronic lymphocytic leukemic B cells (14) and two HSV-seronegative individuals (46). Struyf and coworkers demonstrated that V241I does not alter the HSV entry-receptor activity of HVEM (46), and our own results using similar procedures extend this conclusion to the two HVEM-restricted viruses reported here. These data strongly suggest that the resistance of HT29 cells to K26-gD:A3C/Y38C is not due to an intrinsic defect in HT29 HVEM or to an inability of this receptor to interact with gD:A3C/Y38C. Bender et al. have described heparin-resistant gB binding to a variety of cells and presented evidence that this interaction is required for HSV infection (3). Satoh and coworkers (42) recently identified the immune receptor PILRα (alternatively referred to as FDF03 [15]) as a gB receptor and showed that this receptor is necessary for HVEM-dependent infection of peripheral blood mononuclear cells. It has been reported that HT29 cells do not express FDF03 mRNA (15), raising the possibility that this deficiency is responsible for the resistance of HT29 cells to HVEM-mediated infection. The observation that HT29 cells were readily infected with both nectin-1-restricted HSV and wild-type gD virus would then suggest that HVEM and nectin-1 use different gB receptors to mediate HSV entry. Alternatively, it is possible that HT29 cells express an as-yet-unknown gB receptor that can support infection through both HVEM and nectin-1 but either lack a different activity specifically required for HVEM function in entry or express an HVEM-specific inhibitory activity. We are currently exploring these scenarios.
We selected K26-gD:R222N/F223I for spontaneous variants that could grow on J/C cells in an effort to identify gD residues that may play a role in shaping the nectin-1 binding site. This strategy yielded seven different missense mutations, including three at the mutant positions of the parent gene and four at a distance. Characterization of five of these by transient complementation provided evidence that each mutation suppresses the nectin-1-specific defect of the parental mutant gene. The isolation of rescue mutations at both positions 222 and 223 indicates that these positions act in concert; we did not observe reversion of position 222, which would have required more than a single base change. The two characterized second-site mutations were in regions that affect receptor use. D139N is adjacent to the position of a previously described substitution, S140N, and L25P is within the amino-terminal HVEM-binding region. Both S140N and L25P were initially identified in virus mutants selected for resistance to blocking conditions, either gD expression by the host cells (4, 6) or incubation with neutralizing anti-gD antibodies (33). Both mutations reportedly enable gD receptor-independent cell-to-cell spread (39). L25P, in addition, reduces HVEM-mediated virus infection while enabling the use of nectin-2 as an entry receptor (28, 52). Importantly, the crystal structure of gD indicates that L25 and F223 are in close mutual proximity, contacting the same two residues in the carboxy-terminal region of gD that is displaced by receptor binding (26). Moreover, S140 is positioned on the opposite side near R222 (7, 26). These observations suggest that D139N and L25P may directly compensate for the disruption caused by R222N/F223I of the integrity of a structural element that plays an important role in the nectin-1-mediated liberation of the profusion domain of gD triggering virus entry.
In conclusion, the establishment and characterization of two HVEM-restricted HSV-1 mutant viruses provide new tools with which to examine the functionality of HVEM as an entry receptor on cultured cells and in animals. The ready selection of derivatives that have regained the ability to enter cells through nectin-1 indicates that these viruses are less suitable to determine the contribution of HVEM to virus spread, at least in vivo where spread is difficult to quantify. This limitation may be addressed by new designs based on recent reports clarifying the mechanism of receptor-mediated gD activation (18, 26, 54). Our strategy of mapping the environment of the nectin-1 binding site by the selection of phenotypic revertants can be extended to the A3C/Y38C virus and R222N/F223I second-generation vectors that have been stabilized against the reacquisition of the substitutions identified here. Our study of HVEM function on HT29 cells indicates that HVEM activity in HSV infection is sensitive to one or more positive or negative factors that do not control nectin-1 activity. Together, this study provides new directions for elucidating different receptor-related aspects of HSV-1 entry.
Acknowledgments
We thank Gary Cohen, Patricia Spear, Gabriella Campadelli-Fiume, and David Johnson for antibodies, cell lines, and gene constructs and Prashant Desai for K26GFP. We thank David Clawson, Janet Chan, and Zhanna Hakhverdyan for technical assistance and Marianna Tsvitov for helpful discussions. We also thank Simon Watkins for instruction and the use of his imaging facility.
This work was supported by NIH grants CA119298, NS40923, DK044935, AR050733, and NS44323 to J.C.G.
Footnotes
Published ahead of print on 7 January 2009.
REFERENCES
- 1.Anderson, D. B., S. Laquerre, K. Ghosh, H. P. Ghosh, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2000. Pseudotyping of glycoprotein D-deficient herpes simplex virus type 1 with vesicular stomatitis virus glycoprotein G enables mutant virus attachment and entry. J. Virol. 742481-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babic, N., T. C. Mettenleiter, A. Flamand, and G. Ugolini. 1993. Role of essential glycoproteins gII and gp50 in transneuronal transfer of pseudorabies virus from the hypoglossal nerves of mice. J. Virol. 674421-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, F. C., J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2005. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J. Virol. 7911588-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandimarti, R., T. Huang, B. Roizman, and G. Campadelli-Fiume. 1994. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc. Natl. Acad. Sci. USA 915406-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 622596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume, G., S. Qi, E. Avitabile, L. Foa-Tomasi, R. Brandimarti, and B. Roizman. 1990. Glycoprotein D of herpes simplex virus encodes a domain which precludes penetration of cells expressing the glycoprotein by superinfecting herpes simplex virus. J. Virol. 646070-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8169-179. [DOI] [PubMed] [Google Scholar]
- 8.Cocchi, F., L. Menotti, P. Dubreuil, M. Lopez, and G. Campadelli-Fiume. 2000. Cell-to-cell spread of wild-type herpes simplex virus type 1, but not of syncytial strains, is mediated by the immunoglobulin-like receptors that mediate virion entry, nectin1 (PRR1/HveC/HIgR) and nectin2 (PRR2/HveB). J. Virol. 743909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 729992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connolly, S. A., D. J. Landsburg, A. Carfi, J. C. Whitbeck, Y. Zuo, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2005. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J. Virol. 791282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J. Virol. 778127-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 727563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai, P. J., P. A. Schaffer, and A. C. Minson. 1988. Excretion of non-infectious virus particles lacking glycoprotein H by a temperature-sensitive mutant of herpes simplex virus type 1: evidence that gH is essential for virion infectivity. J. Gen. Virol. 691147-1156. [DOI] [PubMed] [Google Scholar]
- 14.Eling, D. J., P. A. Johnson, S. Sharma, F. Tufaro, and T. J. Kipps. 2000. Chronic lymphocytic leukemia B cells are highly sensitive to infection by herpes simplex virus-1 via herpesvirus-entry-mediator A. Gene Ther. 71210-1216. [DOI] [PubMed] [Google Scholar]
- 15.Fournier, N., L. Chalus, I. Durand, E. Garcia, J. J. Pin, T. Churakova, S. Patel, C. Zlot, D. Gorman, S. Zurawski, J. Abrams, E. E. Bates, and P. Garrone. 2000. FDF03, a novel inhibitory receptor of the immunoglobulin superfamily, is expressed by human dendritic and myeloid cells. J. Immunol. 1651197-1209. [DOI] [PubMed] [Google Scholar]
- 16.Frampton, A. R., Jr., D. B. Stolz, H. Uchida, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2007. Equine herpesvirus 1 enters cells by two different pathways, and infection requires the activation of the cellular kinase ROCK1. J. Virol. 8110879-10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuhara, A., K. Irie, A. Yamada, T. Katata, T. Honda, K. Shimizu, H. Nakanishi, and Y. Takai. 2002. Role of nectin in organization of tight junctions in epithelial cells. Genes Cells 71059-1072. [DOI] [PubMed] [Google Scholar]
- 18.Fusco, D., C. Forghieri, and G. Campadelli-Fiume. 2005. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc. Natl. Acad. Sci. USA 1029323-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 2801618-1620. [DOI] [PubMed] [Google Scholar]
- 20.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313217-220. [DOI] [PubMed] [Google Scholar]
- 21.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 662240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, C., M. Ataai, A. Ozuer, D. Krisky, J. Wechuck, S. Pornsuwan, F. Pourarian, and J. C. Glorioso. 2006. Inactivation of herpes simplex type 1 gene vector on immobilized metal affinity chromatography: oxidative damage by hydroxyl free radicals and its prevention. Biotechnol. Bioeng. 9548-57. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krummenacher, C., F. Baribaud, M. Ponce de Leon, I. Baribaud, J. C. Whitbeck, R. Xu, G. H. Cohen, and R. J. Eisenberg. 2004. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology 322286-299. [DOI] [PubMed] [Google Scholar]
- 25.Krummenacher, C., I. Baribaud, J. F. Sanzo, G. H. Cohen, and R. J. Eisenberg. 2002. Effects of herpes simplex virus on structure and function of nectin-1/HveC. J. Virol. 762424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 244144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 621486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez, M., F. Cocchi, L. Menotti, E. Avitabile, P. Dubreuil, and G. Campadelli-Fiume. 2000. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 741267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manoj, S., C. R. Jogger, D. Myscofski, M. Yoon, and P. G. Spear. 2004. Mutations in herpes simplex virus glycoprotein D that prevent cell entry via nectins and alter cell tropism. Proc. Natl. Acad. Sci. USA 10112414-12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, C. G., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and N. W. Fraser. 2001. Development of a syngeneic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol. Ther. 3160-168. [DOI] [PubMed] [Google Scholar]
- 31.Milne, R. S., A. V. Nicola, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 796655-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87427-436. [DOI] [PubMed] [Google Scholar]
- 33.Muggeridge, M. I., T. T. Wu, D. C. Johnson, J. C. Glorioso, R. J. Eisenberg, and G. H. Cohen. 1990. Antigenic and functional analysis of a neutralization site of HSV-1 glycoprotein D. Virology 174375-387. [DOI] [PubMed] [Google Scholar]
- 34.Nicola, A. V., A. M. McEvoy, and S. E. Straus. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 775324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicola, A. V., and S. E. Straus. 2004. Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J. Virol. 787508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peeters, B., N. de Wind, M. Hooisma, F. Wagenaar, A. Gielkens, and R. Moormann. 1992. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J. Virol. 66894-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peeters, B., J. Pol, A. Gielkens, and R. Moormann. 1993. Envelope glycoprotein gp50 of pseudorabies virus is essential for virus entry but is not required for viral spread in mice. J. Virol. 67170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Rauch, D. A., N. Rodriguez, and R. J. Roller. 2000. Mutations in herpes simplex virus glycoprotein D distinguish entry of free virus from cell-cell spread. J. Virol. 7411437-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rauh, I., and T. C. Mettenleiter. 1991. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J. Virol. 655348-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roller, R. J., and D. Rauch. 1998. Herpesvirus entry mediator HVEM mediates cell-cell spread in BHK(TK−) cell clones. J. Virol. 721411-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satoh, T., J. Arii, T. Suenaga, J. Wang, A. Kogure, J. Uehori, N. Arase, I. Shiratori, S. Tanaka, Y. Kawaguchi, P. G. Spear, L. L. Lanier, and H. Arase. 2008. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 9913-22. [DOI] [PubMed] [Google Scholar]
- 44.Simpson, S. A., M. D. Manchak, E. J. Hager, C. Krummenacher, J. C. Whitbeck, M. J. Levin, C. R. Freed, C. L. Wilcox, G. H. Cohen, R. J. Eisenberg, and L. I. Pizer. 2005. Nectin-1/HveC mediates herpes simplex virus type 1 entry into primary human sensory neurons and fibroblasts. J. Neurovirol. 11208-218. [DOI] [PubMed] [Google Scholar]
- 45.Spear, P. G., S. Manoj, M. Yoon, C. R. Jogger, A. Zago, and D. Myscofski. 2006. Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology 34417-24. [DOI] [PubMed] [Google Scholar]
- 46.Struyf, F., C. M. Posavad, E. Keyaerts, M. Van Ranst, L. Corey, and P. G. Spear. 2002. Search for polymorphisms in the genes for herpesvirus entry mediator, nectin-1, and nectin-2 in immune seronegative individuals. J. Infect. Dis. 18536-44. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, K., H. Nakanishi, M. Miyahara, K. Mandai, K. Satoh, A. Satoh, H. Nishioka, J. Aoki, A. Nomoto, A. Mizoguchi, and Y. Takai. 1999. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J. Cell Biol. 145539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, J. M., E. Lin, N. Susmarski, M. Yoon, A. Zago, C. F. Ware, K. Pfeffer, J. Miyoshi, Y. Takai, and P. G. Spear. 2007. Alternative entry receptors for herpes simplex virus and their roles in disease. Cell Host Microbe 219-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsvitov, M., A. R. Frampton, Jr., W. A. Shah, S. K. Wendell, A. Ozuer, Z. Kapacee, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2007. Characterization of soluble glycoprotein D-mediated herpes simplex virus type 1 infection. Virology 360477-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker, K. W., R. Llull, G. K. Balkian, H. S. Ko, K. M. Flores, R. Ramsamooj, K. S. Black, C. W. Hewitt, and D. C. Martin. 1992. A rapid and sensitive cellular enzyme-linked immunoabsorbent assay (CELISA) for the detection and quantitation of antibodies against cell surface determinants. I. A comparison of cell fixation and storage techniques. J. Immunol. Methods 154121-130. [DOI] [PubMed] [Google Scholar]
- 51.Yoon, M., and P. G. Spear. 2004. Random mutagenesis of the gene encoding a viral ligand for multiple cell entry receptors to obtain viral mutants altered for receptor usage. Proc. Natl. Acad. Sci. USA 10117252-17257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon, M., A. Zago, D. Shukla, and P. G. Spear. 2003. Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2, and 3-O-sulfated heparan sulfate but not with nectin-1. J. Virol. 779221-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhai, Y., R. Guo, T. L. Hsu, G. L. Yu, J. Ni, B. S. Kwon, G. W. Jiang, J. Lu, J. Tan, M. Ugustus, K. Carter, L. Rojas, F. Zhu, C. Lincoln, G. Endress, L. Xing, S. Wang, K. O. Oh, R. Gentz, S. Ruben, M. E. Lippman, S. L. Hsieh, and D. Yang. 1998. LIGHT, a novel ligand for lymphotoxin beta receptor and TR2/HVEM induces apoptosis and suppresses in vivo tumor formation via gene transfer. J. Clin. Investig. 1021142-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, G., and B. Roizman. 2007. Separation of receptor-binding and profusogenic domains of glycoprotein D of herpes simplex virus 1 into distinct interacting proteins. Proc. Natl. Acad. Sci. USA 1044142-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]